Abstract
Much of our current knowledge about the genetic dynamics in range expansions originates from models, simulations and microcosm experiments that need to be corroborated by field data. Here, we report a neutral genetic pattern that matches the predictions of the genetic surfing theory. Genetic surfing occurs when repeated founding events and genetic drift act on the wave of advance of an expanding population, promoting strong spatial structure. In the range expansion of the tortoise Testudo graeca from North Africa to southeastern Spain, we found several genetic signatures consistent with surfing: a decrease of genetic diversity with distance from the initial founder area, clinal patterns in allele frequencies, rare African alleles which have become common at distal sites in the Spanish range, and stronger spatial differentiation in the expanded range than in the original one. Our results provide support for the theory that genetic drift can be an important force in shaping the genetic structure of expanding populations.
Keywords: genetic drift, rare alleles, founding events, isolation-by-distance, spur-thighed tortoise
1. Introduction
The study of genetic processes that occur during range expansions allows a better understanding of the evolutionary consequences of these unique population episodes. Range expansions can generate allele frequency gradients at neutral markers, promote the spread of rare variants into newly occupied territories, induce the genetic structuring of the species over the newly colonized space or even lead to introgression of local genes into the genome of the advancing species (see [1] for a review). Up to now, much of our knowledge about how genetic dynamics vary through time and space in range expansions has been obtained from models, simulations and microcosm experiments. However, empirical studies are needed to corroborate the effects that these processes have in natural populations, which are undoubtedly more complex.
When the dispersal process is more affected by genetic drift than by gene flow, this can lead to surfing. This is a phenomenon in which those alleles arising on the wavefront of advance of an expansion predominantly ride into the newly colonized territory [2]. Because the probability that an allele surfs should be directly proportional to its frequency at the front, clinal allele frequencies or even stable sectors are expected to be found [3,4]. However, this phenomenon is especially notable when initially rare alleles surf by chance, reaching high frequencies in the newly colonized territory [2,5]. Additionally, as the result of repeated founder effects, genetic diversity should decrease with the progression of the wave at the same time as the global genetic differentiation of the population increases [3,6–8].
Here, we report signatures attributable to genetic surfing in the range expansion of the spur-thighed tortoise (Testudo graeca) in southeastern Spain. In previous studies, we have described the time since colonization of Spain and the genetic differentiation within the original North African population of T. graeca [9,10]. In the current study, we explore the dispersal process of T. graeca in southeastern Spain, addressing the main predictions of the genetic surfing theory. We compare the species' spatial genetic pattern in Spain with its original spatial genetic pattern in North Africa. Because T. graeca has low dispersal capability, we expected to find a spatial genetic pattern in Spain more affected by genetic drift than gene flow, consistent with the predictions of genetic surfing.
2. Material and methods
(a). Study species, populations and sampling
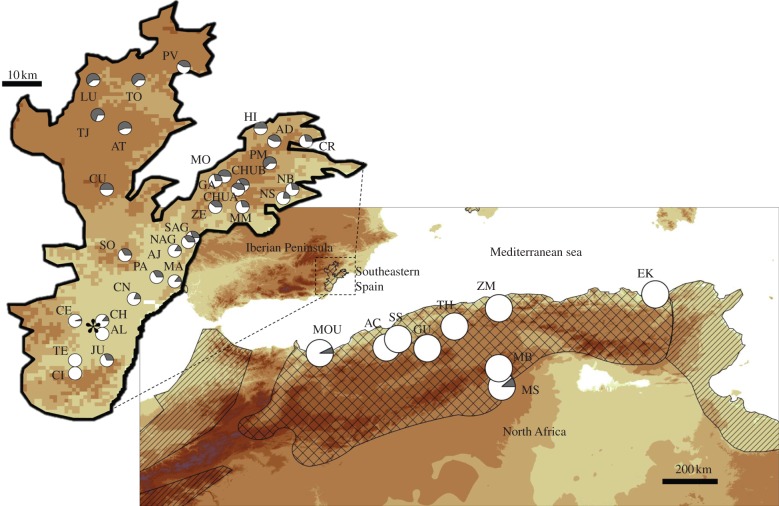
The spur-thighed tortoise (T. graeca) has its western Mediterranean distribution in North Africa, with small and isolated populations on some Mediterranean islands and in the Iberian Peninsula. Most of these European populations are considered to have been introduced by Arabs or Phoenicians (520–3100 years ago) from North Africa. However, for the largest population, located in southeastern Spain and only involving the subspecies Testudo graeca graeca (figure 1), an alternative possibility has been suggested. In a recent study, we found signatures of an ancient bottleneck event approximately 20 000–30 000 years ago [9]. These results indicated that the species could have arrived during the low sea-level conditions of the Mediterranean Basin at the end of the last glacial period. Moreover, the finding of a spatially coherent pattern of genetic variation within the recent population supported the natural dispersal of the species within the newly colonized territory from a single point of entry located in the central-southern part of the current range [9,10].
