Abstract
Objectives
Gene methylation and other epigenetic modifications of gene regulation have been implicated in the growth of ovarian cancer, but the clinical significance of such modifications in the Notch pathway in high-grade serous ovarian cancer (HGS-OvCa) is not well understood. We used The Cancer Genome Atlas (TCGA) data to study the clinical relevance of epigenetic modifications of Notch pathway genes.
Methods
We analyzed the interaction of DNA methylation and miRNAs with gene expression data for Notch family members with the Spearman rank correlation test and explored potential relationships with overall survival (OS) with the log-rank test. We downloaded clinical data, level 3 gene expression data, and level 3 DNA methylation data for 480 patients with stage II-IV HGS-OvCa from TCGA data portal. Patients were randomly divided into training and validation cohorts for survival analyses. In each set, patients were grouped into percentiles according to methylation and microRNA (miRNA) or messenger RNA (mRNA) levels. We used several algorithms to predict miRNA-mRNA interaction.
Results
There were significant inverse relationships between methylation status and mRNA expression for PPARG, CCND1, and RUNX1. For each of these genes, patients with a lower methylation level and higher expression level had significantly poorer OS than did patients with a higher methylation level and lower expression level. We also found a significant inverse relationship between miRNAs and mRNA expression for CCND1, PPARG, and RUNX1. By further analyzing the effect of miRNAs on gene expression and OS, we found that patients with higher levels of CCND1, PPARG, and RUNX1 expression and lower expression levels of their respective miRNAs (502-5p, 128, and 215/625) had significantly poorer OS.
Conclusions
Epigenetic alterations of multiple Notch target genes and pathway interacting genes (PPARG, CCND1, and RUNX1) may relate to activation of this pathway and poor survival of patients with HGS-OvCa.
Keywords: Notch pathway, high-grade serous ovarian carcinoma, epigenetic alterations, High-grade serous ovarian cancer, The Cancer Genome Atlas, Epigenetic modifications of gene regulation
Introduction
Growing evidence from clinical, translational, and genetic studies suggests that epithelial ovarian cancers are heterogeneous with regard to genetic alterations and genomic instability. Even among the same histological sub-type (e.g., serous ovarian cancer), low-grade and high-grade tumors have very different molecular profiles. High-grade serous disease accounts for approximately 70% of all ovarian cancer deaths [1]. Most patients eventually develop chemotherapy-resistant disease. Most cases of high-grade serous ovarian cancer (HGS-OvCa) are characterized by TP53 mutations and inactivation of BRCA1/2 [2-4]. However, the molecular basis of HGS-OvCa is not well understood.
Epigenetic modifications of gene regulation are a prominent feature of many cancers. Epigenetic forms of gene regulation include DNA methylation, histone post-translational modifications, and expression of noncoding RNAs [5, 6]. A common epigenetic modification, DNA methylation, leads to alteration of gene expression and is a hallmark of human cancer [7]. Hypermethylation of CpG islands (i.e., CG-rich regions, usually associated with transcriptionally active genes) is frequently found in tumor suppressor genes, such as BRCA1, p16, MLH, RASSF1, and DARK in ovarian cancer [7]. Recently, an integrative genomic study from,- The Cancer Genome Atlas (TCGA)-, demonstrated that 22% of cases of HGSOvCa exhibited Notch pathway alterations, including amplification, overexpression, and mutations [1]. However, the clinical significance of epigenetic modifications of the Notch pathway in HGS-OvCa is not known.
Here, we used TCGA data to perform an integrated analysis of the clinical relevance of epigenetic modifications of Notch pathway genes. We found that lower gene methylation and microRNA (miRNA) expression levels and higher gene expression levels of multiple Notch family members may be associated with activation of this pathway and with poor outcome of patients with HGS-OvCa.
Materials and Methods
Characterization of HGS-OvCa Patients
We used the TCGA data portal (http://tcga.cancer.gov) to download information on 488 clinically annotated stage II-IV HGS-OvCa patients [1]. We excluded 8 cases (for one patient, there were two samples taken from a primary tumor, and for 6 the overall survival information was missing). The overall survival (OS) duration was defined as the interval (in months) between the date of initial surgical resection to death or last follow-up. Access to the TCGA database was approved by the National Cancer Institute. The University of Texas MD Anderson Cancer Center approved the waiver for performing this survival analysis with de-identified database information.
Notch signaling pathway
We analyzed DNA methylation and miRNA expression data for Notch family members (i.e., its ligands and receptors, targets, and interacting genes, as shown in Supplementary Table S5) and explored their potential relationship with patient OS. We performed survival analyses using expression/methylation levels of Notch family genes, which includes Notch target genes (e.g., CCND1, PPARG), and Notch-interacting genes (e.g., RUNX1).
