Abstract
Soluble adenylyl cyclase (sAC) is an evolutionarily conserved bicarbonate sensor. In mammals, it is responsible for bicarbonate-induced, cAMP-dependent processes in sperm required for fertilization and postulated to be involved in other bicarbonate- and carbon dioxide-dependent functions throughout the body. Among eukaryotes, sAC-like cyclases have been detected in mammals and in the fungi Dictyostelium; these enzymes display extensive similarity extending through two cyclase catalytic domains and a long carboxy terminal extension. sAC-like cyclases are also found in a number of bacterial phyla (Cyanobacteria, Actinobacteria, and Proteobacteria), but these enzymes generally possess only a single catalytic domain and little, if any, homology with the remainder of the mammalian protein. Database mining through a number of recently sequenced genomes identified sAC orthologues in additional metazoan phyla (Arthropoda and Chordata) and additional bacterial phyla (Chloroflexi). Interestingly, the Chloroflexi sAC-like cyclases, a family of three enzymes from the thermophilic eubacterium, Chloroflexus aurantiacus, are more similar to eukaryotic sAC-like cyclases (i.e., mammalian sAC and Dictyostelium SgcA) than they are to other bacterial adenylyl cyclases (ACs) (i.e., from Cyanobacteria). The Chloroflexus sAC-like cyclases each possess two cyclase catalytic domains and extensive similarity with mammalian enzymes through their carboxy termini. We cloned one of the Chloroflexus sAC-like cyclases and confirmed it to be stimulated by bicarbonate. These data extend the family of organisms possessing bicarbonate-responsive ACs to numerous phyla within the bacterial and eukaryotic kingdoms.
Keywords: Chloroflexus aurantiacus, Evolution, Photosynthesis, Cyclic AMP
Introduction
Cyclic adenosine monophosphate (cAMP) is a nearly ubiquitous second messenger that mediates a wide variety of signal-transduction processes in organisms from bacteria through higher eukaryotes. In mammals, two types of adenylyl cyclase (AC) synthesize cAMP: a widely studied family of isoforms, transmembrane ACs (tmACs), and a more recently isolated soluble AC (sAC) (Buck et al. 1999). sAC and tmACs seem to modulate distinct cAMP signaling cascades within mammalian cells (Zippin et al. 2003, 2004). sAC lacks transmembrane domains, is insensitive to G-protein or forskolin regulation (Buck et al. 1999), and is localized to distinct compartments within cells (Zippin et al. 2003), where it is postulated to regulate intracellular targets of cAMP (Wuttke et al. 2001; Zippin et al. 2001, 2004).
Full-length sAC protein is comprised of two N-terminal domains with homology to catalytic domains from all class III cyclases, the largest of the six known classes of nucleotide cyclases, which comprises many bacterial and all known eukaryotic adenylyl and guanylyl cyclases. Similar to tmACs (Tang and Gilman 1995), the two catalytic domains (C1 and C2) are sufficient for enzymatic activity (Buck et al. 1999). These two catalytic domains are followed by a consensus nucleotide-binding P-loop sequence of unknown function and a long carboxy-terminal sequence with little homology to known functional domains (Fig. 1a). A structurally related cyclase was identified in the eukaryotic Fungi, Dictyostelium; SgcA possesses two cyclase catalytic domains, followed by a P-loop consensus sequence (Roelofs et al. 2001). sAC’s catalytic domains are more closely related to catalytic domains found in bacterial ACs than to catalytic domains from other mammalian cyclases (Fig. 1b; Buck et al. 1999; Chen et al. 2000). The sAC-like cyclases described from bacteria (mostly from the phylum Cyanobacteria, but also from Actinobacteria and Proteobacteria) differ from mammalian and Dictyostelium sAC; they possess only a single catalytic domain. It has been assumed that a second cyclase catalytic domain was added in the course of evolution to the eukaryotic enzyme (Roelofs and Van Haastert 2002).