Figure 1.
Increase of a rare allele in an expanded population. Pie charts represent allelic frequencies for one locus (GenBank number: AF517235). Grey shading represents the frequency of a rare North African allele. In the expanded population, its frequency increases with the distance from the most probable site of species arrival (represented by the asterisk, see §3). The striped area indicates the approximate range of T. graeca at the Mediterranean Basin and the grid area indicates the distribution of the subspecies T. g. graeca. The enlarged area in southeastern Spain corresponds to the total range of the species' distribution in this area. Sites are coded as in table 1. (Online version in colour.)
This study augments the characterization of the dispersal process of the species within this range by extending the sampling design of our previous work to a total of 528 genotyped individuals from 31 Spanish and 9 African sites. We used five previously used polymorphic microsatellite markers (GenBank accession numbers: AF517235, AF517229, AF517239, AY822049 and AY822051; table 1), and only those samples coming from sites with at least seven individuals were included in the subsequent analyses [9,10].
Table 1.
Descriptive statistics of sampled sites. Average over all loci: number of individuals (N), number of alleles (Na), number of effective alleles (Ne) and expected heterozygosity (He). At population level: total number of alleles (NA), number of private alleles (NP) and total number of rare alleles (NR). ANOVA tests between North African and southeastern Spanish sampled sites: *F1,38 = 17.55, p < 0.001; **F1,38 = 33.09, p < 0.0001; ***F1,38 = 9.40, p < 0.005. Site codes: AC, Ain Chorfa; AD, Adanes; AJ, Aljife; AL, Alboluncas; AT, Alto Torrecilla; CE, Centinares; CH, Chinas; CHUA, Chuecos Alto; CHUB, Chuecos Bajo; CI, Cintas; CN, Cunas; CR, Crisoleja; CU, Culebras; EK, El Kala; GA, Galera; GU, Guertoufa; HI, Hinojar; JU, Judío; LU, Luchena; MA, Marinica; MB, Moudjbara; MM, Majada del Moro; MO, Montería; MOU, Moulouya; MS, Messad; NAG, North Aguilón; NB, North Bas; PA, Palas; PM, Palomera; PV, Pisadas de la Virgen; SAG, South Aguilón; SB, South Bas; SO, Sotomayor; SS, Saf Saf; TJ, Tejera; TE, Teresa; TH, Theniet el Had; TO, Tova; ZE, Zerrichera; ZM, Zemmouri.
