Abstract
Apaf-1 like molecules assemble into a ring-like platform known as the apoptosome. This cell death platform then activates procaspases in the intrinsic cell death pathway. In this review, crystal structures of Apaf-1 monomers and CED-4 dimers have been combined with apoptosome structures, to provide insights into the assembly of cell death platforms from humans, nematodes and flies. In humans, caspase recognition domains of procaspase-9 and Apaf-1 interact with each other to form a CARD-CARD disk, which interacts with the platform to create an asymmetric proteolysis machine. The disk tethers multiple pc-9 catalytic domains to the platform to raise their local concentration and this leads to zymogen activation. These advances have now set the stage for further studies of this critical activation process on the apoptosome.
The intrinsic cell death pathway
Apoptotic protease activation factor-1 resides in the cytoplasm of healthy cells as an inactive monomer (Liu et al., 1996; Li et al, 1997). Appropriate stimuli trigger Apaf-1 monomers to assemble into a large ring-like platform that activates procaspases in the intrinsic cell death pathway (reviewed by Bratton and Salvesen, 2010; see Zou et al., 1999, 1997; Rodriguez and Lazebnik, 1999; Hu et al., 1999; Srinivasula et al., 1998). Pro-apoptotic stimuli may include developmental cues, growth factor withdrawal, DNA damage and oncogene activation. Apoptosome assembly is required for the activation of apical or initiator procaspases (Yan and Shi, 2005), which clip and activate executioner procaspase dimers, such as pc-3 or pc-7. Activated caspases then target cellular proteins for proteolysis in a process that kills the cell. The controlled termination of unwanted or dangerous cells allows the organism to recycle cellular components without activating inflammation pathways (Green and Evan, 2002; Inohara and Nuñez, 2003); hence, programmed cell death is important to human health. For example, apoptotic pathways in cells affected by cancer and autoimmunity diseases are often down-regulated (Song and Stellar, 1999; Salvesen and Dixit, 1997; Green and Evan, 2002), while cell death may be accelerated in AIDS, ischemic stroke and neuro-degeneration (Danial and Korsmeyer, 2004; Thompson, 1995).
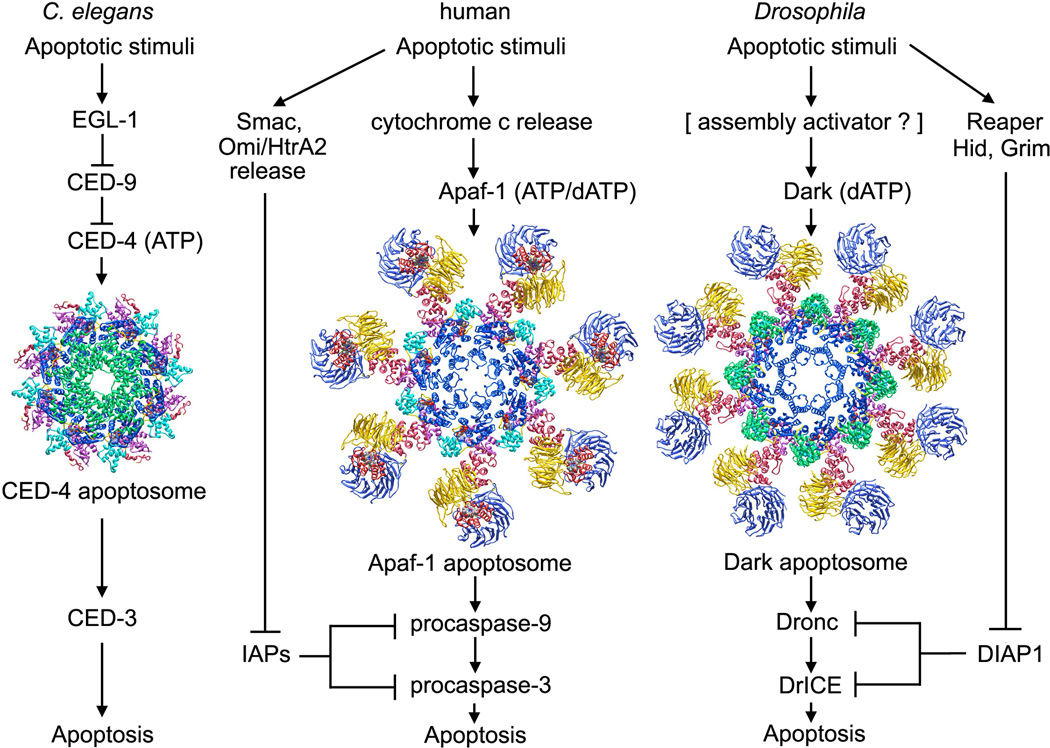
A side-by-side comparison of intrinsic cell death pathways in different “benchmark” metazoans can provide insights into conserved aspects of the molecular machinery, while highlighting mechanistic differences (Danial and Korsmeyer, 2004; Figure 1). In C. elegans, lateral dimers of CED-4 are released from a complex with CED-9 by EGL-1. Four CED-4 dimers then assemble into an apoptosome with quasi 8-fold rotational symmetry (Figure 1, left; Wu et al., 1997; Conradt and Horvitz, 1998; del Peso et al., 1998; Yan et al., 2004). The CED-4 apoptosome binds and activates CED-3, the only procaspase in nematodes, in a step mediated by homotypic interactions between CARDs located at the N-termini of CED-3 and CED-4 (Yang et al., 1998; Seshagiri and Miller, 1997). Intriguingly, CED-3 without the N-terminal CARD has been reported to bind to the underside of the CED-4 apoptosome and this resulted in a 5–10 fold increase in proteolytic activity of CED-3 (Qi et al., 2010). However, the relevance of this observation is not clear given the absence of bona-fide CARD-CARD interactions between the procaspase and CED-4.
Figure 1. The intrinsic cell death pathway in worms, humans and flies. (left) In C. elegans, an apoptosome with quasi 8-fold symmetry assembles from lateral CED-4 dimers after they are released from an inhibited CED-9 complex by EGL-1. (middle) In humans, cytochrome c is released from mitochondria to trigger nucleotide exchange and assembly of a heptameric Apaf-1 apoptosome. (right) In Drosophila, Dark may bind dATP and assemble into active single rings or inactive double rings (not shown).
In humans, cytochrome c is released from binding sites on the inner mitochondrial membrane that contain cardiolipin molecules (reviewed in Gonzalvez and Gottlieb, 2007). In response to appropriate signals, pro-apoptotic Bcl-2 family members are thought to form pores in the outer mitochrondrial membrane through which cytochrome c and other protein effectors flow into the cytoplasm (reviewed in Brunelle and Letai, 2009; Tait and Green, 2010) Cytochrome c then binds to β-propellers in Apaf-1 to trigger nucleotide exchange and stepwise assembly of a heptameric apoptosome (Figure 1, middle; (Hu et al., 1999; Zou et al., 1999). Multiple copies of pc-9 then bind to the Apaf-1 ring to form the holo-apoptosome (Malladi et al., 2009). However, this scenario may be an over-simplification because pc-9 stimulates nucleotide exchange during Apaf-1 assembly (Jiang and Wang, 2000). Thus, ternary complexes comprised of Apaf-1, cytochrome c and pc-9 could form the holo-apoptosome directly. In either case, this process leads to pc-9 activation on the apoptosome. The proteolytically active complex then cleaves loops in pc-3 and pc-7 to create active caspases that execute the death program (reviewed in Riedl and Shi, 2004).
Apoptosis inhibitors, such as XIAP, are modular proteins that contain ~70 residue Baculovirus IAP Repeat (BIR) domains that inhibit procaspases. In XIAP, the BIR3 domain interacts with the clipped p20-p10 linker of processed pc-9 (Shiozaki et al., 2003), while the linker between BIR1 and BIR2 interacts with the active site of caspase-3 (Riedl et al., 2001, Fesik and Shi, 2001). Down-regulation of pc-9 and caspase-3 by IAPs is blocked by pro-apoptotic factors, such as Smac/DIABLO and Omi/HtrA2, which are released from mitochondria along with cytochrome c. These factors interact with their target BIR domains to ensure up-regulation of the pro-apoptotic pathway (Srinivasula et al., 2001, Tait and Green, 2010).
In Drosophila, an Apaf-1 related killer (Ark/Dark; Rodriguez et al., 1999; Zhou et al., 1999; Kanuka et l., 1999;) assembles into an octameric apoptosome (Yu et al., 2006; Yuan et al., 2011a; Figure 1, right). While cytochrome c may not be required for procaspase activation in flies (Dorstyn et al., 2002, 2004; Dorstyn and Kumar, 2008), a role for cytochrome c has been demonstrated in certain tissues (Arama et al., 2006; Mendes et al., 2006; Kornbluth and White, 2005). Dark forms a stable double-ring and does not bind mammalian cytochrome c under in vitro conditions (Yu et al., 2006). Co-assembly of Dark with Dronc, the initiator procaspase (Dorstyn et al. 1999), leads to the formation of a single-ring apoptosome that activates DrICE, the executioner procaspase (Yuan et al., 2011a). The Dark apoptosome is essential for stress induced apoptosis and may account for most of the developmentally-regulated cell death that occurs in flies (Rodriguez et al., 1999; Zhou et al., 1999; Kanuka et al., 1999; Mills et al., 2006; Srivastava et al., 2007; Chew et al., 2004; Daish et al., 2004).
