Summary
Signal transduction pathways ultimately converge upon sequence-specific DNA binding factors to reprogram gene expression and affect necessary changes in cellular growth or behavior. Transcription factors, in turn, team up with chromatin modifying activities, including enzymes that post-translationally modify histones. Chromatin is also dynamically modified to regulate DNA accessibility for replication and repair. However, chromatin is not simply an endpoint for signaling pathways. Histone modifications can relay signals to other proteins to trigger more immediate responses than can be achieved through altered gene transcription. Such signaling might be especially important to time-urgent processes such as the execution of cell cycle check points, chromosome segregation, or exit from mitosis. In addition, histone modifying enzymes often have multiple non-histone substrates, and coordination of activity towards different targets might direct signals both to and from chromatin.
Introduction
Signal transduction classically involves coordinated cascades of protein phosphorylation or dephosphorylation, which in turn alter protein conformation, protein-protein interactions, subcellular protein locations, or protein stability. In many cases these pathways begin at the cell surface and extend into the nucleus, where they alter the interactions of transcription factors and chromatin modifying enzymes with the chromatin template. In some cases, signaling promotes such interactions, while in others, factors are ejected from chromatin in response to incoming signals. Several such pathways have been defined that control developmental fate decisions or response to physiological or environmental changes (for examples, see (Fisher and Fisher, 2011; Long, 2012; Valenta et al., 2012). In these cases, the ultimate endpoint of the signal is often considered to be a modification of chromatin structure to modulate DNA accessibility to control gene expression.
The architecture of chromatin can be altered by a variety of mechanisms, including post-translational modification of histones, alterations in nucleosome locations, and exchange of canonical histones for histone variants. Histone modifications have at least three, non-mutually exclusive effects on chromatin packing (Butler et al., 2012; Suganuma and Workman, 2011). First, modifications such as acetylation or phosphorylation can alter DNA:histone and histone:histone interactions. Second, histone acetylation, methylation, and ubiquitylation can create binding sites for specific protein motifs, thereby directly promoting or inhibiting interactions of regulatory factors with chromatin (Smith and Shilatifard, 2010; Yun et al., 2011). Bromodomains, for example, promote interactions with acetyl-lysines within histones. PHD domains, Tudor domains, and chromo domains can selectively bind particular methylated lysines (Kme). At least one Tudor domain (TDRD3) serves as a reader for methylarginine (Rme) residues (Yang et al., 2010). In contrast, other domains, such as the PhD finger in BHC80 (Lan et al., 2007), are repelled by lysine methylation. Such regulation is enhanced by combining domains to create multivalent ‘readers’ of histone modification patterns (Ruthenburg et al., 2007). Combination of PhD and bromodomains in the TRIM24 protein, for example, creates a motif that specifically recognizes histone H3K23 acetylation in the absence of H3K4 methylation (Tsai et al., 2010). Third, histone modifications also affect the chromatin landscape by influencing the occurrence of other modifications at nearby sites (Lee et al., 2010). Methylation of H3R2, for example, inhibits methylation of H3K4, but not vice versa (Hyllus et al., 2007; Iberg et al., 2008). Such modification ‘cross talk’ can result either from direct affects of a preexisting modification on the ability of a second histone modifying enzyme to recognize its substrate site or from indirect effects on substrate recognition through the recruitment of ‘reader proteins’ that mask nearby modification sites. Binding of the chromodomain in the HP1 protein to H3K9me blocks subsequent phosphorylation of S10 by Aurora kinases, for example (Fischle et al., 2003).
The power of cross talk
Histone modification cross talk can also occur in trans, between sites on two different histones. The most studied example of such cross talk is the requirement of H2B monoubiquitylation for methylation of H3K4 (Shilatifard, 2006). In yeast, the Bre1 E3 ligase ubiquitylates H2BK123 and works together with the Paf1 complex to recruit the Set1 H3K4 methyltransferase complex, often referred to as COMPASS, to gene promoters (Lee et al., 2007). Bre1 mediated-H2B ubiquitylation also stimulates H3K79 methylation by the Dot1 methyltransferase (Nakanishi et al., 2009; Ng et al., 2002). Each of these histone modifications is widely associated with actively transcribed genes and can regulate multiple steps during transcription (Laribee et al., 2007; Mohan et al., 2010; Wyce et al., 2007). These cross talk events are conserved, at least in part, in mammalian systems (Kim et al., 2009; Zhou et al., 2011).