Analysis of methylation and expression data
We found and downloaded level 3 Agilent244K gene expression data for 87 genes related to the Notch signaling pathway and level 3 Affymetrix HG-U133A gene expression data for 79 genes, out of the total of 88 genes considered. In addition, we downloaded level 3 Illumina Infinium DNA methylation data for 164 probes located in the promoter, 5′-untranslated region (UTR), coding sequences, or 3′-UTR (NCBI36/hg18) of genes related to the Notch signaling pathway.
Statistical Analysis
All statistical analyses were performed in the R statistical program (version 2.14.2). The tests were two-sided and statistical significance was defined as a p-value < 0.05. The Spearman's rank-order correlation test was applied to measure the strength of the inverse association between DNA methylation and gene expression. We imposed a cut-off of functional relevance on the Spearman correlation coefficient of -0.2 based on the method published previously [1, (Table S7.1)]. We further checked this level for statistical significance, but given the number of samples involved this was easily met. For survival analysis, the 480 patients were randomly divided into training (two thirds of the set) and validation cohorts (S9). The Log-rank test was employed to determine the relationship between methylation status and OS. The Kaplan-Meier method was used to generate survival curves. For each gene/miRNA/methylated probe, we checked for a relation with survival as follows: Using the training set, we chose a cut-off to optimally split the samples into two groups; using this cut-off, we then examined the results in the validation set, and used the p-value attained here as our estimate.
MiRNA-target interactions
We used several algorithms (miRanda, TargetScan, PITA and microT) to predict miRNAs targets and binding sites. These algorithms are publicly available through http://www.microrna.org, http://www.targetscan.org, http://genie.weizmann.ac.il/pubs/mir07 and http://diana.cslab.ece.ntua.gr/microT, respectively. We also searched miRTarBase http://mirtarbase.mbc.nctu.edu.tw/ for experimentally validating miRNA-target interactions.
Results
Low methylation and high expression of RUNX1, PPARG, and CCND1 predict poor overall survival
To investigate the impact of DNA methylation and gene expression of Notch family members on survival of patients with high grade serous ovarian cancer, we analyzed both DNA methylation and gene expression data and explored potential clinical relationships. We first tested the correlations between DNA methylation and gene expression of Notch family members using the Spearman Rank correlation test, retaining probe/gene pairs with a correlation <-0.2. Our analysis identified significant inverse correlations between gene expression and DNA methylation for 24 methylation probes (Table 1 and S6), interrogating 19 distinct genes. When we considered expression alone, none of these 19 genes showed survival differences in both the training and validation sets. Thus, we were led to consider a more detailed set of comparisons. We first considered just the methylation levels of the probes listed in S6. We saw significant association with poor survival for 2 probes: cg00953256, interrogating CCND1 and cg04632671, interrogating PPARG (Figure 1). We then considered potential methylation/gene expression interactions. For the 24 methylation/gene expression pairs listed in S6, we considered whether the corresponding gene expression level added any information. We did this by first fixing the methylation cut-off level at the optimal point leading to the biggest survival difference in the training set, as chosen above and then varying the gene expression level to choose a second cut-off. For each gene cut-off, this splits the data in four groups corresponding to high/low methylation and expression; we contrasted the two groups linked to a negative association: methylation high and expression low with methylation low and expression high. Again, we chose the gene cutoff using the training set and checked for statistical significance in the validation set. We found improvements in three cases, including both cases where the methylation level alone was significantly associated with survival: 1) probe cg00953256 (methylation cutoff level=0.58), situated in the 3′UTR of CCND1, and CCND1 (gene cutoff level=0.69); 2) probe cg04632671 (methylation cutoff level=0.31), situated in the promoter region of PPARG, and PPARG (gene cutoff level=0.33), and 3) probe cg04632671 (methylation cutoff level=0.33), situated in the promoter region of RUNX1, and RUNX1 (gene cutoff level=0.61) (Figure 2, S1, Figure 3). Since this quadrant-comparison procedure does not use all the samples at each stage, we also tracked the numbers of samples compared in each test to avoid being driven by small sample size artifacts. The smallest number of samples in one of the quadrants contrasted was 21. Since it wasn't immediately clear how to best assess the significance improvement for these pairs in the statistical sense, we focused instead on whether there were marked differences in overall survival between extreme quadrants identified. We noticed a difference in median overall survival of at least 13.4 months in the cases noted above (Figure 2, S1).
Table 1.