The absence of an activator in Dark assembly suggests that inhibitors of apoptosis may play a more prominent role in regulating cell death in flies (Figure 1). For example, the BIR2 domain of Drosophila IAP-1 interacts with a 12 residue linker in Dronc to block activation (Chai et al., 2003), while BIR1 inhibits DrICE (Kaiser et al., 1998; Yan et al., 2004). Importantly, Reaper, Hid and Grim each contain N-terminal IAP binding motifs that interact with BIR2 and BIR1 domains of DIAP1 to reverse the inhibition of Dronc and DrICE, respectively (Yan et al., 2004; Wu et al., 2001; Hawkins et al., 2000; Wang et al., 1999).
Based on this brief introduction, it is clear that divergent evolution has led to profound differences in how apoptosome assembly is regulated. This is mirrored by differences in the conformation and oligomerization state of inactive Apaf-1 like molecules. Divergence has also altered platform symmetry and CARD presentation on worm, human and fly apoptosomes. These points will be explored in more detail in the following sections. Finally, the activation of initiator procaspases is discussed in light of recent structures of pc-9 apoptosomes that revealed an asymmetric proteolysis machine (Yuan et al., 2011b). In a unifying proposal, we suggest that initiator procaspases may be activated in a conserved manner in all metazoans. Thus, a CARD-CARD disk in the holo-apoptosome may tether catalytic domains in close proximity to the platform and regulate the accessibility of platform binding sites that mediate procaspase activation (Yuan et al., 2010, 2011b).
CED-4, Apaf-1 and Dark
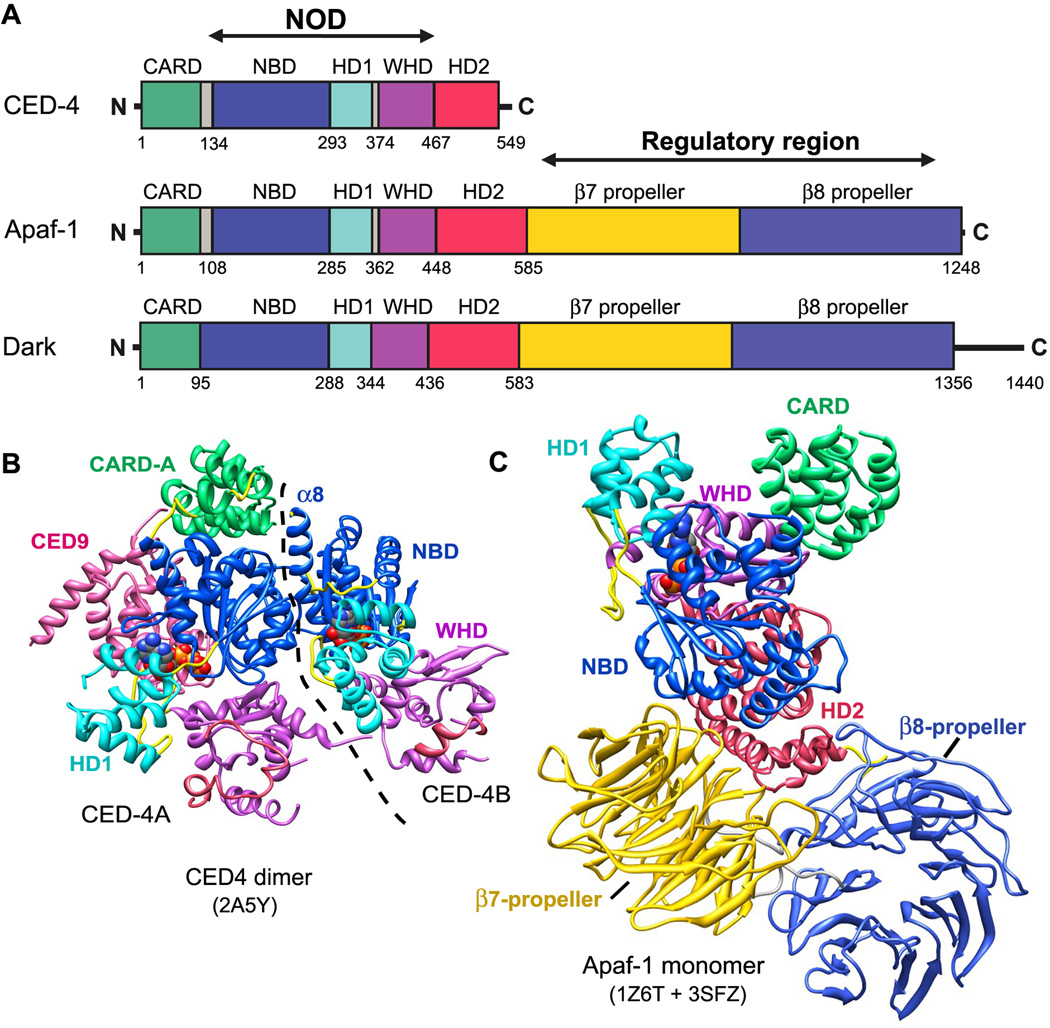
CED-4, Apaf-1 and Dark are signal transduction ATPases with numerous domains (STAND) within the AAA+ super-family (Danot et al., 2009; Leipe et al., 2004). These proteins contain three tandemly-arrayed domains within the nucleotide binding and oligomerization domain (NOD) (Figure 2A). Nucleotide binding domain (NBD) and helix domain 1 (HD1) create a binding pocket for ATP and ADP (or their deoxy analogs). These domains are predicted to oligomerize and form a ring, based on their roles in other AAA+ ATPases (Riedl et al., 2005; Diemand and Lupas, 2006; Danot et al., 2009). A novel winged helix domain (WHD) follows HD1 and also participates in ring formation (Qi et al., 2010; Yuan et al., 2010, 2011b). A moderately conserved helical domain, denoted HD2, is located after the NOD and forms an arm that extends from the central hub to support two β-propellers in human and fly apoptosomes (Figures 1, 2A; Yuan et al., 2010, 2011a; Yu et al., 2005, 2006). The N-terminal effector domain in Apaf-1 like proteins is a CARD that mediates homotypic interactions with a procaspase CARD. In total, there are 23 NOD proteins in humans but only Apaf-1 facilitates apoptosis. Other NOD proteins are predicted to form signaling or activation platforms in inflammation pathways (Proell et al., 2008; Inohara and Nuñez, 2003), as exemplified by the NAIP5-NLRC4 inflammasome (Halff et al., 2012).
Figure 2. Domain architecture and structures of Apaf-1 like proteins in the NOD super-family.
A. Linear domain representations are shown for CED-4, Apaf-1 and Dark. This domain color scheme is used throughout.
B. A crystal structure is shown of the CED-9 inhibited CED-4 lateral dimer (2A5Y). The CARD in monomer B is disordered (Yan et al., 2005) and the boundary between monomers is indicated by a dashed line.
C. A composite model is shown for an inactive Apaf-1 monomer based on two crystal structures (Reidl et al., 2005; Reubold et al., 2011).
The CED-4 molecule is a shorter version of Apaf-1 and Dark, as the latter two proteins have 15 WD40 repeats located directly after HD2 that form tandem 7- and 8-blade β-propellers (Yuan et al., 2010, 2011a; Reubold et al., 2011). Truncation of CED-4 reflects a different strategy for regulating apoptosome assembly in worms, wherein CED-9 forms a complex with a CED-4 lateral dimer to prevent further assembly (Yan et al., 2004, 2005; Figures 1, 2B). In the lateral dimer, both CED-4 monomers bind ATP and adopt an extended conformation, while CED-9 blocks the NBD in monomer A to prevent ring formation. Intriguingly, the N-terminal CARD of CED-4 sits atop the NBD in monomer A, whereas the CARD in monomer B is completely disordered in the crystal. Hence, both CARDs in the CED-4 lateral dimer appear be accessible to other proteins.
Two crystal structures have been determined of inactive Apaf-1, including a C-terminally truncated human molecule that lacks β-propellers (Riedl et al., 2005) and full length mouse Apaf-1 (Reubold et al., 2011). Apaf-1 monomers have a similar conformation in both crystal structures with ADP bound between the NBD and HD1. Latch contacts are present between the WHD, ADP and the NBD, including a salt bridge between His438 and the β-phosphate, and a second bridge between Asp439 and Arg265 (sensor I) (Riedl et al., 2005; Reubold et al., 2011; Danot et al., 2009). In addition, a CARD is present in the human Apaf-1 crystal structure while this domain is disordered in the mouse Apaf-1 structure. Thus, a complete Apaf-1 molecule in the ADP bound conformation was created by merging the two crystal structures (Figure 2C). In previous studies, a truncated Apaf-1 without β-propellers was shown to be constitutively active. This suggested that β-propellers may form a regulatory region in Apaf-1 that shields the CARD to block procaspase binding (Srinivasula et al., 1998; Hu et al., 1998). However, this view may need to be revised in light of recent data.