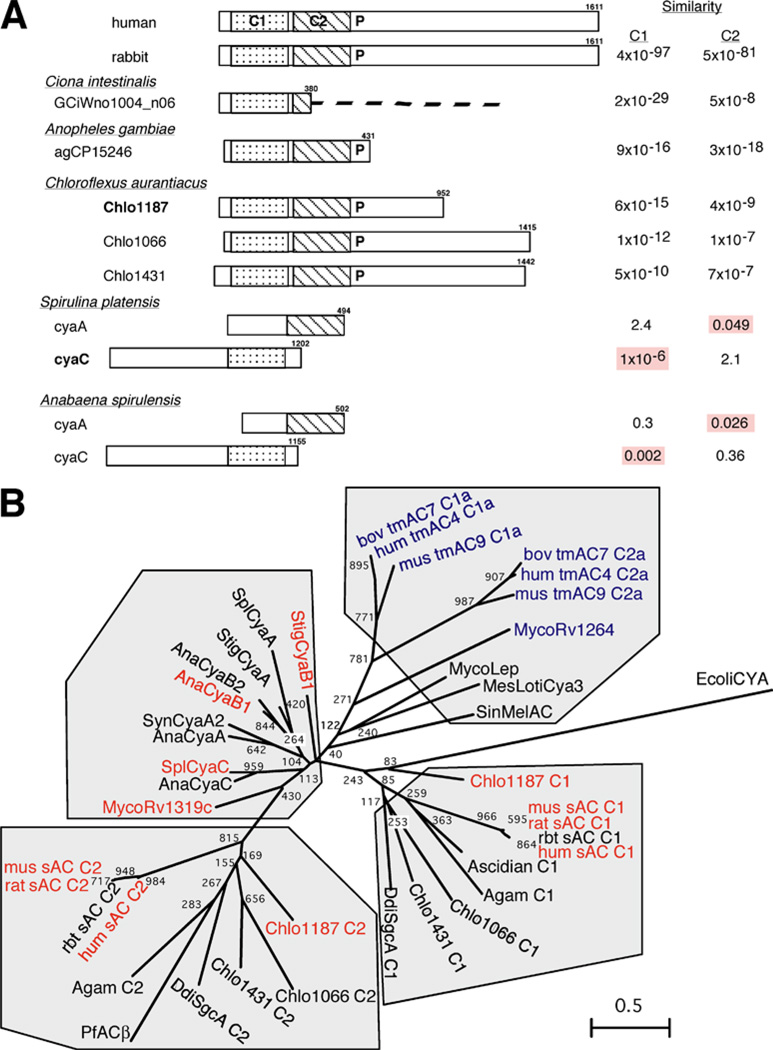
Fig. 1.
Soluble adenylyl cyclase (sAC) orthologues in different species. a Diagram of eukaryotic and Chloroflexusaurantiacus adenylyl cyclases (ACs) along with bacterial ACs. The relative similarities are “expect values” taken directly from a PSI-BLAST search of the human sAC protein vs the non-redundant GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). These values estimate the statistical significance of the match by specifying the number of matches expected to occur by chance. The values indicating greatest similarity are highlighted in red for single-domain bacterial ACs. Relative locations of the catalytic domains within the bacterial ACs are represented as shaded boxes and are aligned under the sAC catalytic domain with greater similarity. P indicates the conserved P-loop nucleotide-binding motif. The number of amino acid residues are also indicated. b Phylogenetic relationship between catalytic domains from a variety of ACs aligned by Clustal W (Higgins et al. 1994) is represented as an unrooted dendrogram constructed by a neighbor-joining plot (Perriere and Gouy 1996). Default values at DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/search/clustalw-j.html) were used with 1,000 bootstrap replications. The numbers indicate bootstrap values, using Escherichia coli CYA as the outgroup. A scale of branch length is shown in the lower right corner. ACs known to be activated by bicarbonate are in red, those known to be bicarbonate insensitive are in blue. Accession numbers for the proteins used: P. falciparum β, NP_704518; C. aurantiacus Chlo1066, ZP_00018085; C. aurantiacus Chlo1187, ZP_00018205; C. aurantiacus Chlo1431, ZP_00018442; Spirulina platensis CyaA, BAA22996; S. platensis CyaC, T17197; Stigmatella aurantiaca CyaA, CAA11549; S. aurantiaca CyaB1, T10905; Synecocystis sp. PCC6803 CyaA2, BAA16969; Anabaena spirulensis CyaA, P43524; A. spirulensis CyaB1, ZP_00018205; A. spirulensis CyaB2, BAA13999; A. spirulensis CyaC, BAA14000; Mycobacterium leprae AC, CAA19149; Sinorhizobium melioti AC, S60684; Mesorhizobium loti Cya3, BAB50205; Dictyostelium discoideum SgcA, AAL92097; A. gambiae, EAA10271; Rattus norvegicus sAC, AF081941; human sAC, NP_060887; Mus musculus sAC, NP_766617; rabbit sAC, AAO38673; Mycobacterium Rv1264, CAB00890; Mycobacterium Rv1319c, Q10632; human tmAC4, AAM94373; bovine tmAC7, CAA89894; Mus musculus tmAC9, CAA90570; Ciona intestinalis AC, GciWno1004_n06; E. coli CYA, CAA47280
Mammalian sAC (Chen et al. 2000; Litvin et al. 2003) and a subset of the bacterial sAC-like ACs (Cann et al. 2003) are directly stimulated by bicarbonate anions. The mechanism of bicarbonate stimulation is conserved between bacterial and mammalian enzymes. Bicarbonate elevates Vmax and alleviates substrate inhibition of both mammalian (Litvin et al. 2003) and cyanobacterial sAC-like enzymes (Litvin 2003). Bicarbonate regulation of mammalian sAC is thought to be responsible for the cAMP-dependent changes in capacitation (Chen et al. 2000) and motility (Esposito et al. 2004) in sperm and the pH-dependent mobilization of proton pumps in the epididymus required for acidification of the lumen (Pastor-Soler et al. 2003). Bicarbonate regulation of cyanobacterial ACs has been postulated to provide a mechanism for sensing availability of inorganic carbon in these photosynthetic bacteria (Cann et al. 2003; Wuttke et al. 2001).
