Abstract
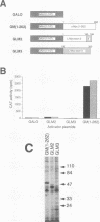
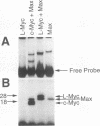
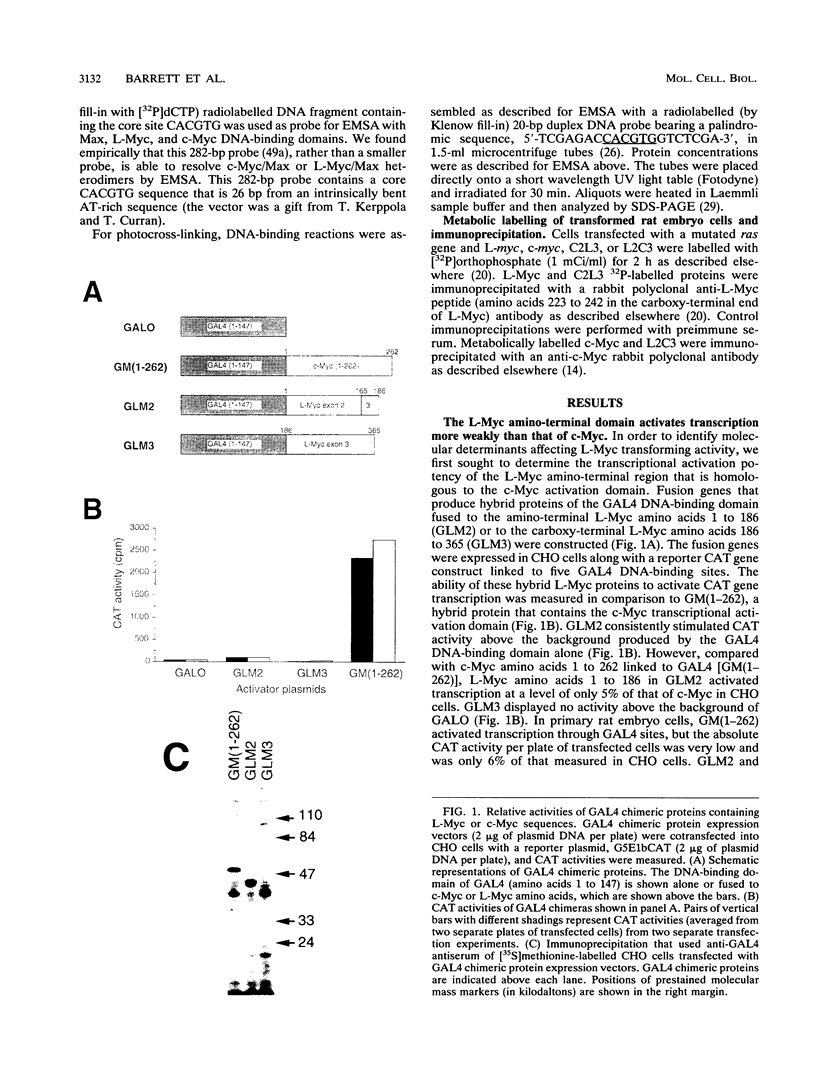
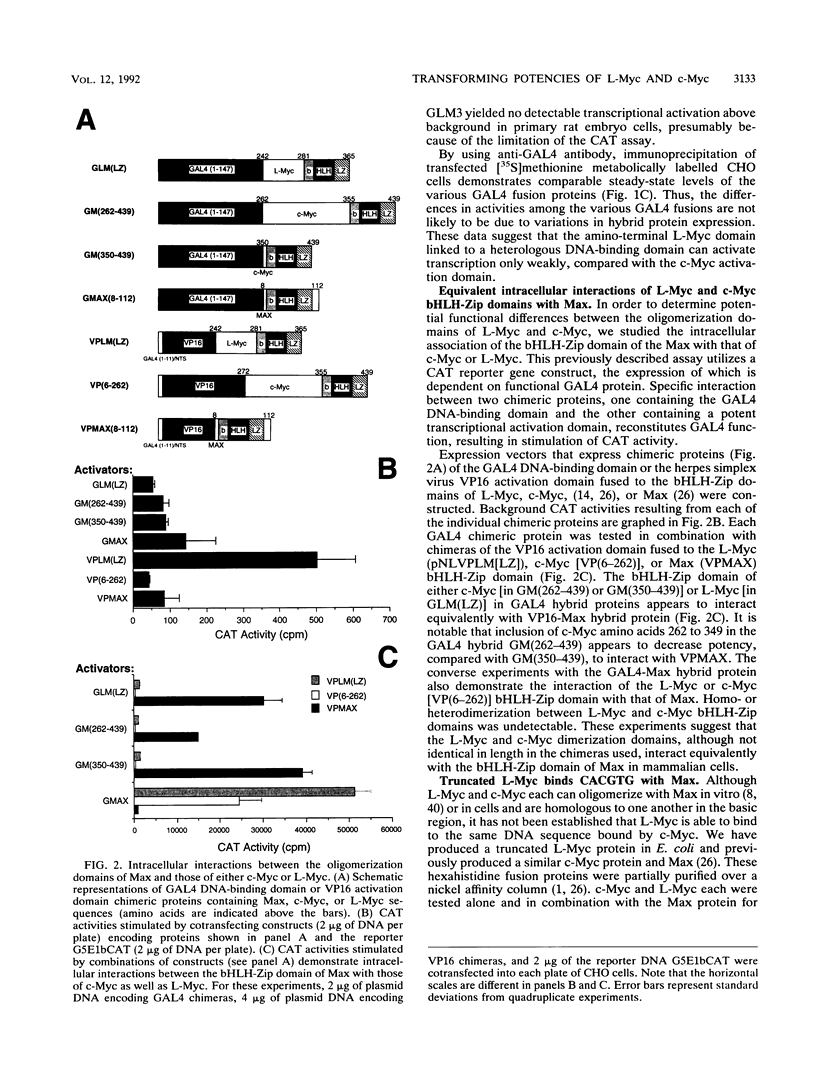
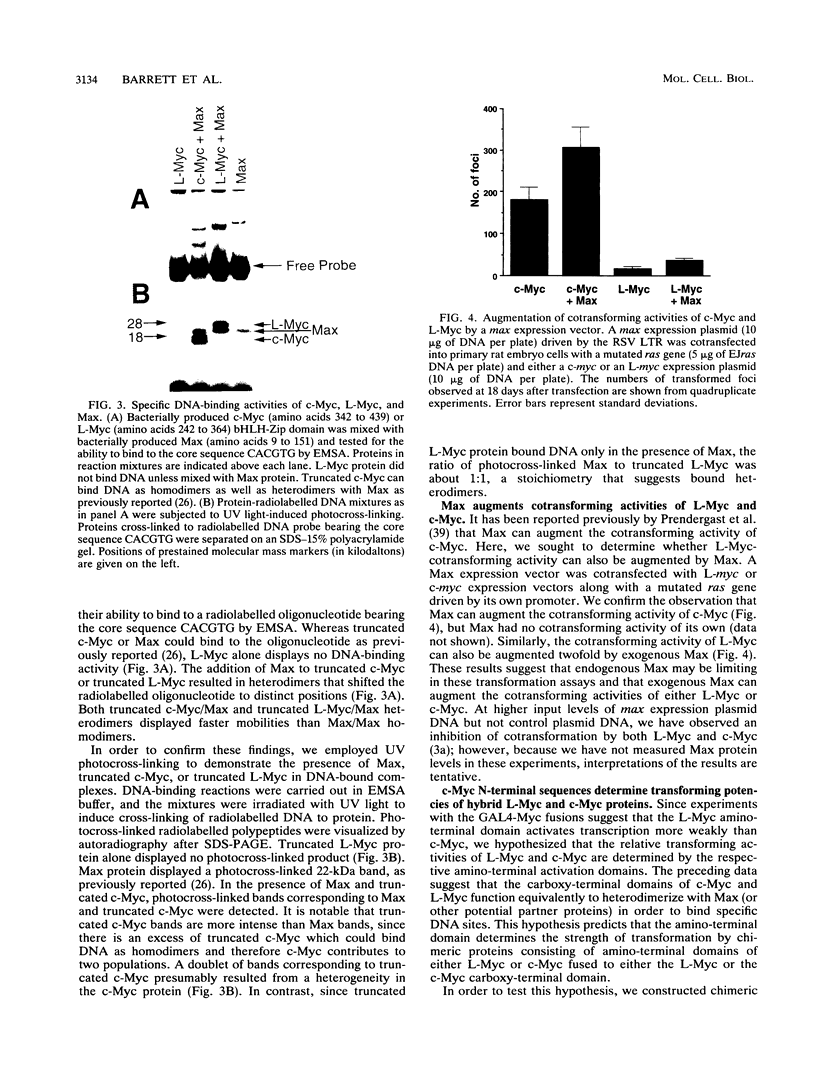
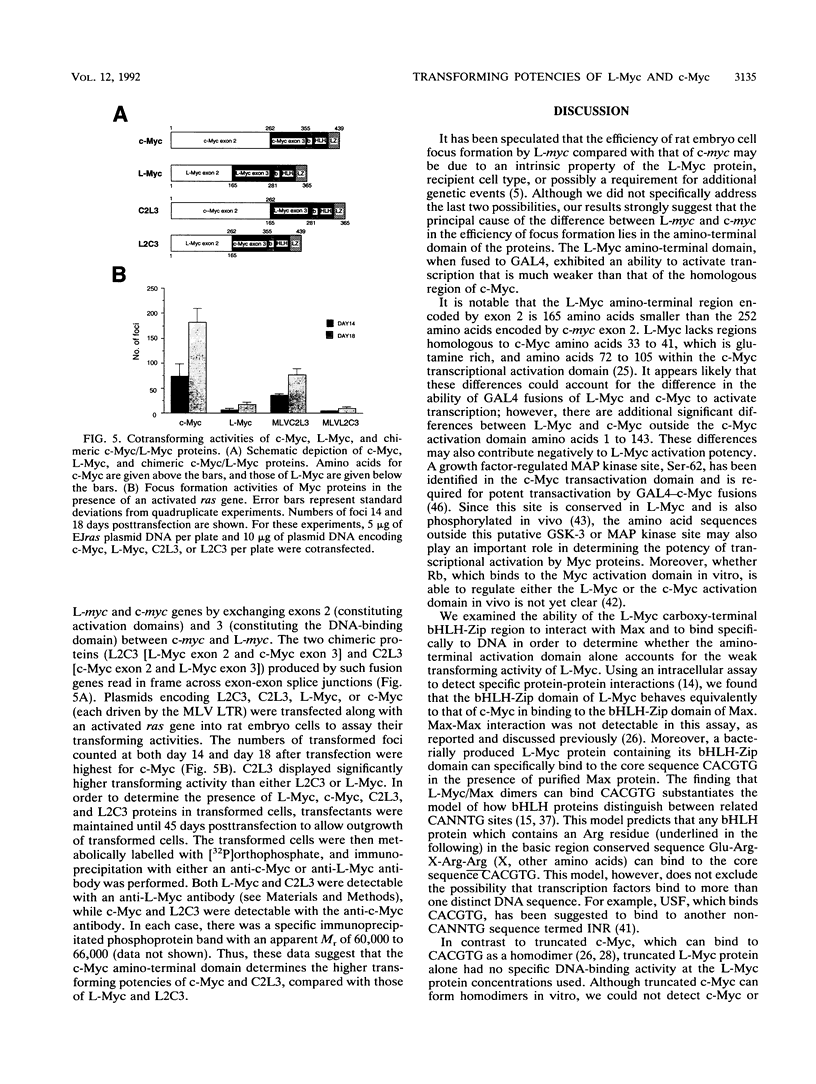
Members of the Myc family of proteins share a number of protein motifs that are found in regulators of gene transcription. Conserved stretches of amino acids found in the N-terminal transcriptional activation domain of c-Myc are required for cotransforming activity. Most of the Myc proteins contain the basic helix-loop-helix zipper (bHLH-Zip) DNA-binding motif which is also required for the cotransforming activity of c-Myc. L-Myc, the product of a myc family gene that is highly amplified in many human lung carcinomas, was found to cotransform primary rat embryo cells with an activated ras gene. However, L-Myc cotransforming activity was only 1 to 10% of that of c-Myc (M. J. Birrer, S. Segal, J. S. DeGreve, F. Kaye, E. A. Sausville, and J. D. Minna, Mol. Cell. Biol. 8:2668-2673, 1988). We sought to determine whether functional differences between c-Myc and L-Myc in either the N-terminal or the C-terminal domain could account for the relatively diminished L-Myc cotransforming activity. Although the N-terminal domain of L-Myc could activate transcription when fused to the yeast GAL4 DNA-binding domain, the activity was only 5% of that of a comparable c-Myc domain. We next determined that the interaction of the C-terminal bHLH-Zip region of L-Myc or c-Myc with that of a Myc partner protein, Max, was equivalent in transfected cells. A Max expression vector was found to augment the cotransforming activity of L-Myc as well as that of c-Myc. In addition, a bacterially synthesized DNA-binding domain of L-Myc, like that o c-Myc, heterodimerizes with purified