| population | geographical place | site code | N | Na* | Ne** | He*** | NA | NP | NR |
|---|---|---|---|---|---|---|---|---|---|
| southeastern Spain | North Lorca | PV | 12 | 3.00 | 1.96 | 0.39 | — | — | — |
| TO | 8 | 2.71 | 1.61 | 0.32 | — | — | — | ||
| LU | 10 | 2.57 | 1.70 | 0.35 | — | — | — | ||
| TJ | 10 | 2.71 | 1.73 | 0.33 | — | — | — | ||
| AT | 8 | 3.14 | 1.90 | 0.38 | — | — | — | ||
| CU | 12 | 3.57 | 2.19 | 0.42 | — | — | — | ||
| Almenara mountain | HI | 15 | 3.57 | 2.06 | 0.40 | — | — | — | |
| CR | 14 | 3.57 | 2.12 | 0.38 | — | — | — | ||
| AD | 11 | 3.14 | 2.16 | 0.39 | — | — | — | ||
| PM | 8 | 3.00 | 2.03 | 0.40 | — | — | — | ||
| MO | 18 | 4.00 | 2.45 | 0.42 | — | — | — | ||
| GA | 25 | 5.71 | 3.18 | 0.49 | — | — | — | ||
| CHUB | 23 | 4.29 | 2.46 | 0.44 | — | — | — | ||
| CHUA | 18 | 4.57 | 2.76 | 0.46 | — | — | — | ||
| NB | 11 | 4.29 | 2.94 | 0.45 | — | — | — | ||
| SB | 23 | 4.57 | 3.21 | 0.47 | — | — | — | ||
| MM | 13 | 3.86 | 2.73 | 0.46 | — | — | — | ||
| ZE | 18 | 4.14 | 2.37 | 0.44 | — | — | — | ||
| Pinos mountain | NAG | 11 | 3.57 | 2.80 | 0.48 | — | — | — | |
| SAG | 17 | 4.29 | 2.91 | 0.46 | — | — | — | ||
| AJ | 16 | 4.86 | 3.21 | 0.50 | — | — | — | ||
| Almagro mountain | SO | 18 | 3.86 | 2.48 | 0.41 | — | — | — | |
| Los Lobos | PA | 15 | 3.57 | 2.70 | 0.47 | — | — | — | |
| MA | 21 | 3.57 | 2.38 | 0.43 | — | — | — | ||
| Vera Basin | CN | 8 | 3.43 | 2.69 | 0.45 | — | — | — | |
| CH | 25 | 5.00 | 3.01 | 0.47 | — | — | — | ||
| CE | 21 | 4.57 | 3.53 | 0.51 | — | — | — | ||
| AL | 8 | 3.00 | 2.58 | 0.42 | — | — | — | ||
| Cabrera mountain | TE | 10 | 3.86 | 2.59 | 0.45 | — | — | — | |
| JU | 9 | 3.71 | 2.48 | 0.46 | — | — | — | ||
| CI | 11 | 3.00 | 2.17 | 0.42 | — | — | — | ||
| overall | 447 | 3.76 | 2.49 | 0.43 | 58 | 1 | 37 | ||
| North Africa | Moulouya Valley | MOU | 7 | 5.14 | 4.16 | 0.54 | — | — | — |
| Oran area | AC | 10 | 5.14 | 3.11 | 0.45 | — | — | — | |
| SS | 9 | 5.71 | 3.81 | 0.54 | — | — | — | ||
| Tiaret | GU | 11 | 5.57 | 4.60 | 0.48 | — | — | — | |
| Ain Defla | TH | 7 | 4.14 | 2.99 | 0.49 | — | — | — | |
| Boumerdes | ZM | 9 | 5.00 | 3.82 | 0.50 | — | — | — | |
| Annaba (El Kala) | EK | 7 | 3.57 | 2.48 | 0.40 | — | — | — | |
| Djelfa | MS | 9 | 4.43 | 3.52 | 0.48 | — | — | — | |
| MB | 14 | 5.86 | 4.46 | 0.48 | — | — | — | ||
| overall | 83 | 4.95 | 3.66 | 0.48 | 80 | 23 | 49 | ||
(b). Genetic data analyses
Genotypic linkage disequilibrium between each pair of microsatellite loci was tested using GENEPOP v. 4.0 [11]. GENALEX v. 6.0 [12] was used to estimate the numbers of alleles, allele frequencies, expected heterozygosity and effective number of alleles for each site, and the numbers of private and rare alleles (with allelic frequencies under 0.05) for each population. In addition, GENEALEX and SMOGD [13] were, respectively, used to obtain pairwise FST and standardized F′ST [14] among sites within and between North Africa and southeastern Spain. The centroid of those southeastern Spanish sites that showed the lowest genetic differentiation with North Africa was considered the probable arrival place of the species. This location in Spain and the African site with the highest genetic diversity were used as starting points in subsequent analyses of the spatial distribution of diversity estimates and allele frequencies, using SPSS v. 20.0 [15]. Finally, to detect the relationship between genetic isolation and geographical distance within the studied ranges, the linearized estimates of pairwise FST and F′ST were correlated with the logarithm of pairwise geographical distances among sites in Mantel tests in GENEALEX.
