Abstract
Tumor-derived exosomes are emerging mediators of tumorigenesis with tissue-specific addresses and messages. We explored the function of melanoma-derived exosomes in the formation of primary tumor and metastases in mouse and human subjects. Exosomes from highly metastatic melanoma increased the metastatic behavior of primary tumors by permanently “educating” bone marrow (BM) progenitors via the MET receptor. Melanoma-derived exosomes also induced vascular leakiness at pre-metastatic sites, and reprogrammed BM progenitors towards a c-Kit+Tie2+Met+ pro-vasculogenic phenotype. Reducing Met expression in exosomes diminished the pro-metastatic behavior of BM cells. Importantly, MET expression was elevated in circulating CD45−C-KITlow/+TIE2+ BM progenitors from metastatic melanoma subjects. RAB1a, RAB5b, RAB7, and RAB27a were highly expressed in melanoma cells and Rab27a RNA interference decreased exosome production, preventing BM education, tumor growth and metastasis. Finally, we identified an exosome-specific “melanoma signature” with prognostic and therapeutic potential, comprised of TYRP2, VLA-4, HSP70, an HSP90 isoform and the MET oncoprotein.
INTRODUCTION
Exosomes are small membrane vesicles (30–100 nm) derived from the luminal membranes of multivesicular bodies, and are constitutively released via fusion with the cell membrane1–5. In addition to diffusible factors, such as cytokines and growth factors, and insoluble factors, such as extracellular matrix molecules, exosomes mediate local and systemic cell communication through the horizontal transfer of information (i.e., mRNAs, microRNAs and proteins) 2,5–15.
It is well recognized that bone marrow-derived cells (BMDCs) are crucial for the generation of a suitable microenvironment for the primary tumor, and the development of metastasis16 through a process called “pre-metastatic niche formation”17,18. Although secreted factors are known contributors to BMDC recruitment to both primary tumor and pre-metastatic niches18–21, the role of exosomes in this process has yet to be evaluated. Exosomes may play a role in the crosstalk between the primary tumor and host cells, leading to homing of both cell types to metastatic organs7,22,23.
Here, we identify tumor-derived exosomes as new factors that promote metastatic niche formation by “educating” BMDCs toward a pro-vasculogenic and pro-metastatic phenotype via upregulation of the MET oncoprotein24–29. Our data show that a specific expression pattern of Ras-related (Rab) proteins30,31 is associated with exosome production in melanoma. Moreover, using circulating exosomes isolated from subjects with Stage IV melanoma, we define a melanoma-specific “exosome signature” comprised of tyrosinase-related protein-2 (TYRP2), very late antigen 4 (VLA-4), heat shock protein 70 (HSP70), an HSP90 isoform, and MET oncoprotein. Importantly, exosomal TYRP2 predicts disease progression in subjects with Stage III melanoma. Finally, we demonstrate that CD45−C-KITlow/+TIE2+ circulating BM progenitors from individuals with metastatic melanoma express high MET receptor levels.
RESULTS
Exosomal proteins for diagnosis in melanoma subjects
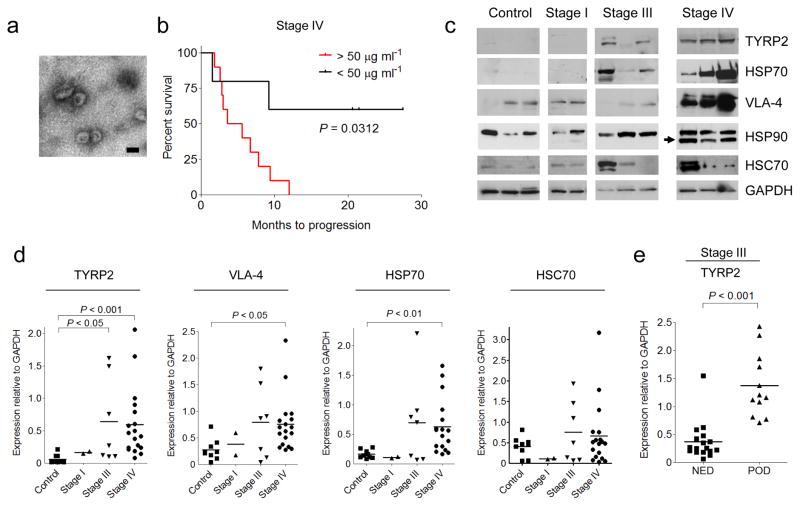
To determine the significance of circulating exosome levels in metastatic melanoma32,33, we prospectively isolated and characterized exosomes from the plasma of human subjects with increasing clinical stage. We isolated exosomes (100nm particles) using standard methods, and confirmed their identity by electron microscopy (Fig. 1a). Known exosome markers including CD63, CD9 and MHC-I were identified (data not shown). Exosome size distribution and number, quantified by NanoSight analysis, did not differ based on clinical stage (Supplementary Fig. 1a). In contrast, exosome protein concentrations were higher in subjects with Stage IV disease compared to all other stages and normal controls (Supplementary Fig. 1b). Furthermore, we found that Stage IV individuals with protein-poor exosomes (< 50 μg ml−1) had a survival advantage compared to those with protein-rich exosomes (> 50 μg ml−1) (Fig. 1b).