Inverse correlation between DNA methylation and gene expression in HGS-OvCa
| Probe | Gene | Affymetrix |
Agilent |
||
|---|---|---|---|---|---|
| Coefficient | FDR | Coefficient | FDR | ||
| cg00953256 | CCND1 | -0.28 | 5.43E-09 | -0.28 | 3.69E-09 |
| cg19836199 | RUNX1 | NS | NS | -0.21 | 4.54E-05 |
| cg04632671 | PPARG | NS | NS | -0.21 | 2.60E-05 |
FDR=False discovery rate; NS= not significant
Figure 1.
Kaplan-Meier survival curves for overall survival among patients in training and validation sets according to methylation status. (A, B) CCND1, (C, D) PPARG.
Figure 2.
Kaplan-Meier survival curves for overall survival among patients in training set according to methylation status and gene expression. (A) PPARG. (B) CCND1. (C) RUNX1.
Figure 3.
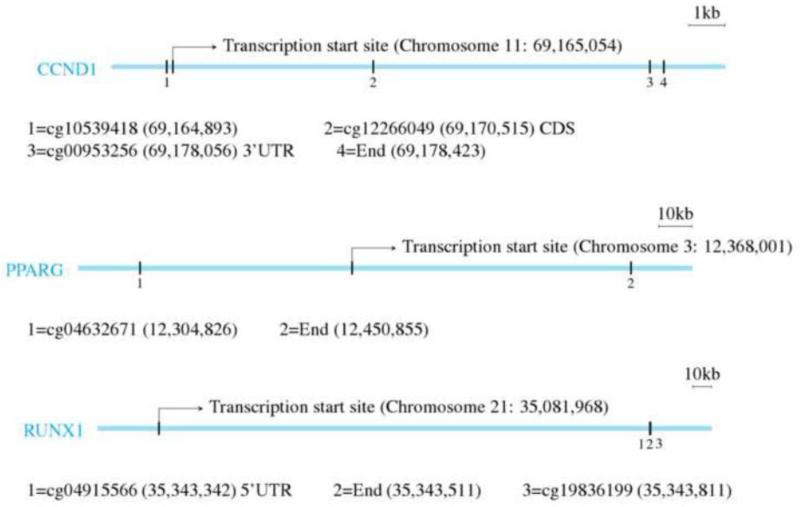
Position of the methylated probes of CCND1, PPARG, and RUNX1 as determined using the University of California Santa Cruz genome browser (Genome Build 36).
Impact of miRNA-mRNA interactions on clinical outcome
To further investigate the impact of miRNA-mRNA interactions on clinical outcome, we used the Spearman Rank correlation test to identify pairs with correlations <-0.2, which are also predicted by all four of the miRNA-target prediction algorithms we used. Taken together, these two tests revealed significant relationships between CCND1, CD44, FZD6, HEYL, LMO2, MNFG, NOTCH3, PPARG, and RUNX1 and several miRNAs (Table 2 and S7). Focusing on our genes of primary interest (Table 2), miR-502-5p targeted CCND1 in the 3′-untranslated region (3′-UTR), miR-128 targeted PPARG within the 3′-UTR, miR-215 targeted RUNX1 within the 3′-UTR, and miR-625 targeted RUNX1 within the 3′-UTR (S2).
Table 2.
Inverse correlation between gene expression and miRNAs predicted by microT, miRanda, Pictar, and TargetScan algorithms to interact with them
| Gene | miRNA | Affymetrix |
Agilent |
||
|---|---|---|---|---|---|
| Coefficient | FDR | Coefficient | FDR | ||
| CCND1 | miR-502-5p | -0.2 | 0.00023 | NS | NS |
| PPARG | miR-128 | -0.24 | 1.13E-05 | -0.31 | 3.49E-09 |
| RUNX1 | miR-215 | NS | NS | -0.23 | 2.01E-05 |
| RUNX1 | miR-625 | -0.22 | 0.00012 | -0.28 | 7.78E-08 |
FDR=False discovery rate; NS=not significant.
For each of the miRNA-gene associations listed in (S7), we checked for improvement in predicting survival as for methylated probe-gene associations discussed above. We found improvement for miR-128 (cut-off used=0.31) and PPARG (cut-off used=0.34), miR-215 (cut-off used=0.45), -625 (cut-off used=0.33) and RUNX1 (cut-off used=0.62), miR502-5p (cut-off used=0.6) and CCND1 (cut-off used=0.7) (Figures 4, S3 and S4). The smallest number of samples in the groups contrasted was 24. We noticed a difference in median overall survival of at least 10.8 months in the cases noted above.
Figure 4.