In the current model, Apaf-1 molecules bind cytochrome c and assemble into a ring-like platform that captures multiple pc-9 zymogens through CARD-CARD interactions (Hu et al., 1999; Zou et al., 1999). Close proximity of initiator procaspases on the apoptosome then leads to activation, although the precise mechanism is being debated (reviewed in Bratton and Salvesen, 2010; Yuan et al., 2011b). Recent crystal structures suggest a different scenario because CARDs in the CED-4 lateral dimer (Yan et al., 2005) and the Apaf-1 monomer (Reubold et al., 2011; Riedl et al., 2005) appear to be accessible (Figures 2B, 2C). Hence, Apaf-1 and CED-4 could interact with pc-9 and CED-3, respectively, either before or during apoptosome assembly. It is important to note that formation of binary pc-9/Apaf-1 complexes and ternary CED-3/CED-4/CED-9 complexes in the cytoplasm would not lead to zymogen activation. Instead, association of Apaf-1 like molecules with initiator procaspases could provide a mechanism for rapid formation of holo-apoptosomes, when assembly is given the green light (Figure 1).
Apoptosome structure and assembly
Accurate apoptosome structures are required to understand assembly and procaspase activation. Until recently, progress towards this goal was limited due to the low stability of apoptosomes, coupled with their large size and tendency to aggregate. However, significant progress has now been made. In particular, a crystal structure was determined of the CED-4 apoptosome at 3.5Å resolution that revealed NOD interactions within an octagonal central hub (Qi et al., 2010). In brief, the NBD-HD1 pair forms an oligomerization domain with 8 copies of two NBD helices (αx12 and α13) forming an inner ring that encircles the central pore (Figure 3A, top). Helix α 12 is known as the Initiator Specific Motif (ISM) and is a novel insert within the NBD (Danot et al., 2009). In addition, HD1 and WHD from each CED-4 subunit form a second ring that encircles the central NBD ring, while the shorter HD2 is somewhat disordered. Subunit NODs have nearly perfect 8-fold symmetry and their lateral contacts are similar to those in the inhibited CED-4 lateral dimer (Figure 4A, left; Qi et al., 2010). Nucleotide exchange is not required for assembly after CED-9 release because ATP molecules are already bound to each molecule in the CED-4 dimer (Yan et al., 2004, 2005; Qi et al., 2010; Danot et al., 2009). Hence, the lateral CED-4 dimer probably represents a trapped assembly intermediate.
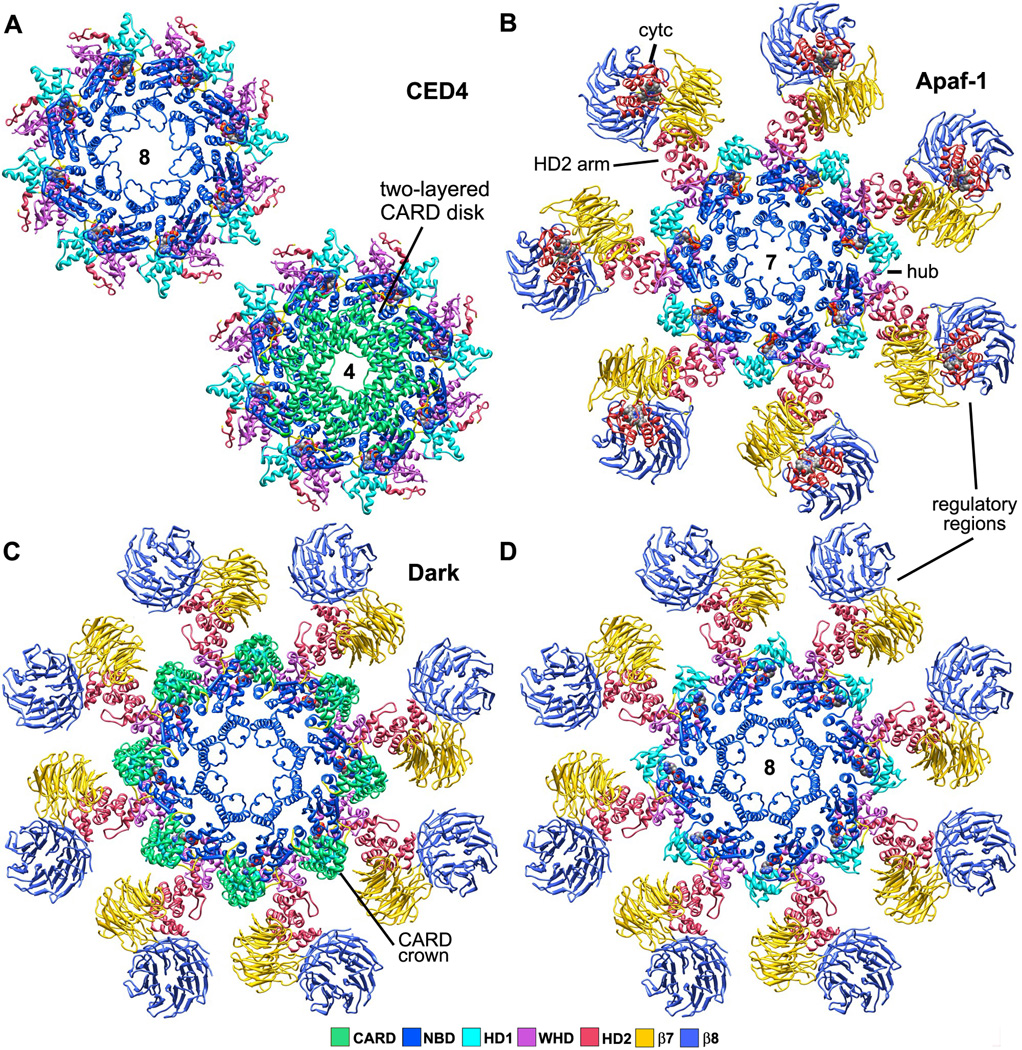
Figure 3. Top views of worm, human and fly apoptosomes.
A. (top left) The central hub of the C. elegans apoptosome has quasi 8-fold symmetry. (lower right) CARDs from the A and B monomers of CED-4 form two tetrameric rings stacked along the 8-fold axis of the hub to create an apoptosome with overall 4-fold symmetry (Qi et al., 2010; 3LQQ).
B. The human apoptosome contains seven Apaf-1 molecules whose CARDs are disordered in the ground state (Yuan et al., 2010; submitted).
C. A Drosophila single ring apoptosome is shown with eight CARDs that bind to the lateral surface of their respective NBDs to form a crown (Yuan et al., 2011a; 1VT4, 3IZ8).
D. The CARD crown has been removed to show the similarity of the octagonal fly apoptosome and the heptameric Apaf-1 apoptosome in top views (compare with panel B). Note that mammalian cytochrome c does not bind to Dark in these complexes.
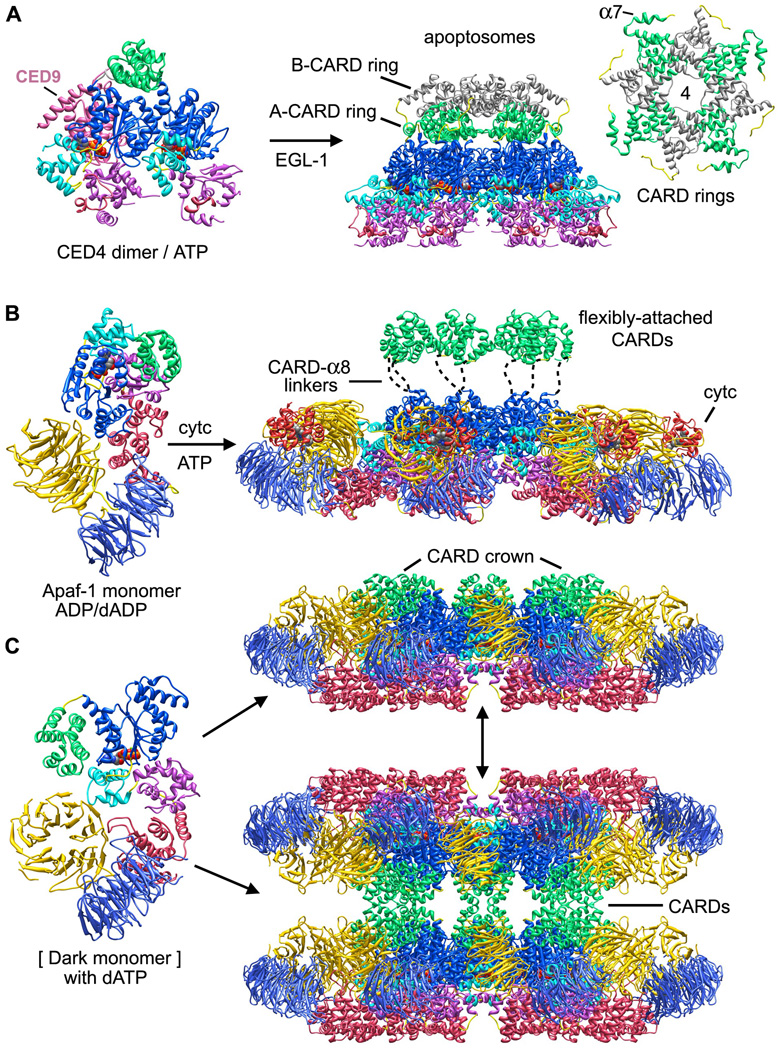
Figure 4. Assembly models for ground state apoptosomes in worms, humans and flies.
A. CED-4 molecules in lateral dimers contain bound ATP and may represent an assembly intermediate blocked by CED-9 (Yan et al., 2005; Qi et al., 2010). EGL-1 interacts with CED-9 to promote further oligomerization of CED-4 to form the apoptosome.