Figure 1. Analysis of protein expression in circulating exosomes from melanoma subjects.
(a) Representative electron microscopic image of exosomes derived from the plasma of a melanoma subject. Scale bar, 100 nm. (b) Kaplan-Meier survival curve showing cumulative probabilities in Stage IV subjects over 42 months of follow-up according to total protein (micrograms) in isolated circulating exosomes per milliliter of plasma analyzed (n = 15). P values were calculated using a log-rank test. (c) Representative Western blot of TYRP2, VLA-4, HSP70, HSP90 and HSC70 proteins in circulating exosomes isolated from the plasma of melanoma subjects (Stages I, III and IV) and healthy controls. Arrow indicates a specific HSP90 isoform found in 70% of melanoma subjects. GAPDH was used as a loading control. (d) Statistical analysis of western blot densitometry of signature proteins in circulating exosomes relative to GAPDH. Controls (n = 9); stage I (n = 2); stage III (n = 7); stage IV (n = 18). P values were calculated by ANOVA. (e) Statistical analysis of western blot densitometry for TYRP2 expression in circulating exosomes relative to GAPDH in a retrospective series of frozen plasma derived from stage III melanoma subjects (n = 29) who had been followed for 4 years to evaluate disease progression (NED = no evidence of disease, POD = progression of disease), (n = 29); P < 0.001 using Mann-Whitney U test.
Exosomes from highly metastatic mouse (B16-F10) and human (SK-Mel28/-202/-265/-35) melanoma cell lines expressed typical exosome markers (Supplementary Fig. 1c). Mass spectrometry led to the identification of additional human and mouse-derived melanoma exosome proteins (Supplementary Table 1) that helped define a potential diagnostic “signature” in melanoma-derived exosomes. TYRP2 (a melanoma-specific protein), VLA-4 and HSP70 levels were increased in the exosomes of subjects with Stage IV disease (Fig. 1c, d) while only TYRP2 was significantly increased (P < 0.05) in Stage III individuals, as compared to control subjects (Fig. 1c, d). Interestingly, an isoform of HSP90, reported to be important in cellular transformation, was found in 70% of Stage IV melanoma exosomes (Fig. 1c)34. We found that exosomal TYRP2 levels were significantly increased (P < 0.001) in exosomes correlated with metastatic progression in a retrospective cohort of individuals with Stage III melanoma (Fig. 1e; data not shown). These data support the use of analyzing circulating exosomes to predict prognosis (Stage III) and outcome (Stage IV) for subjects with melanoma.
Tumor exosomes alter organ tropism and metastatic potential
Exosomes from highly malignant B16-F10, SK-Mel28, and SK-Mel202 melanoma cell lines expressed higher amounts (2–20-fold) of protein than poorly metastatic (B16-F1)35 and non-metastatic cell lines (melan-a) (Fig. 2a). Other less metastatic lung, breast and colon cancer cells secreted lower levels of exosomal proteins than B16-F10 cells (Supplementary Fig. 2a). To analyze the tissue distribution of exosomes, fluorescently-labeled B16-F10 exosomes were injected intravenously into naïve mice and detected in blood vessels of target organs after five minutes (Fig. 2b). After 24 hours, we found B16-F10 exosomes in the interstitium of the lung, BM, (Fig. 2b), liver and spleen (organotropic sites for B16-F10 metastasis) but no longer in the circulation (Supplementary Table 2, data not shown). To analyze vascular leakiness properties in the lung, an early event in pre-metastatic niche formation36, we injected B16-F10 exosomes followed by fluorescently-labeled dextran perfusion 24 hours later. B16-F10 exosomes enhanced lung endothelial permeability as judged by the presence of extravasated dextran when compared to conditioned media, control particles (Fig. 2c) and exosomes from non-metastatic cell lines (Supplementary Fig. 2b).
Figure 2. Role of tumor-derived exosomes in metastasis.
(a) Measurement of the total protein per million cells in exosomes isolated from human and mouse melanoma cells in culture. Error bars represent s.e.m. (b) Confocal microscopic analysis of B16-F10 exosome tissue distribution (green) 5 min (lung, left panel) or 24 h (lung and BM, right panels) after tail vein injection. Scale bar, 50 μm. (c) Analysis of lung endothelial permeability following fluorescently-labeled dextran perfusion (red) 24 h after tail vein injection of B16-F10 exosomes, conditioned medium or control particles. Scale bar, 50 μm. (d) Analysis of primary tumor growth (left panel) after subcutaneous flank injection of B16-F10mCherry cells in WT mice treated with B16-F10 exosomes for 3 weeks (n = 6 mice per group; error bars represent s.e.m.; * P < 0.05 by ANOVA). Red arrows indicate exosome injections. The black arrow denotes the timepoint (day 19) at which lung micrometastatic lesions were analyzed. Lung micrometastases (mCherry+, middle panels, scale bar, 50 μm) were quantified by immunofluorescence (right panel). (e) Analysis of primary tumor growth after subcutaneous flank injection of B16-F10-luciferase cells in mice pre-treated with 5μg of B16-F10 and B16-F1 exosomes three times a week for 28 d. (f) Metastatic burden was quantified by luciferin photon flux at 21 d post-tumor injection (left panel). Scale bar, 200 μm. Quantification of total photon flux in lungs and bones (right panel), n = 10 mice per group; error bars represent s.e.m.; *P < 0.05 by ANOVA.