3. Results
No significant linkage disequilibrium was found for any of the 10 pairwise locus combinations. North African sites had significantly higher levels of allelic diversity and expected heterozygosity than those obtained from southeastern Spain (all p < 0.005; table 1). Moreover, out of the 81 alleles found, 23 were private for North Africa, whereas only one was private for southeastern Spain. Pairwise FST between sites within southeastern Spain and North Africa ranged from less than 0.01 to 0.27 and from 0.01 to 0.19, respectively (see the electronic supplementary material, S1).
Both FST and F′ST revealed that the three lowest levels of differentiation across the Mediterranean Sea were found between Saf (SS), a site located on the east coast of Algeria (near Oran), and the sites Teresa (TE), Centinares (CE) and Cunas (CN), located in the south-centre of the distribution of the species in southeastern Spain (figure 1). The geographical distances of southeastern Spanish sites from the centroid of these three southeastern Spanish locations were used in the subsequent correlations: first, both the number of effective alleles (Ne) and the expected heterozygosity (He) significantly decreased with increasing distances to the centroid (both p < 0.001; table 1). Second, 13 allele frequencies of all loci significantly varied in relation to their distance from this point (six increased, whereas seven decreased; table 2 and see figure 1 as an example). It is especially remarkable that although four out of these 13 alleles were rare in North Africa, they showed high frequencies in some places within southeastern Spain. On the other hand, within North Africa, we failed to find any significant correlation between Ne or He and the increasing distance to Moudjbara (MB), the site with the highest genetic diversity. In addition, despite the higher number of detected alleles and the lower number of sampled sites, which would facilitate the finding of clinal patterns, only four allele frequencies were significantly correlated with the distance to MB.
Table 2.
North African and southeastern Spanish allele frequencies showing spatial clines. Southeastern Spanish allele frequencies were correlated with the distance to the putative origin of this expansion, while North African frequencies were correlated with the distance to the site with the highest genetic distance (MB). For each locus the total number of detected alleles (NA) is given.