While H2B ubiquitylation is observed in the bodies of all actively transcribed genes, knockdown of the mammalian homolog of Bre1, ring finger protein 20 (RNF20), affects the expression of only a small subset of genes (Shema et al., 2008). Interestingly, RNF20 depletion not only led to the repression of some genes but also caused the upregulation of other genes. Genes negatively regulated by RNF20 and H2B ubiquitylation include several proto-oncogenes, such as c-MYC and c-FOS, as well as other positive regulators of cell proliferation. On the other hand, depletion of RNF20 and reduction in H2B ubiquitylation reduced the expression of the p53 tumor suppressor gene and impaired the activation of p53 in response to DNA damage. Consistent with these selective changes in gene expression, RNF20 depletion elicited a number of phenotypes associated with oncogenic transformation. The suggestion that RNF20 may function as a tumor suppressor is further supported by the finding of decreased levels of RNF20 and H3K79 methylation in testicular seminomas (Chernikova et al., 2012) and the observation that the RNF20 promoter is hypermethylated in some breast cancers (Shema et al., 2008).
A more concrete link between these histone modifications and human cancer comes from leukemias bearing translocations of the Mixed Lineage Leukemia (MLL) gene. MLL is a H3K4 methyltransferase related to the yeast Set1 protein found in the COMPASS complex. A number of different gene partners are found to be translocated to the MLL locus and this invariably creates an MLL fusion protein that lacks H3K4 methyltransferase activity. Interestingly, many of the translocation partners are part of a ‘super elongation complex’ that stimulates progress of the polymerase through gene bodies (Mohan et al., 2010; Smith et al., 2011). Recent data suggests that at least some of these oncogenic MLL fusion proteins alter the expression of select target genes, such as HOXA, by increasing H3K79 methylation (Okada et al., 2005). Knockdown of Dot1 reduced H3K79 methylation at these targets and inhibited oncogenic transformation by MLL fusion proteins. These examples demonstrate how deregulation of cross talk among different histone modifications can contribute to diseases such as cancer.
Not just for histones
Just as in histones, modifications in non histone proteins are subject to regulatory cross talk and serve as platforms for binding of ‘reader’ proteins. For example, a yeast kinetochore protein, Dam1, is methylated at K233 by the Set1 methyltransferase, an ortholog of mammalian MLL proteins (Zhang et al., 2005). The functions of Dam1, like those of other kinetochore proteins, are highly regulated by Aurora kinase mediated phosphorylation (Lampson and Cheeseman, 2011). At least some of these phosphorylation events are inhibited by prior methylation of Dam1, creating a phospho-methyl switch that impacts chromosome segregation (Zhang et al., 2005).
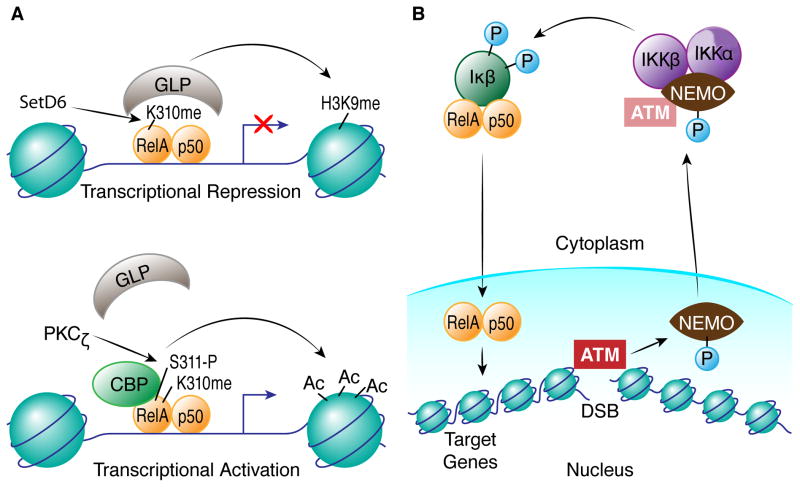
Another, more complicated example of a ‘phospho-methyl’ regulatory cassette occurs in the RelA subunit of NF-κB (Levy et al., 2011). RelA is monomethylated by SETD6 at K310, and this modification inhibits RelA functions in transcriptional activation through recruitment of another methyltransferase, G9a-like protein (GLP). GLP binds to K310me1 in RelA and induces a repressive histone modfication, H3K9me, in RelA target genes. Phosphorylation of the adjacent S311 in RelA, however, blocks GLP association with RelA and instead promotes the recruitment of CREB binding protein (CBP) to activate transcription of NF-κB targets (Duran et al., 2003) (Figure 1A).