Kaplan-Meier survival curves for overall survival among patients in training set according to miRNA-mRNA expression. (A) miR-128-PPARG. (B) miR-215-RUNX1. (C) miR-625-RUNX1. (D) miR-502-5p-CCND1.
In addition to investigating the role of methylation and miRs in regulating gene expression, we also examined the relationship between HDACs and gene expression using the Spearman Rank correlation test. Here, we used the mRNA expression levels of a set of genes (HDACs) as a surrogate for the acetylation levels of histones. We looked for inverse correlations between expression levels of the HDAC genes and the 19 genes identified as being associated with methylation above. We found expression levels of HDAC7A and HDAC2 were inversely correlated with expression of CD44, PPARG and RUNX1 (S8).
Discussion
In this study, we identified a significant inverse relationship between methylation status and mRNA expression for PPARG, CCND1, and RUNX1. For these three genes, patients with lower methylation status and higher expression level had significantly worse OS. In addition, patients with lower expression of miR-128, miR-215/625, and miR-502-5p and higher expression of respective PPARG, RUNX1 and CCND1 had significantly poorer OS, suggesting that epigenetic modification of these genes may be associated with activation of the Notch pathway and with poor outcome of patients with HGS-OvCa.
Whether peroxisome proliferator-activated receptor gamma (PPARG) promotes or suppresses HGS-OvCa is unknown. PPARG is a member of the nuclear hormone receptor superfamily of ligand-activated transcription factors. PPARG regulates lipid metabolism, affecting inflammation and cancer [8, 9]. It has been reported that PPARG agonists inhibit the growth of many types of cancers, including ovarian cancer [10]. Variations in PPARG expression or gene mutations have consistently been reported to be associated with tumorigenesis [11-13]. However, conflicting results have been reported and raised the question of whether PPARG promotes or suppresses tumorigenesis. It has been also reported that epigenetic silencing of PPARG is correlated with colorectal cancer progression and adverse patient outcome [13].
The exact biology and potential mechanisms of hypermethylation of PPARG in HGS-OvCa are not well understood[7]. The hypermethylation might be explained by the increased DNA methyltransferase activity and altered HDACs in this disease because histone acetylation is typically associated with increased transcription and HDAC alterations are also common in ovarian cancer. In our study, HDAC7A and HDAC1 were inversely correlated with the expression of PPARG. Other studies have shown that PPARG promoter hypermethylation correlates with reduced gene transcription, the presence of H3K9me3 and H3K27me3, and concomitant recruitment of HDAC1, MeCP2, and EZH2 [7]. Conversely, epigenetic treatment with 5-aza-2′-deoxycytidine induces PPARG [7]. Thus, whether PPARG is crucial in promoting or suppressing HGS-OvCa needs further exploration.
Another epigenetic aberration, via miRNA, was also notably seen in HGS-OvCa by using TCGA database information and target prediction analysis. We showed that miR-128, miR-502-5p, and miR-215/625 were significantly inversely correlated with PPARG, CCDN1, and RUNX1 expression, respectively. The miRNA-target prediction revealed that these three genes were direct targets of these miRNAs. Low expression of miR128, but not miR502-5p or miR-215/625 [14], significantly correlated with poor patient OS in both the training set and the validation sets, which is consistent with other reports for breast and colon cancers [15, 16]. Moreover, patients with higher expression of CCND1 and lower expression of miR-502-5p had significantly poorer OS than did patients with lower CCND1 expression and higher miR502-5p expression. Whether these miRNA alterations are specific for GS-OvCa is unknown. A miRNA signature has been reported for ovarian cancer [17]: miR-200a, miR-141, miR-200b, and miR-200c were reported to be upregulated in ovarian cancer, whereas miR-199a, miR-140, miR-145, miR-125bl, and miR-let7i were among the most downregulated [7]. Our findings that miR-215/624, miR-502-5p, and miR-128 were downregulated had not previously been reported for HGSOvCa.
The exact biology of miR-502-5p remains unclear. It has been reported to inhibit tumor growth in colon cancer [18]. Another study reported that a polymorphism at the miR-502 binding site in the 3′-UTR of the SET8 gene was associated with risk for epithelial ovarian cancer [19]. How miR-502-5p and CCND1 could be manipulated with respect to HGS-OvCa is not yet known. Whether this miRNA could function as a potential tumor suppressor and a potential candidate for developing miRNA-based therapeutic strategies requires further investigation.
In summary, epigenetic alterations of multiple Notch target genes and interacting genes (PPARG, CCND1, and RUNX1) may be associated with activation of this pathway and with poor patient survival. Targeting epigenetic modifications of the Notch pathway might hold potential for novel therapies against HGS-OvCa.
Supplementary Material
Highlights.