B. Inactive Apaf-1 monomers (Reidl et al. 2005; Reubold et al., 2011) bind cytochrome c and undergo nucleotide exchange to promote assembly of a heptameric apoptosome. CARDs are flexibly tethered to NBDs in the central hub by CARD-α8 linkers (Yuan et al., 2010).
C. An extended Dark monomer was extracted from the apoptosome model and is shown on the left. In vitro assembly requires dATP to form single and double rings, as shown on the right.
The CED-4 apoptosome has global 4-fold symmetry because eight CARDs are packed within two tetrameric rings that sit on top of the central hub (Figure 3A, bottom; Figure 4A, right panel and inset). CARDs in the lower A-ring have well ordered CARD-NBD linkers that form an additional α-helix (α7). This conformation was also observed in monomer A of the CED-4 lateral dimer (Yan et al. 2005; Qi et al., 2010). The CARD B-ring stacks on top of the A-ring (Figure 4A, right) and this packing arrangement reflects the structural plasticity of CARD-NBD linkers, with helix α7 being unwound in B-subunits. Interestingly, helix α7 is not present in the CARD-NBD linker of Apaf-1 or Dark. Finally, the two CARD tetramers are rotationally offset from each other by ~45° and their approximate 4-fold symmetry axes are co-linear with the central 8-fold axis.
The human apoptosome has proven inimical to crystallization; hence, structural analysis has proceeded in a stepwise manner using cryo-EM, single particle methods and modeling (Acehan et al., 2002; Yu et al., 2005; Yuan et al., 2010, 2011b; submitted). Structures of Apaf-1 apoptosomes have now been obtained in the ground state and with bound pc-9 CARDs at 12.5 and 9.5Å resolution. The highest resolution map was determined using pc-9 CARD apoptosomes in which a thrombin site in the CARD-p20 linker of pc-9 was cleaved by protease after assembly. Under these conditions, catalytic domains (p20-p10) of pc-9 were released quantitatively from active complexes by a single clip in each procaspase. A model of the heptameric apoptosome was assembled in silico with rigid body docking of appropriate domains (Yuan et al., 2010). Structures of CED-4 and Apaf-1 apoptosomes were solved independently, yet the models provide a similar picture of the central hub, even though they have different rotational symmetries (quasi-8 versus 7-fold; Figures 3A top and Figure 3B; Yuan et al., 2010; Qi et al., 2010).
In the human apoptosome, HD2 in each subunit extends radially to form an arm that links the central hub to tandem 7- and 8-blade β-propellers, while cytochrome c sits in a V-shaped cleft between the two propellers (Figure 2A; Yuan et al., 2010; Yu et al., 2005; Acehan et al., 2002). A recent crystal structure of full length Apaf-1 (Reubold et al., 2011) and improved docking software provided the impetus to create a more complete picture of the human apoptosome. First, homology models of human 7- and 8-blade β-propellers were docked into the regulatory region along with cytochrome c. Second, the NOD and HD2 arm of Apaf-1 were flexibly modeled within the platform (Yuan et al., submitted). This model is used in figures throughout this review.
The domain architecture of the Apaf-1 platform is similar in active and ground state apoptosomes. However, the presentation of Apaf-1 CARDs differs dramatically in the two states. CARDs are disordered in the ground state but remain attached to their respective NBDs through flexible linkers between helices α6 and α8 (Figure 4B, right; Yuan et al., 2010). This was confirmed with protease protection experiments (Yuan et al., 2011b). In the active apoptosome, Apaf-1 and pc-9 CARDs form a novel disk like structure that sits above the central hub. The disk is somewhat blurred in a map of the pc-9 CARD apoptosome. Hence, linkers that tether the CARD-CARD disk to NBDs in the central hub may retain some flexibility (Yuan et al., 2010).
To complete the apoptosome trifecta, the structure of a Dark double-ring was determined at ~6.9Å resolution (Yuan et al., 2011a). This allowed a detailed homology model of the Dark apoptosome to be built. A single-ring extracted from the final model displays all the hallmarks of an apoptosome, while differences in the central hub include an altered packing of the α12 and α13 helices around the central pore (Figures 3C, 3D). To accommodate 8 Dark subunits, the angle of the HD2 arm has been altered along with the radial position and tilt of β-propellers, relative to their positions in the Apaf-1 apoptosome (Figures 4B, 4C, rightmost panels). Intriguingly, the Dark monomer is 192 residues longer than Apaf-1 and these residues are located in the C-terminal half of the molecule (Figure 2A). These residues could not be modeled at this stage (Yuan et al., 2011a). However, some of the missing residues may form large flexible loops that emanate from the β-propellers and could also form an extended bridge that links β-propellers in adjacent subunits.
The disposition of CARDs in the Dark apoptosome differs significantly from that observed in worm and human cell death platforms. In the Dark apoptosome eight CARDs interact with lateral surfaces of their respective NBDs to create a CARD crown on the central hub (Figure 3C, 4C). The CARD-NBD interaction is facilitated by a short linker between the CARD and helix α8. Indeed, some residues that form the CARD-NBD linker in Apaf-1 may have formed a coil to extend helix α8 in Dark. It also remains to be determined whether residues in helix α8 unwind during Dronc binding to facilitate formation of a CARD-CARD disk in the active Dark apoptosome (Yuan et al., 2010; 2011b).
We do not have a crystal structure of the inactive Dark monomer. However, newly synthesized Dark molecules may bind dATP directly and adopt an extended conformation that promotes ring assembly (Figure 4C, left). In this case Dark would not require an intrinsic ATPase activity. While the active Dark apoptosome is probably a single-ring (Yuan et al., 2011a), double-rings could form spontaneously in the absence of Dronc within the cytoplasm, due to CARD-CARD interactions between opposing single-rings. Hence, single and double-rings may exist in a dynamic equilibrium that is dependent upon the Dronc concentration (Figure 4C, right). In this scenario, procaspase activation would be controlled by Dronc degradation mediated by DIAP-1 rather than a regulated Dark assembly (Chai et al., 2003; Hawkins et al., 2000; Yuan et al., 2011a). This would explain why cytochrome c is not required for assembly in vitro (Yu et al., 2006). However, a possible role for cytochrome c in fly apoptosis requires further study.
An analysis of worm, human and fly apoptosomes suggests that novel ISM helices may play similar roles in nucleating assembly at the center of their respective hubs (Danot et al., 2009). In addition, variations in domain interactions within the central hub are responsible for different ring symmetries. From a functional point of view, local changes in domain packing may not impinge on the global process of procaspase activation, so long as platform assembly remains efficient. Importantly, the domain architecture of apoptosomes can be used to explain gain and loss of function mutations in NOD proteins (Qi et al., 2010; Yuan et al., 2010; 2011a).
The oligomerization state of the CED-4 lateral dimer, Apaf-1 monomer and Dark rings may be intimately linked to strategies that regulate assembly and procaspase activation in their respective cell death pathways (Figures 1, 4). While Apaf-1 and Dark both have extended HD2 arms and β-propellers, the use of pre-assembled oligomers may be common to CED-4 and Dark. At the same time, inactive CED-4 lateral dimers and Apaf-1 monomers have exposed CARDs that may facilitate interactions with procaspase CARDs during apoptosome assembly and a similar process could also occur in Drosophila. Thus, a common ancestral Apaf-1 like molecule has been tailored by divergent evolution to create different assembly paths with a common end point, the activation of procaspases. In addition, CARDs are exposed on the top surface of the apoptosome in a different manner in flies, humans and worms (Qi et al., 2010; Yuan et al., 2010, 2011a). This reflects a remarkable plasticity of the linker between the CARD and helix α8 and this linker could play a role in the assembly of CARD-CARD disks.
Nucleotide exchange
Apoptosome structures have provided an accurate sequence alignment for NODs (Sup_Figure 1) that is useful in understanding the role of particular residues in nucleotide binding and apoptosome assembly. For example, canonical Walker A and B motifs are well conserved in Apaf-1 like molecules, along with an arginine (sensor I) that may interact with the γ-phosphate of nucleotide triphosphate (Arg265 in Apaf-1; Sup_Figure 1; Hanson and Whiteheart; 2005). An activating arginine finger that is normally provided by an adjacent subunit in AAA+ ATPases (Danot et al., 2009) is absent in assembled apoptosomes, which suggests a rationale for why these complexes do not support a robust ATPase activity (Jiang and Wang, 2000; Reubold et al., 2009). In addition, a low ATPase activity has been measured for the Apaf-1 monomer (Jiang and Wang, 2000; Kim et al., 2005; Riedl et al., 2005; Reubold et al., 2009), while CED-4 lateral dimers bind ATP but do not display ATPase activity (Yan et al. 2005).
Inactive Apaf-1 monomers can be purified with bound dATP (Jiang and Wang, 2000; Kim et al., 2005) or ADP (Riedl et al., 2005; Bao et al., 2007; Reubold et al., 2009). This may reflect differences in the physiological states of cells that were used to express these proteins and the disparity remains to be explained. Either dATP or ATP will support Apaf-1 assembly in the presence of cytochrome c, but the concentration of these NTPs in cells is ~10 uM and 1–2 mM, respectively. Hence, it seems likely that ADP and ATP will be major nucleotide co-factors for Apaf-1 (Reubold et al., 2009). In the case of Dark only dATP will support assembly in vitro (Yu et al., 2006). The requirement for dATP over ATP is not understood, nor is it known which NTP may be used by Dark in vivo. However, catalytic residues in the Walker B motif of Apaf-1 (Asp243-Asp244) and CED-4 (Asp250-Asp251) are replaced by a leucine-asparagine pair in Dark (Leu245-Asn246; Sup_Figure 1). Hence, Dark monomers and rings are expected to have very low ATPase activity.