We explored the evolutionary conservation of sAC-like enzymes across several phyla in the eukaryal and bacterial kingdoms, using database mining and molecular cloning. We detected additional sAC orthologues in rabbit, sea squirt, mosquito, and a very well conserved sAC orthologue in Chloroflexi eubacteria. Chloroflexusaurantiacus is a thermophilic, anoxygenic, green, phototrophic eubacterium that is found in alkaline hot springs at temperatures up to 70°C (Pierson and Castenholz 1974). C. aurantiacus represents the earliest branch of eubacteria capable of photosynthesis, and many of its characteristics can be found in diverse groups of phototrophic bacteria. Chloroflexus sAC is unique among bacterial orthologues; its domain organization is more similar to eukaryotic enzymes than other bacterial sAC-like enzymes.
Materials and methods
Recombinant DNA
Rabbit sAC was cloned from rabbit testis (Pel-Freez Biologicals, Rogers, Ark., USA). Total RNA was isolated with TRIzol (Invitrogen), and a cDNA template was prepared by using the 3′ rapid amplification of cDNA ends (RACE) system (Invitrogen). Using 20 pmol of each primer and 5 µl of the synthesized cDNA, polymerase chain reaction (PCR) amplifications were performed for 35 cycles (30 s at 94°C, 120 s at 55°C, and 120 s at 72°C) with 2.5 U of Ex-Taq DNA polymerase (TaKaRa, Kyoto). Primer sets used are as follows: LRL1295, LRL1296, LRL866, LRL741, LRL1319, LRL786, LRL1339, and LRL788. LRL1339–AUAP and LRL787–AUAP primer combinations were used for 3′ RACE reactions. The 5′ end of the mRNA was determined using 5′ RACE system (Invitrogen), using primer LRL1317 for synthesizing RNA. LRL577–AUAP and LRL1318–AUAP combinations were used for 5′ RACE reactions. Amplified fragments were cloned into a pCRII-TOPO vector (Invitrogen). Sequences of the primers were:
| LRL577: 5′-CTCCACCAGCTGCTCGGC-3′ |
| LRL741: 5′-CCGCTCGAGTCAGTTCTTTAGTCTGACACCACT-3′ |
| LRL786: 5′-GGGCCAGAGGCAAGATGACAA-3′ |
| LRL787: 5′-GGCGTGTGGTTCAAATATGAA-3′ |
| LRL788: 5′-GTCCCATTCCTGAAGTCTGGC-3′ |
| LRL866: 5′-GACCTGCGACTCTGTCACCTAC-3′ |
| LRL1295: 5′-TAGCTGCTCACTTACCAGACCTCAT-3′ |
| LRL1296: 5′-ACGCCATACCAAACATGACTTTCTC-3′ |
| LRL1317: 5′-CGCACTGATGTAGTAGTTGAGGATCTCCAC-3′ |
| LRL1318: 5′-GTACATGGCTGTGCTGAACTTCTC-3′ |
| LRL1319: 5′-AAGAACCTCGACCACCACAGGGTG-3′ |
| LRL1339: 5′-GGAAGACATCATCCCTCTGGAATC-3′ |
Chloroflexus sAC (Chlo1187) was cloned from C. aurantiacus (American Type Culture Collection #29366). Cells were freeze-dried in liquid nitrogen, homogenized with mortar and pestle, dissolved in water, and used as a template for PCR. After denaturing at 94°C for 5 min, using 20 pmol each of primers LRL1351 (5′-CACCATGGACATTCCAAGACGCAGCGG-3′) and LRL1352 (5′-AGCTAATCATCAGTGATGTTGAGCACCG-3′), PCR amplification was performed for 35 cycles (30 s at 94°C, 60 s at 55°C, and 210 s at 72°C) with 2.5 U of Ex-Taq DNA polymerase (TaKaRa). The amplified fragment was cloned into a pCRII-TOPO vector (Invitrogen). The plasmid was digested with EcoRI and the 1.4-kb fragment cloned into pGSX-3T vector (Amersham Biosciences). Sequences were verified using the Rockefeller University DNA sequencing resource center.