| locus (GenBank accession no.) | NA | allele | North Africa |
southeastern Spain |
||
|---|---|---|---|---|---|---|
| frequency | spatial correlation (r) | frequency | spatial correlation (r) | |||
| Test 71 (AY822051) | 6 | 128 | 0.77 | n.s. | 0.52 | 0.49** |
| 130 | 0.06 | n.s. | 0.47 | −0.45* | ||
| Test 21 (AY822049) | 8 | 200 | 0.41 | −0.82** | 0.17 | −0.80** |
| 204 | 0.44 | n.s. | 0.72 | 0.56** | ||
| 214 | 0.01 | n.s. | 0.11 | 0.65** | ||
| GmuD16 (AF517235) | 21 | 198 | 0.02 | n.s. | 0.32 | 0.74** |
| 218 | 0.04 | n.s. | 0.19 | −0.48** | ||
| 230 | 0.13 | n.s. | 0.03 | −0.46** | ||
| 234 | 0.10 | 0.67* | 0.10 | n.s. | ||
| GmuB08 (AF517229) | 18 | 212 | 0.05 | n.s. | 0.08 | −0.47** |
| 227 | 0.13 | n.s. | 0.02 | −0.57** | ||
| 233 | 0.08 | n.s. | 0.22 | −0.61** | ||
| GmuD51 (AF517239) | 28 | 139 | 0.06 | −0.81** | 0.00 | — |
| 159 | 0.04 | 0.78* | 0.00 | — | ||
| 239 | 0.12 | n.s. | 0.31 | 0.73** | ||
| 163 | 0.01 | n.s. | 0.37 | 0.39* | ||
*p < 0.05; **p < 0.01
The Mantel tests yielded significant correlations between genetic (pairwise FST) and geographical distance within both ranges (southeastern Spain: p < 0.001; r = 0.49; North Africa: p < 0.05; r = 0.51). Similar differentiation levels were described for both areas, although mean inter-population distance was more than 10 times larger in North Africa than in southeastern Spain (southeastern Spain: mean FST = 0.07, mean inter-population distance = 35.29 km; North Africa: mean FST = 0.07, mean inter-population distance = 413 km). Similar results were obtained when using F'ST, except that the correlations were slightly weaker (southeastern Spain: p < 0.001, r = 0.47, mean F'ST = 0.17; North Africa, p = 0.07; r = 0.45 mean F'ST = 0.28).
4. Discussion
Our results reveal a neutral genetic pattern matching with the recent arrival of T. graeca in southeastern Spain, in which the finding of an isolation-by-distance pattern suggests the natural expansion of the species within the colonized area from a single arrival point located in the central-southern part of the current range, as proposed by Graciá et al. [9,10]. In this range expansion context, we find a spatially structured genetic pattern within the Spanish range that shows several signatures predicted by genetic surfing: (i) genetic diversity significantly decreases with increasing distance to the probable arrival area for the species; (ii) more clinal patterns of allele frequencies involving different unlinked loci are present in the expanded population; (iii) some of these clinal patterns correspond to African rare alleles that are frequent in some areas of southeastern Spain; and (iv) spatial differentiation is stronger in the more recently established range than in the original one. Furthermore, Graciá et al. [9,10] found regions of relative genetic uniformity within southeastern Spain, both using mitochondrial and microsatellite markers. Notably, some mitochondrial haplotypes appeared fixed in different regions of the studied area. All of these properties could be caused by strong genetic drift acting on the front of advance during the expansion within southeastern Spain, and are concordant with previous experimental and simulation studies in which founding stable demes with low diversity originated from a few individuals [3,4,7]. With low dispersal rates, the fate of a few individuals can determine the genetic composition of newly founded demes, promoting the structuring of the population into regions of nearly fixed alleles and, in consequence, increasing its global differentiation [3,4,6,7], which is even maintained with the presence of migration among populations [8].
Two important alternative scenarios may result in spatial patterns similar to those reported here: selection and introgression between two independent colonizations. The possibility of selection as the structuring force is highly unlikely given that similar spatial patterns (allele clines and stable sectors) were reported from multiple unlinked and presumably neutral loci. In addition, the finding of a strong isolation-by-distance pattern in southeastern Spain suggests the main natural dispersal of the species from a single area, as reported by Graciá et al. using a clustering approach [8]. Moreover, introgression from other populations would not predict a decrease in genetic diversity with increasing distance from our derived single introduction site.
Characteristics such as landscape heterogeneity, low density of individuals in the front of the wave and low dispersal ability promote the strong genetic structuration of a population on short time scales [16]. Our study system fits these characteristics, and our results give support for the theory that genetic drift can be an important force causing the neutral genetic patterns of some expansions [7].
Acknowledgements
Financial support was granted by the Spanish Ministry of Education and Science (projects: CGL2009-08251 and CGL2012-35232, with support from the ERDF). E.G. was financially supported by a FPI grant from the Regional Government of the Community of Valencia, and this article is part of her PhD (BFPI/2007/294). We thank Castillo de Chuecos and Global Nature foundations for allowing us to work in their reserves. We appreciate the interest of the Regional Governments of Murcia and Andalucía in the conservation of this tortoise. We thank the anonymous reviewers for their valuable comments on an earlier version of this manuscript.