Nucleotide exchange is required for assembly of Apaf-1 with ATP or dATP replacing a bound nucleoside diphosphate (Kim et al., 2005). In addition, PHAPI, cellular apoptosis susceptibility protein (CAS) and Hsp70 may accelerate nucleotide exchange while preventing aggregation of Apaf-1/cytochrome c complexes (Kim et al., 2008). Apaf-1 can be contrasted with the lateral dimer of CED-4, which is in an ATP bound conformation and does not require nucleotide exchange to promote assembly (Yan et al., 2005; Qi et al., 2010). However, ATP may bind to newly synthesized CED-4 molecules to form lateral dimers whose assembly is blocked by CED-9. Since there are pools of ATP/ADP and dATP/dADP in cells, it seems likely that Apaf-1 like molecules may bind to di- or triphosphate nucleosides as they fold. Thus, inappropriately bound ADP/dADP may undergo a facile nucleotide exchange in CED-4 (for ATP) and Dark (for dATP), which would lead to assembly of lateral dimers and rings, respectively. Conversely, bound ATP or dATP in Apaf-1 may be hydrolyzed to the nucleoside diphosphate by an endogenous ATPase activity to form the inactive monomer. Nucleotide hydrolysis would be critical to maintain Apaf-1 in an inactive conformation, until cytochrome c is released in response to the appropriate cell death signal (Figure 1). Since the intrinsic ATPase activity of Apaf-1 is rather low, it is possible that an unknown ATPase activating factor may be required to ensure that newly synthesized Apaf-1 molecules adopt an ADP bound conformation. This step would block the ability of Apaf-1 to assemble or form aggregates.
Conformational dynamics in Apaf-1 assembly
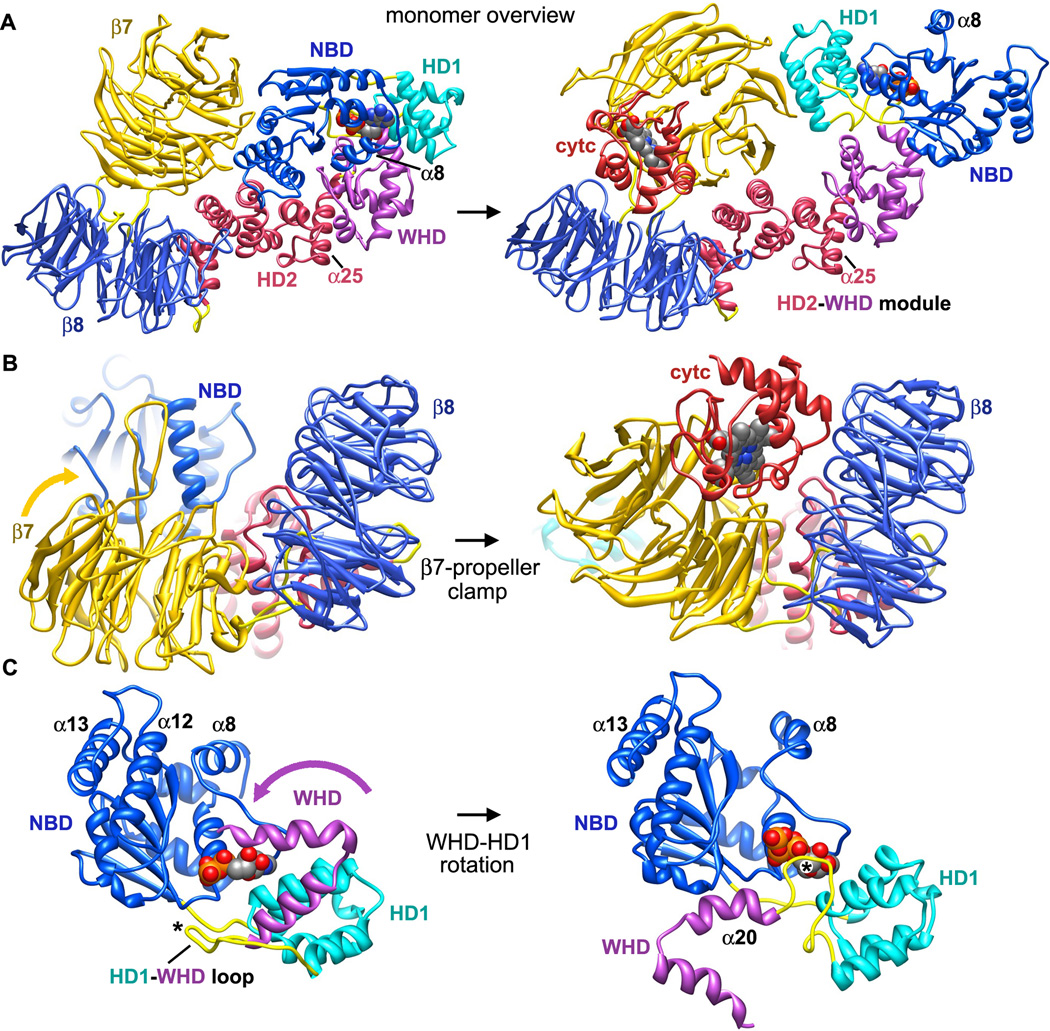
The critical role of the intrinsic cell death pathway in human disease, makes it of particular interest to understand how Apaf-1 assembles. A structure of mouse Apaf-1 (Reubold et al., 2011) and a model of the apoptosome have provided insights into a plausible pathway for Apaf-1 assembly (Reubold and Eschenburg, 2012; Yuan et al., submitted). When cytochrome c is released from mitochondria, this basic protein binds to a cleft between β-propellers to trigger changes in the relative orientation of these Apaf-1 domains (Figure 5A). This includes a large rotation of the β 7 propeller, coupled with smaller movements of the β8 propeller to clamp cytochrome c between them (Figure 5B). The cytochrome c-β7 propeller interaction would also break salt bridges between the β7 propeller, NBD and HD2, allowing the NOD to become more flexible.
Figure 5. Conformational transitions in Apaf-1 during apoptosome assembly.
A. (left) A homology model of an inactive human Apaf-1 monomer with bound ADP is shown without the N-terminal CARD. (right) Cytochrome c binding to β-propellers induces a conformational change that may promote nucleotide exchange and assembly. Apaf-1 monomers have been aligned on their WHD-HD2 modules. The direction of view is approximately along the 7-fold axis of the apoptosome for the molecule on the right.
B. The β7-propeller acts like a clamp to pin cytochrome c between the propellers.
C. A close-up is shown of the rotation of WHD relative to the NBD and HD1 pair that occurs upon nucleotide exchange. The associated HD2 arm and β-propellers are not shown. The HD1-WHD loop (marked with an asterisk) with an S/TxYxY motif flips to interact with the ribose region of the bound nucleotide during, while the WHD-HD2 arm rotates. Helices α8, α12 and α13 also move during apoptosome assembly.
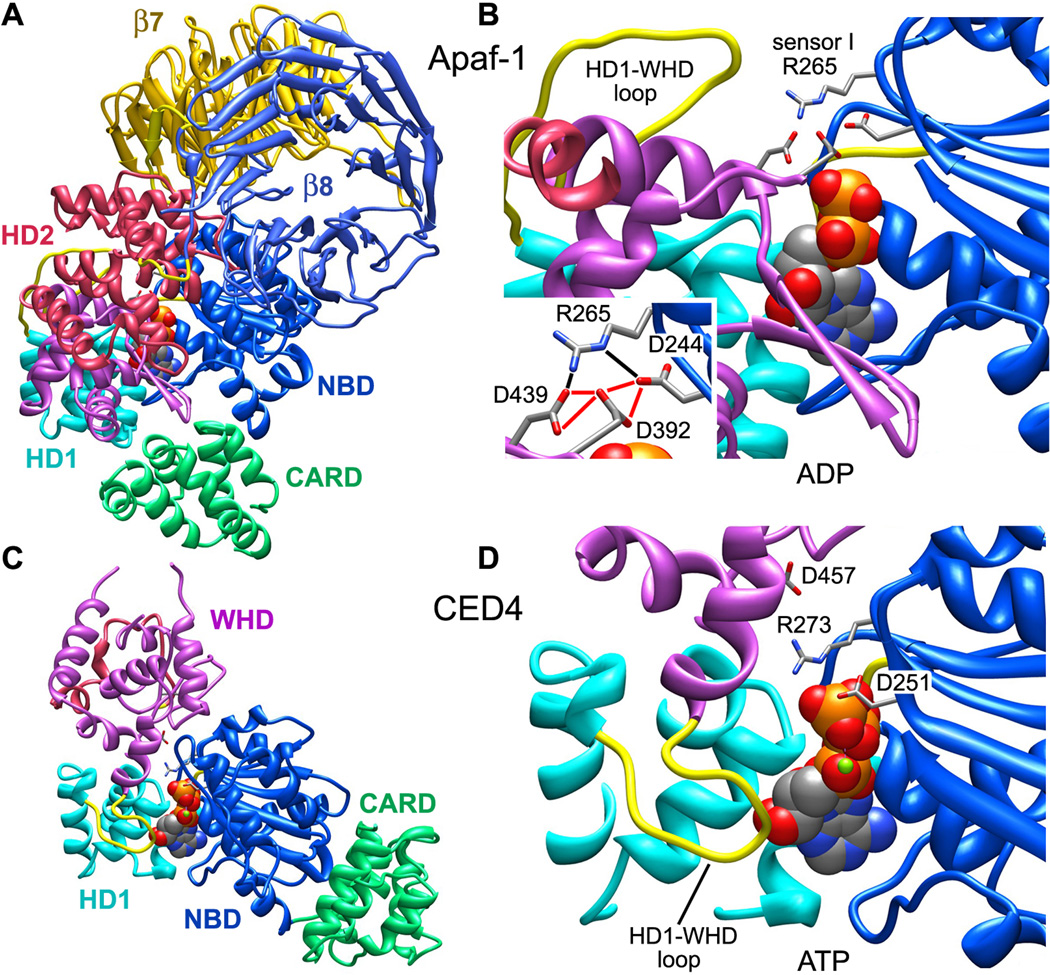
To avoid a clash between the β7 propeller and NBD, cytochrome c binding may also induce a concerted re-orientation of the NBD-HD1 pair to facilitate nucleotide exchange (Figure 5C). This may cause sensor I (Arg265) to rotate away from a position where it stabilizes a repulsive triangle formed by carboxyl side chains of Asp244, 392 and 439 in the inactive monomer (Reubold et al., 2011; icon view in Figure 6A and expanded view in Figure 6B). This movement probably re-positions Arg265 so that it can interact with the γ-phosphate of ATP, because a similar interaction occurs in CED-4 apoptosomes and lateral dimers (icon view in Figure 6C and larger view in Figure 6D; Qi et al., 2010; Yan et al., 2005). These changes may drive formation of an assembly competent Apaf-1. In line with these ideas, calcium is known to inhibit Apaf-1 assembly by preventing nucleotide exchange (Bao et al, 2007). Perhaps Ca+2 ions interact with the carboxyl triad to stabilize the closed conformation of Apaf-1, thereby preventing a breathing motion that may be required for nucleotide exchange.