Expression and purification of recombinant AC proteins
Chloroflexus protein was expressed in Escherichia coli BL21(DE3) as a GST fusion protein. Bacteria were grown at 37°C in Luria Broth containing ampicillin (100 µg/ml) until the A595 was 0.5. Isopropyl-1-thio-β-d-galactopyranoside was added to the final concentration of 0.5 mM, and the expression was induced for 3 h at room temperature. Cells were pelleted, and protein was purified through a glutathione Sepharose 4B column as previously described (Litvin et al. 2003). The sAC portion of the fusion protein was excised with thrombin for 20 h at 16°C, according to the manufacturer’s instructions (Amersham Biosciences). Protein was purified using DE52 anion exchange chromatography (Whatman, Clifton, N.J., USA).
Cyclase assay
Cyclase assays were performed in a final volume of 100 µl, using ~10 ng of purified cyclase protein in the presence of 50 mM Tris-HCl (pH 7.4), 10 mM ATP, and either 5 mM MnCl2, MgCl2, CoCl2, ZnCl2, or CaCl2, as indicated. Reactions were incubated at 30°C for 30 min unless otherwise noted and were stopped by adding 100 µl 0.2 N HCl. cAMP formed was measured using a Correlate-EIA Direct cAMP Enzyme Immunoassay Kit (Assay Designs), and kinetic analyses were performed using the program EnzymeKinetics, version 1.11 (Trinity Software, Plymouth, N.H., USA).
Results
Identification of sAC orthologues in several organisms
The sAC gene was first identified in rat (Buck et al. 1999), and orthologues have been described from human (Jaiswal and Conti 2001; Litvin et al. 2003) and mouse (Esposito et al. 2004; Jaiswal and Conti 2001). In an attempt to identify sAC orthologues in other species, we used PCR and degenerate oligonucleotide primers derived from rat, mouse, and human sAC genes and searched several databases, using the human sAC catalytic region as the query. We were able to PCR amplify a sAC orthologue from rabbit mRNA (GenBank accession number AY212921), and we found sAC-like sequences in Arthropod and Chordates in the completed genomes of the mosquito (Anopheles gambiae), and the sea squirt (Ciona intestinalis) (Fig. 1a). A putative sAC-like cyclase, with tandemly arranged catalytic domains, was recently detected within a hypothetical open reading frame from the malaria parasite Plasmodium falciparum (Muhia et al. 2003), but its complete domain organization is not yet known. We also found a number of interesting sAC-like sequences in the genome of the thermophilic bacterium, C. aurantiacus. These Chloroflexus sAC-like genes were structurally more similar to mammalian sAC than they were to other bacterial sAC-like ACs (Fig. 1a).
Phylogenic analysis of sAC proteins
Three genes from Chloroflexus (GenBank accession numbers ZP_00018205, ZP_00018085, and ZP_00018442) possess two sAC-like catalytic domains, followed by a conserved P-loop nucleotide-binding motif (Fig. 1a). The deduced amino acid sequence of the most similar of these, Chlo1187 (GenBank accession number ZP_00018205), received a BLAST score of 173 over its entire length and shares 30% identity within its catalytic domains when compared with human sAC. Not only does its domain structure more closely resemble mammalian sAC, but BLAST (Fig. 1a) and phylogenetic analysis (Fig. 1b) reveal its individual catalytic domains are more similar to those of mammalian sAC than are any of the bacterial ACs containing only a single catalytic domain.