Max protein to bind the core DNA sequence CACGTG. To determine the region of L-Myc responsible for its relatively diminished cotransforming activity, we constructed chimeras containing exons 2 (constituting activation domains) and 3 (constituting DNA-binding domains) of c-Myc fused to those of L-Myc. The cotransforming potencies of these chimeras were compared with those of full-length L-Myc of c-Myc in rat embryo cells. The relative cotransforming activities suggest that the potencies of the activation domains determine the cotransforming efficiencies for c-Myc and L-Myc. This correlation supports the hypothesis that the Myc proteins function in neoplastic cotransformation as transcription factors.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Luk D., Gentz R., Rauscher F. J., 3rd, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Koskinen P., Mäkelä T. P., Saksela K., Sistonen L., Winqvist R. myc oncogenes: activation and amplification. Biochim Biophys Acta. 1987 Apr 20;907(1):1–32. doi: 10.1016/0304-419x(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Asker C., Steinitz M., Andersson K., Sümegi J., Klein G., Ingvarsson S. Nucleotide sequence of the rat Bmyc gene. Oncogene. 1989 Dec;4(12):1523–1527. [PubMed] [Google Scholar]
- Beckmann H., Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991 Jun;5(6):1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- Birrer M. J., Segal S., DeGreve J. S., Kaye F., Sausville E. A., Minna J. D. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol Cell Biol. 1988 Jun;8(6):2668–2673. doi: 10.1128/mcb.8.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Molecular themes in oncogenesis. Cell. 1991 Jan 25;64(2):235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Cole M. D. Myc meets its Max. Cell. 1991 May 31;65(5):715–716. doi: 10.1016/0092-8674(91)90377-b. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Dolde C., Gillison M. L., Kato G. J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988 Oct;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Dang C. V. c-myc oncoprotein function. Biochim Biophys Acta. 1991 Dec 10;1072(2-3):103–113. doi: 10.1016/0304-419x(91)90009-a. [DOI] [PubMed] [Google Scholar]
- De Greve J., Battey J., Fedorko J., Birrer M., Evan G., Kaye F., Sausville E., Minna J. The human L-myc gene encodes multiple nuclear phosphoproteins from alternatively processed mRNAs. Mol Cell Biol. 1988 Oct;8(10):4381–4388. doi: 10.1128/mcb.8.10.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Dosaka-Akita H., Rosenberg R. K., Minna J. D., Birrer M. J. A complex pattern of translational initiation and phosphorylation in L-myc proteins. Oncogene. 1991 Mar;6(3):371–378. [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Predicted structural similarities of the DNA binding domains of c-Myc and endonuclease Eco RI. Science. 1992 Jan 24;255(5043):464–466. doi: 10.1126/science.1734524. [DOI] [PubMed] [Google Scholar]
- Ingvarsson S., Asker C., Axelson H., Klein G., Sümegi J. Structure and expression of B-myc, a new member of the myc gene family. Mol Cell Biol. 1988 Aug;8(8):3168–3174. doi: 10.1128/mcb.8.8.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kaye F., Battey J., Nau M., Brooks B., Seifter E., De Greve J., Birrer M., Sausville E., Minna J. Structure and expression of the human L-myc gene reveal a complex pattern of alternative mRNA processing. Mol Cell Biol. 1988 Jan;8(1):186–195. doi: 10.1128/mcb.8.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff E., Bister K., Klempnauer K. H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nau M. M., Brooks B. J., Battey J., Sausville E., Gazdar A. F., Kirsch I. R., McBride O. W., Bertness V., Hollis G. F., Minna J. D. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985 Nov 7;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Roy A. L., Meisterernst M., Pognonec P., Roeder R. G. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991 Nov 21;354(6350):245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- Rustgi A. K., Dyson N., Bernards R. Amino-terminal domains of c-myc and N-myc proteins mediate binding to the retinoblastoma gene product. Nature. 1991 Aug 8;352(6335):541–544. doi: 10.1038/352541a0. [DOI] [PubMed] [Google Scholar]
- Saksela K., Mäkelä T. P., Hughes K., Woodgett J. R., Alitalo K. Activation of protein kinase C increases phosphorylation of the L-myc trans-activator domain at a GSK-3 target site. Oncogene. 1992 Feb;7(2):347–353. [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B., Sheen J. Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988 Jul 30;67(2):271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- Seth A., Alvarez E., Gupta S., Davis R. J. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991 Dec 15;266(35):23521–23524. [PubMed] [Google Scholar]
- Skeiky Y. A., Iatrou K. Synergistic interactions of silkmoth chorion promoter-binding factors. Mol Cell Biol. 1991 Apr;11(4):1954–1964. doi: 10.1128/mcb.11.4.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. J., Charron-Prochownik D. C., Prochownik E. V. The leucine zipper of c-Myc is required for full inhibition of erythroleukemia differentiation. Mol Cell Biol. 1990 Oct;10(10):5333–5339. doi: 10.1128/mcb.10.10.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A., Cziepluch C., Hamann U., Schürmann J., Schwab M. The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells. EMBO J. 1991 Dec;10(12):3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]