Figure 6. The role of sensor I in Apaf-1 monomer and CED4 apoptosomes.
A. An icon view is shown of the Apaf-1 monomer.
B. Stabilizing interactions of sensor I (R265) with an acidic triangle of carboxylate groups in the inactive Apaf-1 monomer are shown (see inset). Note the distal position of the HD1-WHD loop.
C. An icon view is shown of a CED4 monomer extracted from the assembled apoptosome.
D. Sensor I (R273) interactions are shown with the γ-phosphate of ATP in the CED-4 apoptosome. The HD1-WHD loop is in close proximity to the ribose of ATP.
Conformational changes during assembly are distributed over the entire Apaf-1 molecule. When cytochrome c binds to Apaf-1, lysines in the heme protein (especially K7, K25, K39 and K72) may interact with acidic residues in the 7- and 8-blade β-propellers (Yu et al., 2001). Cytochrome c binding also places the heme group in a solvent protected environment between β-propellers (Purring-Koch and McLennon, 2000; Yuan et al. submitted). The order of cytochrome c binding and nucleotide exchange during assembly has not been established experimentally. However, ADP is deeply buried between the NBD and HD1 of the inactive Apaf-1 molecule, with the WHD also helping to shield nucleotide from solution (Riedl et al., 2005; Reubold et al., 2011). This suggests that cytochrome c binding may occur first, consistent with its role as an activator that is specifically released from mitochondria to trigger assembly.
Beta-propellers in the regulatory region do not make extensive contacts with the NBD, HD1, WHD and CARD in the closed form (Reubold et al., 2011). Thus, it is not clear why Apaf-1 remains inactive in the absence of cytochrome c. Perhaps the stability of the inactive form is governed by a series of kinetic barriers that control the dynamics of this multi-domain protein. In this scenario, cytochrome c binding would tip the energetics of Apaf-1 towards the assembly competent form by causing local changes in the relative orientation of the β7-propeller, along with the loss of three salt bridges to HD2 and the NBD (Reubold et al., 2011).
The WHD-HD2 interface is quite extensive in crystal structures (Riedl et al., 2005; Reubold et al. 2011) and this domain pair may act as a rigid module during conformational changes that occur at either end of the Apaf-1 molecule. At the HD1-WHD interface, HD1 rotates about 12° when ATP is bound relative to the NBD. At the same time, a relative rotation of about 160° occurs between the WHD-HD2 module and NBD-HD1 pair (Figure 5C). The overall effect of these changes is to bring β-propellers from a position behind the NBD, into an extended position at the end of the HD2 arm (Figure 7). During this process the HD1-WHD loop, which points away from bound nucleotide in the inactive form, rotates about 60° when viewed from the underside of the central hub. This positions the S/TxYxY motif to interact with the ribose ring of ATP (Figure 5C; Yuan et al., 2011a) and a similar interaction occurs in the CED-4 apoptosome (Qi et al., 2010; Figure 6D).
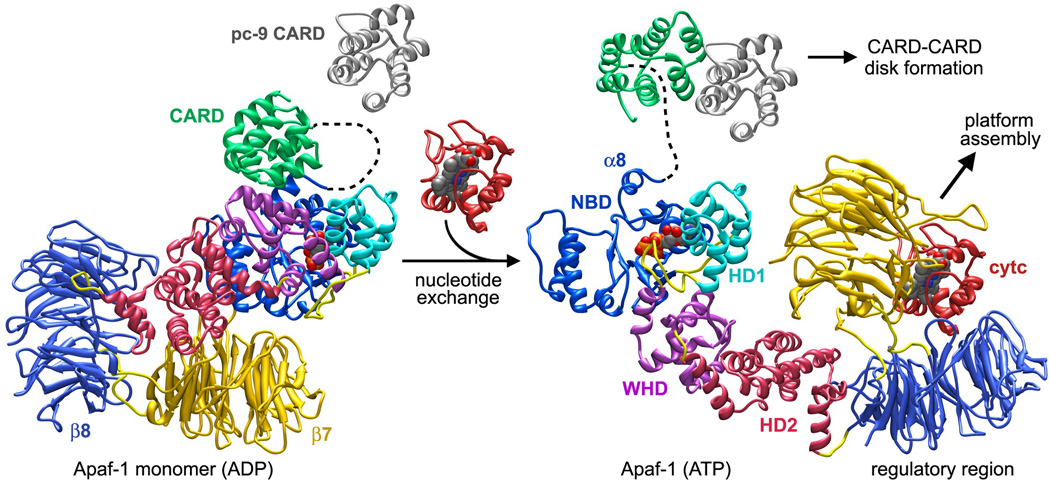
Figure 7.
Possible conformational changes that occur during Apaf-1 assembly are shown with inactive and active conformations aligned on the NBD. Cytochrome c binding may trigger inter-domain rearrangements and motions that lead to nucleotide exchange. An extensive rearrangement of the NBD-HD1 module occurs relative to the rest of Apaf-1. This creates an extended and assembly competent conformation. Procaspase-9 may interact with the CARD in the Apaf-1 monomer during nucleotide exchange leading directly to the formation of a CARD-CARD disk as the platform assembles.
Further stepwise association of monomers into lateral dimers may stabilize the extended conformation of Apaf-1 with bound ATP, and lateral binding of additional subunits to this “seed” may culminate in ring closure. The loss of entropy by individual Apaf-1 subunits during assembly would be offset by the large number of contacts formed between NODs in the apoptosome. Thus, nucleotide exchange would create an equilibrium in which an extended Apaf-1 conformation is favored that can be used as a building block in assembly, rather than being funneled into an abortive pathway that leads to aggregation (Danot et al., 2009).
In Apaf-1 the global effects of conformational changes induced by cytochrome c and nucleotide exchange can be described as follows (Figure 7). Large inter-domain movements expose the lateral surface of the NBD so that it can associate with NBDs in other subunits to form a ring within the central hub. During this process, the a12-α13 helix pair become accessible and form a picket fence of α-helices that defines the central pore. These movements also entail rearrangements of helix α8 to help form the interface between adjacent NBDs. This step is intriguing because the α8-α9 loop of the NBD cradles the ribose ring of bound nucleotide. Additional rotations of HD1 and the WHD-HD2 module ensure that the HD1-WHD ring can be formed on the hub periphery. This large conformational change positions the HD2 arm so that it extends from the hub to support the β-propellers. Large movements of the WHD relative to the NBD-HD1 module may release the bound CARD, which remains attached to the NBD by a flexible linker. Alternatively, Apaf-1/pc-9 complexes could be formed in the presence of pc-9, either before or during assembly. In either case, assembly would lead directly to the formation of an active apoptosome.