As expected, rabbit sAC and the putative Anopheles sAC also possess this domain architecture, and although information regarding its C terminus is missing, the Ciona sAC-like gene also appears to be similarly organized. A recent phylogenetic analysis of a number of class III cyclases in bacteria and archaebacteria revealed only one other species that possessed a putative AC sharing this domain structure (Shenoy and Visweswariah 2004). Two genes within Leptospira interrogans (GenBank accession numbers YP_003109 and NP_714188) share overall structural similarity and a BLAST similarity score of 120 with mammalian sAC.
Enzymatic characterization of Chloroflexus sAC
The enzymatic properties of a number of bacterial sAC-related cyclases have been explored and their bicarbonate responsiveness investigated (Cann et al. 2003; Chen et al. 2000). However, the cyclases selected for these analyses all consisted of a single catalytic domain. Due to its structural similarity and conserved sequence, the sAC-like cyclase from L. interrogans was predicted to be bicarbonate responsive (Shenoy and Visweswariah 2004), but this was never tested. We PCR-amplified the sAC-like cyclase, Chlo1187, from Chloroflexus genomic DNA. Because the P-loop containing, C-terminal portion of mammalian sAC has been purported to have an inhibitory effect on catalytic activity (Buck et al. 1999; Jaiswal and Conti 2001; Jaiswal and Conti 2003), we heterologously expressed and purified from E. coli the portion of Chlo1187 containing its two catalytic domains (Fig. 2a).
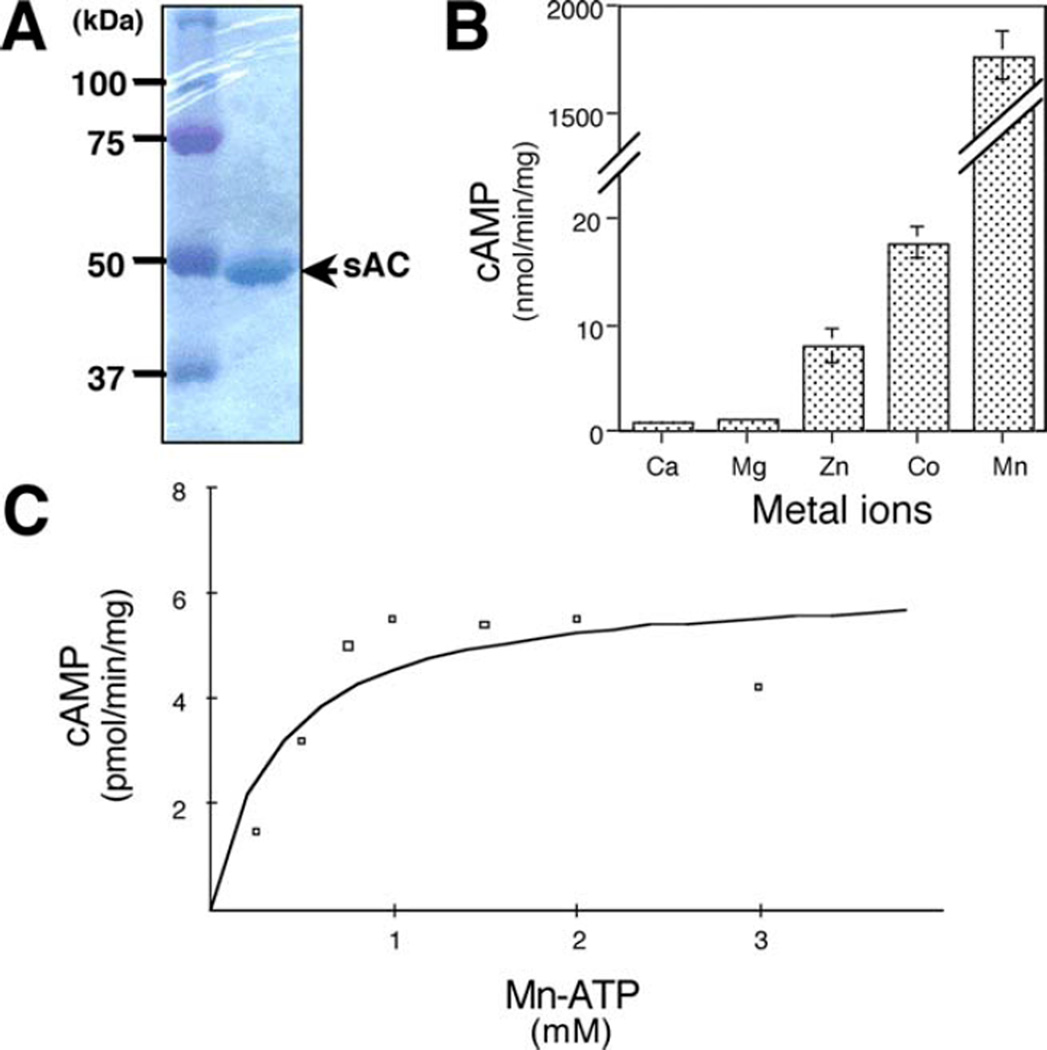
Fig. 2.