Figure 1. Regulation of RelA/NF-κB by a phospho-methyl switch and in response to DNA damage.
A) Methylation of RelA K310 by SETD6 creates a binding site for GLP, which in turn methylates H3K9 at NF-κB target genes to inhibit transcription. Phosphorylation of RelA at S311 by PKCζ blocks binding of GLP to RelA (Levy et al., 2011) and, along with other RelA modifications not shown, promotes its interaction with CBP, leading to histone acetylation and activation of NF-κB target genes (Duran et al., 2003). B) Phosphorylation of NEMO by ATM in response to a DSB promotes its export from the nucleus. In the cytoplasm, NEMO activates the IKK complex, leading to IκB phosphorylation and degradation and NF-κB (RelA-p50) translocation to the nucleus where it can activate transcription as shown in (A). Note that some ATM may translocate with NEMO to the cytoplasm and participate in IKK activation.
These two examples, in yeast and in mammalian cells, likely foreshadow the discovery of many additional regulatory ‘switches’ created by modification cross talk. The p53 tumor suppressor is a prime candidate for such regulation, as it harbors several diverse modifications. Moreover, many kinase consensus sites contain arginine or lysine residues, providing a high potential for phospho-methyl, phospho-acetyl, or phospho-ubiquitin switches (Rust and Thompson, 2011).
The induction of H3K9me by recruitment of GLP via a methylation event in RelA illustrates how a signaling pathway, in this case mediated by NF-κB, can transduce a signal to chromatin. However, signaling can also occur in the other direction, i.e. a histone modification can affect the modification state of a non-histone protein. Methylation of Dam1, for example, requires ubiquitylation of histone H2B (Latham et al., 2011). Most likely H2Bub recruits the Set1 complex to centromeric nucleosomes, positioning it for methylation of Dam1 at the kinetochore. Thus, transregulation of post-translational modifications can occur both between histones (e.g. H2Bub and H3K4me) and between histones and non-histones (e.g. H2Bub and Dam1K233me), providing a platform for bidirectional signaling from chromatin.
Signaling to and from chromatin in response to DNA damage
Signaling to and from chromatin impacts other important cellular processes as well. DNA repair involves coordination among the repair machinery, chromatin modifications, and cell cycle checkpoint signaling. At the apex of the DNA damage response are three kinases related to the PI3 kinase family, ataxia telangiectasia mutated (ATM), ATM and Rad3 related protein (ATR) and DNA-PK (Jackson and Bartek, 2009; Lovejoy and Cortez, 2009). DNA-PK is activated when its regulatory subunit Ku70/80 binds to the end of a DNA double-strand break (DSB). ATR activation involves recognition of single-stranded DNA coated with replication protein A (RPA) by the ATR interacting protein, ATRIP, as well as direct interaction with topoisomerase IIβ binding protein (TopBP1) (Burrows and Elledge, 2008). Like DNA-PK, ATM is also activated in response to DSBs but rather than recognition of broken DNA ends, ATM appears to be activated in response to large-scale changes in chromatin structure caused by a DSB (Bakkenist and Kastan, 2003). How alterations in chromatin structure are signaled to ATM is at present unclear.
One of the earliest events in the DNA damage response is the phosphorylation of a variant of histone H2A, H2AX, by ATM, DNA-PK, and/or ATR (Rogakou et al., 1998). Phosphorylated H2AX (γH2AX) provides a mediator of DNA damage signaling directed by these kinases, and this modification is found in flanking chromatin regions as far as one megabase from a DNA DSB. This phosphorylation event creates a binding motif for the mediator of DNA damage checkpoint (MDC1) protein, which in turn recruits other proteins, such as Nijmegen breakage syndrome 1 (NBS1) and RNF8, to sites of DSBs through additional phospho-specific interactions (Chapman and Jackson, 2008; Kolas et al., 2007; Stucki and Jackson, 2006). NBS1 is part of the MRN complex that also contains Mre11 and Rad50 and is involved in DNA end processing for both the homologous recombination and non-homologous end joining pathways of DSB repair (Zha et al., 2009). In addition, NBS1 functions as a co-factor for ATM by stimulating its kinase activity and recruiting ATM to sites of DSBs where many of it substrates are located (Lovejoy and Cortez, 2009; Zha et al., 2009). ATM also phosphorylates effector proteins that only transiently localize to DSBs. One of these proteins is the checkpoint 2 (Chk2) kinase, which can be activated by ATM-mediated phosphorylation at sites of damage but then spreads throughout the nucleus to phosphorylate and regulate additional proteins as part of the DNA damage response (Bekker-Jensen et al., 2006). ATM also phosphorylates transcription factors, such as p53 and E2F1, to regulate the expression of numerous genes involved in the cellular response to DSBs (Banin et al., 1998; Biswas and Johnson, 2011; Canman et al., 1998; Lin et al., 2001). These events again illustrate that signals to chromatin, in this case resulting in H2AX phosphorylation, can be relayed to other proteins, both on and off the chromatin-DNA template.