A CARD-CARD disk in the human apoptosome
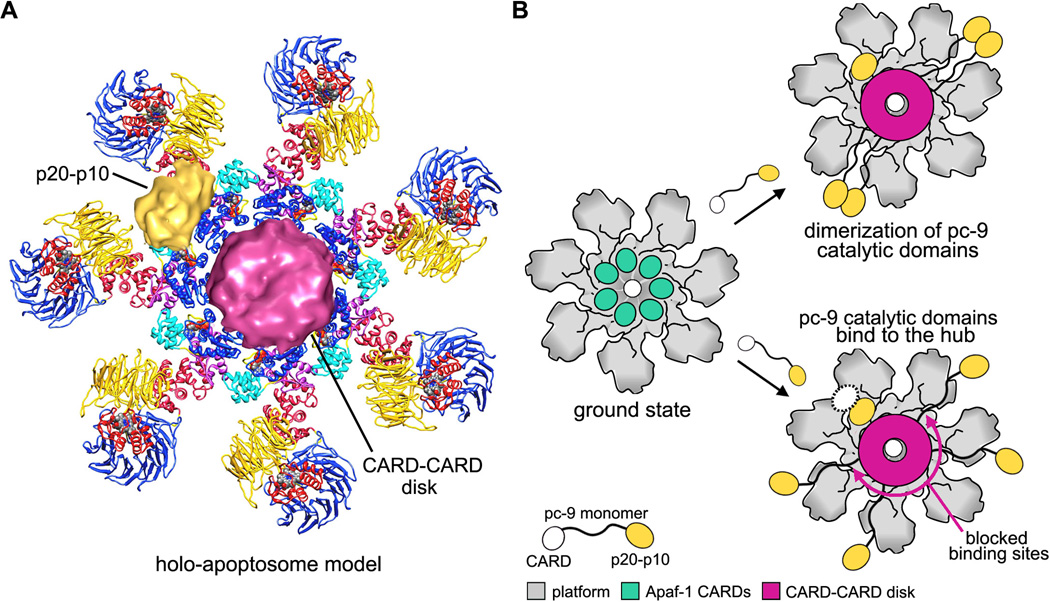
The goal of Apaf-1 assembly is to form a platform that can recruit and activate procaspases. Recent data suggests that Apaf-1 and pc-9 CARDs co-assemble to create a disk that sits on top of the central hub (Figure 8A). The CARD-CARD disk is attached to NBDs by linkers that retain a limited degree of flexibility since the disk is blurred in 3D maps (Yuan et al., 2010; 2011b). The disk is also located in an off-center position relative to the central hub (Figure 8A) and has a tilted appearance in maps calculated without imposing 7-fold symmetry during refinement (Yuan et al., 2011b). Thus, assembly of the holo-apoptosome creates an asymmetric proteolysis machine.
Figure 8. Models of procaspase-9 activation on the human apoptosome.
A. A composite model is shown for the holo-apoptosome, based on cryo-EM structures for the platform and active complex (Yuan et al. submitted).
B. The platform is shown in grey and domains of pc-9 are color coded in a cartoon model of pc-9 activation. (top right) A proximity-induced dimerization model is shown with flexibly tethered pc-9 molecules. (bottom right) The proximity-induced association/allosteric model is shown. One p20-p10 catalytic domain (in gold) is bound to the hub and a possible second catalytic domain is shown as a dashed white oval, to indicate that it may be disordered.
As shown previously, worm, human and fly apoptosomes display their CARDs quite differently (Figure 3). However, the ground state may exist only transiently or not at all in activated cells, as procaspases with CARDs may co-assemble with Apaf-1 like molecules during ring formation (Figure 7). We hypothesize that CARD-CARD disks may be formed in apoptosomes from all three organisms, when they bind procaspases to form active complexes. This would require a significant reorganization of CARDs, based on their preferred modes of association with platforms in CED-4 and Dark apoptosomes. However, disk assembly may be facilitated by structural plasticity of CARD-NBD linkers (Qi et al., 2010; Yuan et al., 2010; Riedl et al., 2005; Reubold et al., 2011).
The nature of the CARD-CARD disk remains mysterious. However, Death Domain family members contain 6-helix bundles that form large complexes with appropriate family members (Park et al., 2007b). Crystal structures of signaling platforms have shown that Death Domains form truncated helices or lock washer ensembles, rather than rings, with at least three distinct interactions involving ~10–14 domains. These novel structures include the Myddosome, comprised of MyD88, IRAK4 and IRAK2 (Lin et al., 2010), the PiDDosome formed by PIDD and RAIDD (Park et al., 2007a) and the FAS-FADD death disc (Wang et al., 2010; reviewed in Ferrao and Wu, 2012). Since CARDs are members of the Death Domain family they might be expected to form similar complexes.
The number of CARD carrying procaspases that are bound to active apoptosomes has been the subject of some debate. Measurements with CARD-less CED-3 suggest that two procaspases are bound to the CED-4 apoptosome (Qi et al., 2010), in agreement with estimates for the number of pc-9s bound to the Apaf-1 apoptosome (Malladi et al., 2009). However, a recent study suggests that 5–7 pc-9 molecules may be bound to the holo-apoptosome (Yuan et al., 2011b). N-terminal CARDs should be equally accessible on all Apaf-1 like molecules during assembly. Thus, the number of CARDs available from the apoptosome and the preferred structure of the CARD-CARD disk will determine how many procaspases are recruited. Additional studies are needed to clarify the number of initiator procaspases bound to apoptosomes and to visualize the disk structure.
Physical dimensions of the CARD-CARD disk and its likely composition (Yuan et al., 2011b) suggest that this assembly may be more akin to the FAS-FADD disc formed in the extrinsic cell death pathway, which contains 10–12 Death Domains. In this scenario, the Apaf-1/pc-9 CARD heterodimer in crystals may represent a particularly stable pairing (Qin et al., 1999) and additional interactions would be needed to form the disk. Importantly, disk assembly would tether catalytic domains to the apoptosome, creating a high local concentration to facilitate activation. The timing of pc-9 activity in cells (Malladi et al., 2009) might also be controlled by disk assembly and disassembly on the apoptosome.
Procaspase-9 activation
Until recently, much of the evidence for pc-9 activation favored a model in which catalytic domains are brought into close proximity on the apoptosome to dimerize. Proximity-induced dimerization would induce a reorientation of appropriate loops in pc-9 to complete the active sites (Renatus et al., 2001; Boatright et al., 2003; Figure 8B, upper path). This model is supported by a number of observations and experiments. First, inactive initiator procaspases are monomers in solution, while executioner procaspases are constitutive dimers (Boatright et al., 2003; Chao et al., 2005; Renatus et al., 2001; reviewed in Li and Yuan, 2008). Hence, it seems reasonable that dimerization would trigger activation of initiator procaspases. Second, crystallization of pc-9 catalytic domains in the presence of a peptide inhibitor revealed a dimer with an unusual interface, in which inhibitor was bound to only one “active” monomer (Renatus et al., 2001). Third, chaotrophic salts were shown to drive dimerization of pc-9 molecules and induce activity (Pop et al., 2006). Fourth, a chimeric zymogen with a pc-9 CARD and CARD-p20 linker attached to catalytic domains of pc-8 (p20-p10) was activated on the apoptosome (Pop et al., 2006). Fifth, pc-9 catalytic domains connected by a short linker (~6–7 residues) to the leucine zipper dimerization motif of GCN4 were activated (Yin et al., 2006).
Recent studies of the pc-9 apoptosome have provided data to support an alternate hypothesis, first described as an allosteric mechanism, wherein pc-9 catalytic domains interact directly with the platform to become active (Chao et al., 2005). This model was based primarily on an experiment in which pc-9 molecules with a re-engineered interface were forced to dimerize. In this case, dimers were shown to be much less active than native pc-9 in the holo-apoptosome. This suggested that catalytic domains might bind to the platform to be activated. In addition, experiments with a GCN4/pc-9 chimera showed that these molecules had significantly less activity towards procaspase-3 than the pc-9 apoptosome. In this case, it was suggested that holo-apoptosomes may contain bound pc-9 dimers (Yin et al., 2006).
The allosteric model can be described as a proximity-induced association model because a high local concentration of catalytic domains is critical for activation (Yuan et al., 2011b). Support for the allosteric model was provided by a symmetry free reconstruction of pc-9 apoptosomes. This 3D map showed density on the central hub with the right size and shape for a single p20-p10 catalytic pair (Yuan et al., 2011b; Figure 8A and 8B, lower right). This suggested that catalytic domains from a single pc-9 may bind to the hub and be activated. Alternatively, a dimer of catalytic domains could be bound with one p20-p10 pair being disordered (see dashed oval in Figure 8B, lower right). However, the dimer interface in a pc-9 crystal structure is quite extensive (Renatus et al., 2001), so this type of flexibility may not be possible. It should also be noted that this hypothetical geometry, with one stably bound pc-9 subunit and one flexibly bound subunit, would probably activate a single p20-p10 pair within the putative dimer. This is reminiscent of what was seen in the crystal structure of a pc-9 dimer (Renatus et al., 2001).
Further support for the allosteric activation model is provided by a number of observations. First, truncations of the CARD-p20 linker in pc-9 led to a significant loss of proteolytic activity with the active apoptosome. This suggested a role for the CARD-p20 linker in pc-9 binding to the platform and is consistent with studies which showed that phosphorylation of Thr125 in the linker reduced activation. This post-translational modification may interfere with catalytic domain binding to the hub (Allan et al., 2003, 2007; Reubold and Eschenburg, 2012). Second, protease protection experiments on pc-9 apoptosomes showed that a thrombin site within the CARD-NBD linker of Apaf-1 was partially protected. When a naturally activated caspase-3 was present on the pc-9 apoptosome, it provided even greater protection than pc-9 by itself. These data suggest that pc-9 and caspase-3 may share overlapping binding sites on the central hub, since the thrombin sites were located between the disk and the platform (Yuan et al., 2011b). Third, proteolytic activation of pc-3 by the pc-9 apoptosome resulted in a significant down-regulation of pc-9 activity, as processed caspase-3 increased during the time course of the reaction. Strikingly, pc-9 activity was lost even though catalytic domains of pc-9 remained tethered to the disk in these complexes (Yuan et al., 2011b). It was also verified that active caspase-3 dimers bind directly to the apoptosome (Yin et al., 2006; Yuan et al., 2011b), whereas previous studies suggested that caspase-3 binding may be mediated by XIAP (Bratton et al., 2001; Hill et al. 2004). In addition, unprocessed pc-3 dimers were bound only weakly, if at all, to the apoptosome (Yuan et al., 2011b). When taken together, the data provides additional support for the idea that pc-9 catalytic domains may be activated when they bind to the apoptosome (Yuan et al., 2011b). While a physiological role for caspase-3 binding to the apoptosome remains to be determined, this feedback mechanism could inactivate pc-9 apoptosomes.