Characterization of Chloroflexus sAC activity. a Coomassie Blue-stained 10% SDS-PAGE demonstrating the purity of Chloroflexus sAC protein used in this study. bChloroflexus sAC activity measured in the presence of different cations. cChloroflexus sAC activity measured as a function of substrate ATP-Mn2+ in the presence of excess MnCl2 (100 mM) for 10 min. The Km value of 0.7 mM ATP-Mn2+ was determined using non-linear regression analysis. Graphs shown are representative of three independent experiments performed in triplicates
We first tested cyclase activity of recombinant sAC in the presence of the divalent cations present in Chloroflexus growth medium (Fig. 2b). As is the case with mammalian sAC, Chlo1187 has higher activity in the presence of ATP-Mn2+ compared to ATP-Mg2+ (Chen et al. 2000; Jaiswal and Conti 2001; Litvin et al. 2003). Also similar to mammalian sAC, cobalt also supports catalytic activity, and like mammalian sAC (Braun 1975; Goh and White 1988; Hyne and Garbers 1979; Litvin 2003), ATP-Co2+ supports more activity than ATP-Mg2+ but less than ATP-Mn2+ (Fig. 2b). Interestingly, zinc was found to have opposite effects on mammalian and Chloroflexus sAC-like enzymes. Zinc supported approximately ten times greater activity of Chloroflexus sAC-like cyclase relative to ATP-Mg2+ (Fig. 2b), while it is a potent inhibitor of the mammalian enzyme (Braun 1975; Litvin 2003). The Km for ATP-Mn2+ of Chloroflexus sAC-like cyclase was 0.7 mM (Fig. 2c), which is very similar to the value of 0.8 mM reported for the mammalian enzyme (Litvin et al. 2003).
Chloroflexus grow optimally at 55°C. The in vitro cyclase activity of Chlo1187 was unchanged by incubation at 55°C (data not shown), revealing that a class III cyclase domain is capable of functioning at this elevated temperature.
Bicarbonate activation of Chloroflexus sAC
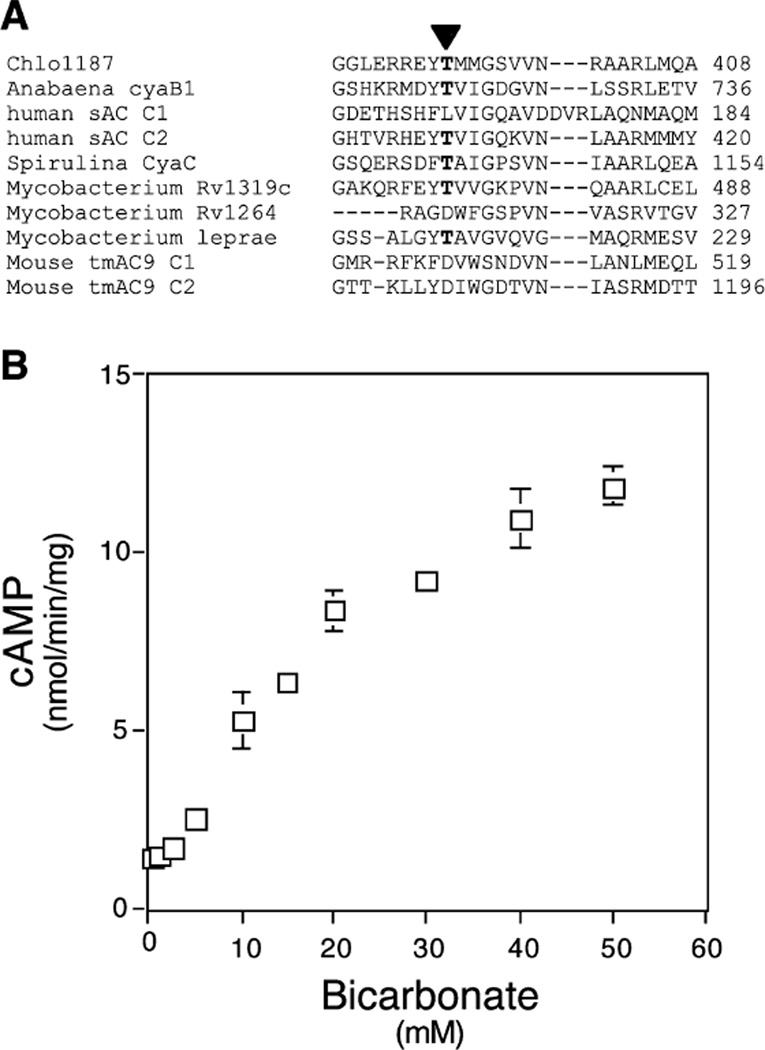
Chlo1187 possesses a threonine (Fig. 3a) at the position thought to be predictive of bicarbonate responsiveness (Cann et al. 2003). We confirmed that heterologously expressed and purified Chlo1187 could be stimulated by bicarbonate (Fig. 3b). We measured bicarbonate responsiveness in the presence of ATP-Mn2+, which is more commonly used to reflect the substrate of bacterial cyclases during growth under natural conditions (Cann et al. 2003; Chen et al. 2000). Activation was dose responsive, leading to nearly tenfold stimulation of activity with an EC50 of 25 mM NaHCO3 (Fig. 3b).