Bi-directional signaling is illustrated even further by another branch of the ATM-mediated DNA damage response that involves activation of NF-κB. NF-κB is normally sequestered in an inactive state in the cytoplasm though its association with IκB. Following ATM activation by a DNA DSB, ATM phosphorylates NF-κB essential modulator (NEMO) in the nucleus (Wu et al., 2006), which promotes additional modifications to NEMO and export from the nucleus to the cytoplasm. Once in the cytoplasm, NEMO participates in the activation of the canonical inhibitor of NF-κB (IKB) kinase (IKK) complex that targets IκB for degradation, leading to NF-κB activation. NF-κB then translocates to the nucleus where it regulates the expression of genes important for cell survival following DNA damage. In this case, a change in chromatin structure caused by a DSB initiates a signal that travels to the cytoplasm and back to the nucleus to activate transcription of NF-κB target genes by modifying chromatin structure (Figure 1).
Multiple roles for H2B ubquitylation
In addition to phosphorylation of H2A/H2AX, a number of other histone modifications are induced at sites of DSBs in yeast and mammalian cells. One such modification is H2Bub, the same mark involved in regulating transcription as described above. As with transcription, the Bre1 ubiquitin ligase (RNF20-RNF40 in mammalian cells) is responsible for H2Bub at sites of DNA damage (Game and Chernikova, 2009; Moyal et al., 2011; Nakamura et al., 2011). Moreover, H2Bub is required for and promotes H3K4 and H3K79 methylation at sites of damage, similar to its role at actively transcribed genes. These histone modifications are important for altering chromatin structure to allow access to repair factors involved in DNA end-resection and processing (Moyal et al., 2011; Nakamura et al., 2011). Moreover, H2Bub and H3K79me are not only required for DNA repair but are also important for activating the Rad53 kinase and imposing subsequent cell cycle checkpoints (Giannattasio et al., 2005). Blocking H2B ubiquitylation or H3K79 methylation in response to DSBs inhibits Rad53 activation and impairs the G1 and intra S phase checkpoints.
Bre1-mediated H2B ubiquitylation and subsequent methylation of H3K4 by Set1 and H3K79 by Dot1 are also involved in regulating mitotic exit in yeast. The Cdc14 phosphatase controls mitotic exit by dephosphorylating mitotic cyclins and their substrates during anaphase (D’Amours and Amon, 2004). Prior to anaphase, Cdc14 is sequestered on nucleolar chromatin through interaction with its inhibitor, the Cf1/Net1 protein. Two pathways, FEAR (Cdc Fourteen Early Anaphase Release) and MEN (Mitotic Exit Network), control the release of Cdc14 from ribosomal DNA (rDNA) in the nucleolus. Upon inactivation of the MEN pathway, H2B ubiquitylation and methylation of H3K4 and H3K79 are necessary for FEAR pathway-mediated release of Cdc14 from the nucleolus (Hwang and Madhani, 2009). It appears that alteration of rDNA chromatin structure induced by these modifications is important for this process.
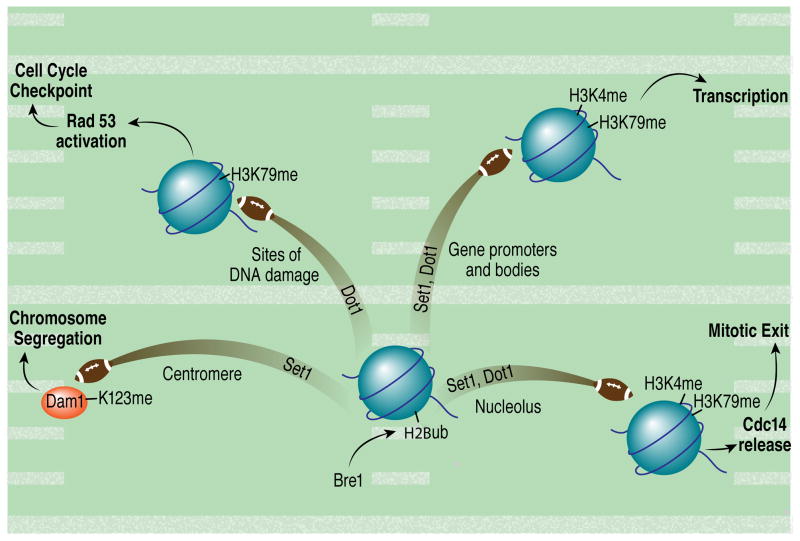
Thus, depending on its chromosomal location, H2Bub can regulate gene transcription, DNA repair and checkpoint signaling, mitotic exit, and chromosome segregation (Figure 2). The ability of this modification to affect methylation of both histone (H3K4 and H3K79) and non-histone proteins in trans highlights its potential to serve as a nexus of signals coming into and emanating from chromatin.