➢ Patients with lower DNA methylation status and higher expression of PPARG, CCND1, or RUNX1 had significantly worse overall survival.
➢ Patients with lower microRNAs 502-5p, 128, 215/625 expression and higher PPARG, CCND1, RUNX1 expression had worse overall survival.
Acknowledgement
We thank Elizabeth Hess in the Department of Scientific Publications for helpful editing. This work was supported, in part by the NIH (CA 109298, P50 CA083639, P50 CA098258, CA128797, RC2GM092599, U54 CA 151668), the Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), the DOD (OC073399, OC093146, OC 100237, BC085265), CPRIT (RP110595) the Zarrow Foundation, the Marcus Foundation, the RGK Foundation, the Gilder Foundation, and the Betty Anne Asche Murray Distinguished Professorship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflicts of interest were disclosed.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Integrated genomic analyses of ovarian carcinoma. Nature. l2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landen CN, Jr., Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovariancancer. J Clin Oncol. l2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 3.Saad AF, Hu W, Sood AK. Microenvironment and pathogenesis of epithelial ovarian cancer. Horm Cancer. l2010;1:277–90. doi: 10.1007/s12672-010-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodurka DC, Deavers MT, Tian C, Sun CC, Malpica A, Coleman RL, Lu KH, Sood AK, Birrer MJ, Ozols R, Baergen R, Emerson RE, Steinhoff M, Behmaram B, Rasty G, Gershenson DM. Reclassification of serous ovarian carcinoma by a 2-tier system: A Gynecologic Oncology GroupStudy. Cancer. l2012;118:3087–94. doi: 10.1002/cncr.26618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. l2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 6.Asadollahi R, Hyde CA, Zhong XY. Epigenetics of ovarian cancer: from the lab to the clinic. Gynecol Oncol. l2010;118:81–7. doi: 10.1016/j.ygyno.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Seeber LM, van Diest PJ. Epigenetics in ovarian cancer. Methods Mol Biol. l2012;863:253–69. doi: 10.1007/978-1-61779-612-8_15. [DOI] [PubMed] [Google Scholar]
- 8.Davidson B, Hadar R, Stavnes HT, Trope CG, Reich R. Expression of the peroxisomeproliferator-activated receptors-alpha, -beta, and -gamma in ovarian carcinoma effusions isassociated with poor chemoresponse and shorter survival. Hum Pathol. l2009;40:705–13. doi: 10.1016/j.humpath.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, Lee JJ, Heo DS. PPARgamma ligands induce growth inhibition and apoptosis through p63 and p73 in human ovarian cancer cells. Biochem Biophys Res Commun. l2011;406:389–95. doi: 10.1016/j.bbrc.2011.02.052. [DOI] [PubMed] [Google Scholar]
- 10.Yang YC, Tsao YP, Ho TC, Choung IP. Peroxisome proliferator-activated receptor-gammaagonists cause growth arrest and apoptosis in human ovarian carcinoma cell lines. Int J Gynecol Cancer. l2007;17:418–25. doi: 10.1111/j.1525-1438.2006.00866.x. [DOI] [PubMed] [Google Scholar]
- 11.Sarraf P, Mueller E, Smith WM, Wright HM, Kum JB, Aaltonen LA, de la Chapelle A, Spiegelman BM, Eng C. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol Cell. l1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 12.Sabatino L, Casamassimi A, Peluso G, Barone MV, Capaccio D, Migliore C, Bonelli P, Pedicini A, Febbraro A, Ciccodicola A, Colantuoni V. A novel peroxisome proliferator-activated receptor gamma isoform with dominant negative activity generated by alternative splicing. J Biol Chem. l2005;280:26517–25. doi: 10.1074/jbc.M502716200. [DOI] [PubMed] [Google Scholar]
- 13.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers:complex stories. Nat Rev Cancer. l2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Li C, Nie H, Lv X, Qu Y, Yu B, Su L, Li J, Chen X, Ju J, Yu Y, Yan M, Gu Q, Zhu Z, Liu B. Down-regulated miR-625 suppresses invasion and metastasis of gastric cancer by targeting ILK. FEBS Lett. l2012;586:2382–8. doi: 10.1016/j.febslet.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 15.White NM, Khella HW, Grigull J, Adzovic S, Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, Evans AJ, Gabril M, Yousef GM. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. l2011;105:1741–9. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, Burke S, Shroyer K, Ju J. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. l2011;10:340–7. doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li SD, Zhang JR, Wang YQ, Wan XP. The role of microRNAs in ovarian cancer initiation and progression. J Cell Mol Med. l2010;14:2240–9. doi: 10.1111/j.1582-4934.2010.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.