Fig. 3.
Bicarbonate activation of Chlo1187. a Sequence alignment of a portion of the catalytic domain of Chlo1187 with the homologous region of other cyclases. Arrowhead indicates the conserved threonine residue suggested to be responsible for bicarbonate activation. bChloroflexus sAC activity was assayed in the presence of 10 mM ATP and 5 mM MnCl2 and indicated concentrations of NaHCO3. Values represent averages of triplicate determinations, with error bars indicating SD from the means
Discussion
Upon cloning mammalian sAC, we postulated that the mammalian enzyme originated from an evolutionary fusion of distinct bacterial AC genes. Each species of Cyanobacteria or Myxobacteria possessing a related sAC-like cyclase was found to have at least two genes encoding sAC-like cyclases, each with a single catalytic domain; one cyclase was always more similar to mammalian sAC C1, with the other being more similar to sAC C2 (Buck et al. 1999). Not surprisingly, the known eukaryal sAC-like cyclases contain this two catalytic domain organization; however, we now demonstrate tandem catalytic domains are also found in eubacterial (Chloroflexi or green nonsulfur bacteria) cyclases as well.
Eukaryotic sAC-like cyclases, including those found in mammals, Arthropods, Chordates, and slime molds, also possess a third conserved domain: a consensus P-loop nucleotide-binding motif, not previously found in any of the bacterial ACs, was found downstream of the second catalytic domain. This domain structure is found in eubacteria as well. Leptospira (Shenoy and Visweswariah 2004) and Chloroflexus contain cyclases with tandem catalytic domains, followed by a consensus P-loop binding motif. We selected the Chloroflexus cyclase most similar to sAC for enzymatic characterization. Similar to mammalian sAC (Buck et al. 1999), the P-loop consensus was not needed for catalytic activity. This P-loop binding motif remains of unknown function, but its conservation from bacteria to man suggests an important modulatory role. Also similar to mammalian sAC, the two catalytic domains were sufficient for bicarbonate-responsive enzymatic activity.
C. aurantiacus was originally found in hot springs and is considered to be one of the most ancient lineages of phototrophs. We found Chloroflexus sAC to be equally active at 55°C as it was at 30°C, demonstrating that class III cyclase domains are capable of functioning at elevated temperatures. Sequences related to class III cyclase domains are found in archaebacteria (Shenoy and Visweswariah 2004); our data demonstrating activity of Chloroflexus AC suggests that these archaeal genes may, in fact, encode functional ACs. Functioning ACs in archaea would establish that this kingdom also utilizes cAMP as a second messenger.
The existence of bicarbonate-regulated cAMP signal-transduction pathways in two of the earliest photosynthetic classes of organisms, blue-green algae and green nonsulfur bacteria, suggest a link between the evolution of photosynthesis and carbon dioxide sensing. The conservation of this signaling paradigm in mammals reveals its importance to life.
Acknowledgements
This work was supported in part by the Uehara Memorial Foundation (to M.K.), National Institutes of Health grants GM62328 and HD42060 (to J.B.) and HD38722 (to L. R.L.), and by the Ellison Medical Foundation (to J.B.). We thank members of the Levin/Buck laboratory for discussion.