In the emerging model, pc-9 zymogens bind to the apoptosome to create a CARD-CARD disk that is located off-center relative to the central hub. The size of the disk and a local mismatch between the disk and the hub may block many of the possible binding sites for pc-9 catalytic domains. Hence, the resulting asymmetry may leave only a single binding site available at any given time for pc-9 catalytic domains (Figure 8B, lower right). It is hypothesized that the interaction of pc-9 catalytic domains with NBDs may induce zymogen loops to rearrange and complete the active site. Thus, pc-9 would remain active only while catalytic domains are bound to the central hub. Moreover, processing of the linker between catalytic domains (p20-p10 linker) may actually enhance binding to the platform and facilitate activation (Yuan et al., 2011b). In this scenario, pc-9 activation may be dynamic with catalytic domains of different pc-9 molecules coming on and off the platform binding site. At first glance this mechanism seems rather complicated. However, multiple steps would ensure that robust pc-9 activation could only occur in the context of a fully assembled holo-apoptosome.
Published data show that pc-9 dimerization can be induced by various manipulations to create an active procaspase (Yin et al., 2006; Pop et al., 2006; Boatright et al., 2003; Chao et al., 2005). However, the question remains: “are these experiments relevant to what occurs on the apoptosome?” For example, activation of a chimeric pc-9/pc-8 zymogen can be explained because the pc-9 CARD in this construct provides a normal binding mode to the apoptosome though disk formation, while the CARD-p20 linker of pc-9 may have helped to form the binding site for pc-8 catalytic domains on the platform (Yuan et al., 2011b). When taken together, these processes could have led to activation of pc-8 catalytic domains, due to their similarity with pc-9 (Pop et al., 2006), and this may have occurred with monomers or dimers, depending upon which model is correct. In principle, pc-9 catalytic domains flexibly-attached to the disk could dimerize and become active, due to their close proximity on the apoptosome, as occurred with the GCN4/pc-9 chimera (Yin et al., 2006). However, active caspase-3 binding to the apoptosome significantly reduced pc-9 activity (Yuan et al., 2011b). This activity loss would not be expected if flexibly-attached pc-9 catalytic domains were the active species in the apoptosome.
Concluding remarks and future directions
Divergent evolution has tailored the assembly of cell death platforms in worms, humans and flies. This is reflected by differences in the oligomeric state of inactive Apaf-1 like molecules, by different mechanisms of assembly regulation, and by variations in platform symmetry. In addition, apoptosome assembly creates platforms in which CARDs are displayed quite differently in three benchmark metazoans. In humans, apoptosome assembly may proceed by a series of steps that create a favorable equilibrium between inactive and assembly competent Apaf-1 molecules and accessory proteins may facilitate the process in vivo. Formation of lateral dimers and complete rings may trap Apaf-1 like molecules in an extended conformation with bound ATP/dATP. In a recurring theme, domain linkers play important roles in apoptosome assembly and procaspase activation. Thus, the role of the CARD-p20 linker in pc-9 binding and activation remains to be clarified. Higher resolution studies of human and fly apoptosomes are now needed to improve our understanding of assembly and to reveal the conformation of β -propellers in the presence (Apaf-1) and absence of cytochrome c (Dark).
Finally, procaspase activation may depend on the formation of a CARD-CARD disk which sits above the central hub. Disk formation may concentrate catalytic domains of procaspases near the platform to enhance productive binding and activation. The CARD-CARD disk is positioned acentrically to create an asymmetric proteolysis machine. Perhaps the disk may act as a spatial filter to insure that only a single binding site for pc-9 catalytic domains is accessible at a time. Recent data support an allosteric model of pc-9 activation, with the suggestion that catalytic domains from a single procaspase may be bound to the platform. However, many experiments have shown that enforced dimerization of pc-9 leads to catalytic activity. Thus, further tests are needed to distinguish between these models of pc-9 activation.
Supplementary Material
Acknowledgements
Due to space considerations, we could not cite all papers that deal with intrinsic cell death and pc-9 activation. We apologize for any oversights in this regard. This work was supported by an NIH grant to C. Akey (R01 GM63834).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding and activation. Mol. Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell. Biol. 2003;5(7):647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell. 2007;26:301–310. doi: 10.1016/j.molcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Lu W, Rabinowitz JD, Shi Y. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol. Cell. 2007;25:181–192. doi: 10.1016/j.molcel.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell. Sci. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM. Recruitment, activation and retention of caspases-9 and −3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nature Struct. Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3(6):183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev. Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase Dronc is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insights into signaling by STAND proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- del Peso L, González VM, Núñez G. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J. Biol. Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J. Struct. Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Kumar S. A biochemical analysis of the activation of the Drosophila caspase DRONC. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. 2008;16:461–470. doi: 10.1038/sj.cdd.4402288. [DOI] [PubMed] [Google Scholar]
- Dorstyn L, Mills K, Lazebnik Y, Kumar S. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 2004;167:405–410. doi: 10.1083/jcb.200408054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 2002;156:1089–1098. doi: 10.1083/jcb.200111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesik SW, Shi Y. Structural biology. Controlling the caspases. Science. 2001;294:1477–1478. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes and Develop. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work., Nature Rev. Molec. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23:2134–2145. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ding L, Spencer DM, Núñez G. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- Hu Y, Benedict MA, Ding L, Nunez G. Role of Cytochrome c and dATP/ATP hydrolysis in Apaf-1 mediated caspase-9 activation and apoptosis. The EMBO J. 1999;13:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Vucic D, Miller LK. The Drosophila inhibitor of apoptosis DIAP1 suppresses cell death induced by the caspase drICE. FEBS Letters. 1998;440:243–248. doi: 10.1016/s0014-5793(98)01465-3. [DOI] [PubMed] [Google Scholar]
- Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1 a Drosophila Apaf-1/CED-4 related caspase activator. Mol. Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- Kim H-E, Du F, Fang M, Wang X. Formation of an apoptosome is initiated by cytochrome c induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. U SA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HE, Jiang X, Du F, Wang X. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol. Cell. 2008;30:239–247. doi: 10.1016/j.molcel.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO reports. 2006;7:933–939. doi: 10.1038/sj.embor.7400773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K, Daish T, Harvey KF, Pfleger CM, Hariharan IK, Kumar S. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J. Cell Biol. 2006;172:809–815. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007a;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yan g J.K, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu, Rev, Immunol. 2007b;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol. Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3(4):e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purring-Koch C, McLendon G. Cytochrome c binding to Apaf-1: the effects of dATP and ionic strength. Proc. Natl. Acad. Sci. U S A. 2000;97:11928–11931. doi: 10.1073/pnas.220416197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, Luo G, Li H, Wang J, Yan N, Shi Y. Crystal Structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of Procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase-9. Proc. Natl. Acad. Sci(USA) 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J. Biol. Chem. 2009;284:32717–32724. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. Crystal structure of full-length apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure. 2011;19:1074–1083. doi: 10.1016/j.str.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S. A molecular view on signal transduction by the apoptosome. Cell Signal. 2012;24:1420–1425. doi: 10.1016/j.cellsig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural Basis for the Inhibition of Caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. Caspase-9 and Apaf-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nature Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Miller LK. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr. Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Song Z, Steller H. Death by design: mechanism and control of apoptosis. Trend. Cell Biol. 1999;12:49–52. [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, Bergmann A. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1 mediated oligomerization. Mol. Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Saleh A, Hedge R, Datta P, Shiozaki E, Chai J, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. Nature. 2001;409:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, Robinson CV, Siegel RM, Walz T, Wu H. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wallen HD, Inohara N, Nuñez G. Interaction and regulation of the Caenorhabditis elegans death protease CED-3 by CED-4 and CED-9. J. Biol. Chem. 1997;272:21449–21454. doi: 10.1074/jbc.272.34.21449. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Yan N, Shi Y. Mechanisms of apoptosis through structural biology. Annu. Rev. Cell Dev. Biol. 2005;21:35–56. doi: 10.1146/annurev.cellbio.21.012704.131040. [DOI] [PubMed] [Google Scholar]
- Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nature Struct. Molec. Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, Xue D, Shi Y. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1997;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J. Biol. Chem. 2001;276:13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure. 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Dorstyn L, Kumar S, Ludtke SJ, Akey CW. Structure of the Drosophila apoptosome at 6.9Å resolution. Structure. 2011a;19:128–140. doi: 10.1016/j.str.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW. The holo-apoptosome: activation of procaspase-9 and interactions with procaspase-3. Structure. 2011b;19:1084–1096. doi: 10.1016/j.str.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An Apaf-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.