References
- 1.Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40, 481–501 10.1146/annurev.ecolsys.39.110707.173414 (doi:10.1146/annurev.ecolsys.39.110707.173414) [DOI] [Google Scholar]
- 2.Klopfstein S, Currat M, Excoffier L. 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490 10.1093/molbev/msj057 (doi:10.1093/molbev/msj057) [DOI] [PubMed] [Google Scholar]
- 3.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19 926–19 930 10.1073/pnas.0710150104 (doi:10.1073/pnas.0710150104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallatschek O, Nelson DR. 2009. Life at the front of an expanding population. Evolution 64, 193–206 (doi:10.1111/j.1558–5646.2009.00809.x) [DOI] [PubMed] [Google Scholar]
- 5.Edmonds CA, Lillie AS, Cavalli-Sforza LL. 2004. Mutations arising in the wave front of an expanding population. Proc. Natl Acad. Sci. USA 101, 975–979 10.1073/pnas.0308064100 (doi:10.1073/pnas.0308064100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallatschek O, Nelson DR. 2008. Gene surfing in expanding populations. Theor. Popul. Biol. 73, 158–170 10.1016/j.tpb.2007.08.008 (doi:10.1016/j.tpb.2007.08.008) [DOI] [PubMed] [Google Scholar]
- 7.Excoffier L, Ray N. 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 23, 347–351 10.1016/j.tree.2008.04.004 (doi:10.1016/j.tree.2008.04.004) [DOI] [PubMed] [Google Scholar]
- 8.Slatkin M, Excoffier L. 2012. Serial founder effects during range expansion: a spatial analog of genetic drift. Genetics 191, 171–181 10.1534/genetics.112.139022 (doi:10.1534/genetics.112.139022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graciá E, Giménez A, Anadón JD, Harris DJ, Fritz U, Botella F. 2013. The uncertainty of Late Pleistocene range expansions in the western Mediterranean: a case study of the colonization of southeastern Spain by the spur-thighed tortoise, Testudo graeca. J. Biogeogr. 40, 323–334 10.1111/jbi.12012 (doi:10.1111/jbi.12012) [DOI] [Google Scholar]
- 10.Graciá E, Giménez A, Anadón JD, Botella F, García-Martínez S, Marín M. 2011. Genetic patterns of a range expansion: the spur-thighed tortoise Testudo graeca in southeastern Spain. Amphibia Reptilia. 32, 49–61 10.1163/017353710X542985 (doi:10.1163/017353710X542985) [DOI] [Google Scholar]
- 11.Rousset F. 2007. Genepop 4.0 for Windows and Linux. Montpellier, France: Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université Montpellier II [Google Scholar]
- 12.Peakall R, Smouse PE. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 10.1111/j.1471-8286.2005.01155.x (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford NG. 2010. SMOGD: software for the measurement of genetic diversity. Mol. Ecol. Res. 10, 556–557 10.1111/j.1755-0998.2009.02801.x (doi:10.1111/j.1755-0998.2009.02801.x) [DOI] [PubMed] [Google Scholar]
- 14.Hedrick PW. 2005. A standardized genetic differentiation measure. Evolution 59, 1633–1638 10.1554/05-076.1 (doi:10.1554/05-076.1) [DOI] [PubMed] [Google Scholar]
- 15.SPSS 2011. SPSS advanced statistics 20.0. Chicago, IL: SPSS [Google Scholar]
- 16.Short KH, Petren K. 2011. Fine-scale genetic structure arises during range expansion of an invasive gecko. PLoS ONE 6, e26258. 10.1371/journal.pone.0026258 (doi:10.1371/journal.pone.0026258) [DOI] [PMC free article] [PubMed] [Google Scholar]