Footnotes
The nucleotide sequence of rabbit sAC has been deposited (GenBank accession number AY212921)
References
- Braun T. The effect of divalent cations on bovine spermatozoal adenylate cyclase activity. J Cyclic Nucleotide Res. 1975;1:271–281. [PubMed] [Google Scholar]
- Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann MJ, Hammer A, Zhou J, Kanacher T. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J Biol Chem. 2003;278:35033–35038. doi: 10.1074/jbc.M303025200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- Esposito G, Jaiswal BS, Xie F, Krajnc-Franken MA, Robben TJ, Strik AM, Kuil C, Philipsen RL, Van Duin M, Conti M, Gossen JA. Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect. Proc Natl Acad Sci USA. 2004;101:2993–2998. doi: 10.1073/pnas.0400050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PP, White IG. Control of ram sperm adenylate cyclase by divalent cations. Aust J Biol Sci. 1988;41:377–385. doi: 10.1071/bi9880377. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyne RV, Garbers DL. Regulation of guinea pig sperm adenylate cyclase by calcium. Biol Reprod. 1979;21:1135–1142. doi: 10.1095/biolreprod21.5.1135. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Conti M. Identification and functional analysis of splice variants of the germ cell soluble adenylate cyclase. J Biol Chem. 2001;276:31698–31708. doi: 10.1074/jbc.M011698200. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Conti M. Calcium regulation of the soluble adenylyl cyclase expressed in mammalian spermatozoa. Proc Natl Acad Sci USA. 2003;100:10676–10681. doi: 10.1073/pnas.1831008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvin TN. Pharmacology. New York: Weill Medical College and Graduate School of Cornell University; 2003. A novel mechanism of adenylyl cyclase activation conserved from cyanobacteria to man; p. 129. [Google Scholar]
- Litvin TN, Kamenetsky M, Zarifyan A, Buck J, Levin LR. Kinetic properties of “soluble” adenylyl cyclase. Synergism between calcium and bicarbonate. J Biol Chem. 2003;278:15922–15926. doi: 10.1074/jbc.M212475200. [DOI] [PubMed] [Google Scholar]
- Muhia DK, Swales CA, Eckstein-Ludwig U, Saran S, Polley SD, Kelly JM, Schaap P, Krishna S, Baker DA. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J Biol Chem. 2003;278:22014–22022. doi: 10.1074/jbc.M301639200. [DOI] [PubMed] [Google Scholar]
- Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003;278:49523–49529. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Pierson BK, Castenholz RW. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- Roelofs J, Van Haastert PJ. Deducing the origin of soluble adenylyl cyclase, a gene lost in multiple lineages. Mol Biol Evol. 2002;19:2239–2246. doi: 10.1093/oxfordjournals.molbev.a004047. [DOI] [PubMed] [Google Scholar]
- Roelofs J, Meima M, Schaap P, Van Haastert PJ. The Dictyostelium homologue of mammalian soluble adenylyl cyclase encodes a guanylyl cyclase. EMBO J. 2001;20:4341–4348. doi: 10.1093/emboj/20.16.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy AR, Visweswariah SS. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 2004;561:11–21. doi: 10.1016/s0014-5793(04)00128-0. [DOI] [PubMed] [Google Scholar]
- Tang WJ, Gilman AG. Construction of a soluble adenylyl cyclase activated by Gs alpha and forskolin. Science. 1995;268:1769–1772. doi: 10.1126/science.7792604. [DOI] [PubMed] [Google Scholar]
- Wuttke MS, Buck J, Levin LR. Bicarbonate-regulated soluble adenylyl cyclase. J Pancreas. 2001;2:154–158. [PubMed] [Google Scholar]
- Zippin JH, Levin LR, Buck J. CO(2)/HCO(3)(−)-responsive soluble adenylyl cyclase as a putative metabolic sensor. Trends Endocrinol Metab. 2001;12:366–370. doi: 10.1016/s1043-2760(01)00454-4. [DOI] [PubMed] [Google Scholar]
- Zippin JH, Chen Y, Nahirney P, Kamenetsky M, Wuttke MS, Fischman DA, Levin LR, Buck J. Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains. FASEB J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- Zippin JH, Farrell J, Huron D, Kamenetsky M, Hess KC, Fischman DA, Levin LR, Buck J. Bicarbonate-responsive “soluble” adenylyl cyclase defines a nuclear cAMP microdomain. J Cell Biol. 2004;164:527–534. doi: 10.1083/jcb.200311119. [DOI] [PMC free article] [PubMed] [Google Scholar]