Figure 2. H2Bub passes signals to different receivers to regulate different cellular processes dependent on its chromosomal location.
In yeast, Bre1-mediated ubiquitylation of H2B promotes H3K4 and Dam1 methylation by Set1 and H3K79 methylation by Dot1. Depending on its location, H2Bub can participate in the regulation of transcription, chromosome segregation, cell cycle checkpoints, and mitotic exit.
Unanswered questions and summary
The roles of H2B ubiquitylation and H3K4 and H3K79 methylation in regulating non-transcriptional processes are well established in yeast. An unanswered question is whether these histone modifications regulate similar cellular processes in humans. If so, then defects in chromatin signaling, independent of transcription, could contribute to diseases associated with alterations in histone modifying enzymes. At present, studies aimed at understanding the oncogenic properties of MLL fusion proteins have focused on their abilities to regulate transcription. Likewise, the putative tumor suppressor function of RNF20 is assumed to be due to selective regulation of certain genes (Shema et al., 2008). However, it is possible that defects in the DNA damage response or chromosomal segregation might contribute to the oncogenic properties of MLL fusion proteins or participate in the transformed phenotype associated with depletion of RNF20. Indeed, RNF20 was recently shown to localize to sites of DNA DSBs to promote repair and maintain genome stability, a function that is apparently independent of transcriptional regulation.
The importance of chromatin organization, and reorganization, for the regulation of gene expression and other DNA-templated processes is unarguable. Defining how such changes are triggered by incoming signals is clearly important for understanding how cells respond to changes in their environment, developmental cues, or insults to genomic integrity. However, emerging studies indicate that chromatin is not simply an obstacle to gene transcription or DNA repair. Rather, it is an active participant in these processes that can provide real time signals to facilitate, amplify, or terminate cellular responses. Given the regulatory potential of modification cross talk within histones and between histone and non-histone proteins, coupled with ongoing definitions of vast networks of protein methylation, acetylation, and ubiquitylation events, our current views of signaling pathways as ‘one-way streets’ that dead end at chromatin are likely soon to be converted into a view of chromatin as an information hub that directs multi-layered and multi-directional regulatory networks. Defining these networks will not only provide a greater understanding of biological processes but will also provide whole new game plans for combating complex human diseases that result from inappropriate signal transduction.
Acknowledgments
We thank Becky Brooks for preparation of the manuscript, Chris Brown for graphics and Mark Bedford and Boyko Atanassov for suggestions and insightful comments. This research is supported in part by grants from the National Institutes of Health CA079648 to D.G.J., GM096472 and GM067718 to S.R.D. and through MD Anderson’s Cancer Center Support Grant CA016672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. The Journal of cell biology. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas AK, Johnson DG. Transcriptional and Nontranscriptional Functions of E2F1 in Response to DNA Damage. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-2196. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, Chernikova DA, Kwok S, Brooks JD, Bailey SM, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012;72:2111–2119. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Amon A. At the interface between signaling and executing anaphase--Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- Duran A, Diaz-Meco MT, Moscat J. Essential role of RelA Ser311 phosphorylation by zetaPKC in NF-kappaB transcriptional activation. The EMBO journal. 2003;22:3910–3918. doi: 10.1093/emboj/cdg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair. 2009;8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. The Journal of biological chemistry. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Madhani HD. Nonredundant requirement for multiple histone modifications for the early anaphase release of the mitotic exit regulator Cdc14 from nucleolar chromatin. PLoS Genet. 2009;5:e1000588. doi: 10.1371/journal.pgen.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. The Journal of biological chemistry. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- Latham JA, Chosed RJ, Wang S, Dent SY. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM- dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair. 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. The Journal of cell biology. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. The Journal of biological chemistry. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rust HL, Thompson PR. Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem Biol. 2011;6:881–892. doi: 10.1021/cb200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. The EMBO journal. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Boboila C, Alt FW. Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]