To investigate the molecular pathways affected by exosome delivery, we performed gene expression profiling of lung tissue 24 and 48 hours after B16-F10 exosome tail vein injection (Supplementary Fig. 2c). We found 130 differentially expressed genes, many related to ECM remodeling and inflammation, such as heat-shock proteins, S100a8 and S100a9 (Supplementary Table 3), which are known effectors of pre-metastatic niche formation37; these findings were confirmed by QRT-PCR (Supplementary Fig. 2d). TNF-α was upregulated 24 hours post-injection (Supplementary Fig. 2d), suggesting that TNF-α could be involved in early events (i.e. vascular permeability)38.
To further examine the role of tumor exosomes in primary tumor growth and metastasis, we injected B16-F10-derived exosomes intravenously three days a week, starting seven days following orthotopic injection of B16-F10mCherry cells. In contrast to controls, mice treated with exosomes exhibited lung micrometastasis at day 19 (Fig. 2d). Primary tumor growth was only enhanced later in the time course (Fig. 2d). To analyze the differences between exosomes from highly versus poorly metastatic melanomas, we injected equal concentrations of physiologically relevant B16-F10 and B16-F1-derived exosomes intravenously (Supplementary Fig. 3a) into mice three times a week over 28 d followed by subcutaneous implants of B16-F10-luciferase cells (See methods). Exosome treatment did not affect primary tumor growth (Fig. 2e). However, mice injected with B16-F10 exosomes had greater metastatic burden in the lung (240—fold increase measured by luciferase intensity; Fig. 2f) compared to control-treated mice or mice treated with low metastatic B16-F1-derived exosomes (Fig. 2f). Furthermore, B16-F10 exosome-treated animals developed metastasis with increased tissue distribution, including bone (Fig. 2f) and brain (data not shown), compared to either control-treated or B16-F1-treated mice. These data suggest that qualitative differences in exosome content can mediate metastatic potential and organotropism.
Tumor-derived exosomes enhance metastasis by BM education
Given the importance of BMDCs in metastatic progression17,39, we determined whether tumor-derived exosomes could “educate” BMDCs and influence metastatic progression (Fig. 3a; see Methods). We transplanted lethally irradiated C57Bl/6 mice with BM from GFP mice treated with B16-F10 exosomes (BM-educated) for 28 d versus synthetic exosomes or PBS treated mice (BM-control). After BM reconstitution, mice were implanted subcutaneously with B16-F10mCherry cells. Exosome-educated BM mice had greater tumor growth compared to BM-control mice (Fig. 3b), indicating that pre-education of BM cells with tumor exosomes was sufficient to accelerate tumor growth. Recruitment of BMDCs and tumor vascular density were enhanced by approximately 4.5—fold and 3—fold, respectively, in primary tumors of BM-educated mice compared to controls (Fig. 3c).
Figure 3. Role of tumor-derived exosomes in BM cell education and metastasis.
(a) Schematic of the experiment performed to analyze the influence of tumor exosomes on BM cell education and metastasis (GFP+= green fluorescent protein). (b) Analysis of primary tumor growth after subcutaneous flank injection of B16-F10mCherry cells in mice transplanted with B16-F10 exosome-educated BM (BM-educated). BM derived from mice treated with control particles (BM-control) was used in parallel, n = 5 mice per group; error bars represent s.e.m.; ***P < 0.001 by ANOVA. (c) Confocal microscopic analysis of BMDCs (GFP+) and vasculature (lectin-red) in primary tumors from BM-educated mice and controls (top panels). Scale bar, 200 μm. Quantification of vasculature and total BMDCs is shown below (lower panels), n = 5 mice per group; error bars represent s.e.m. P value by ANOVA. (d) BMDCs (GFP+) and tumor B16-F10 cells (mCherry+-red) in lung metastatic lesions at 28 d post-tumor injection (top panels). Scale bar, 50 μm. Quantification of metastatic area, tumor burden and total BMDCs is shown in the lower panels (left), n = 5 mice per group; error bars represent s.e.m. P value by ANOVA. Macroscopic analysis of lung metastases at day 35 is shown in the right panel. Scale bar, 200 mm. (e) Flow cytometric analysis of indicated BM progenitor cell populations in mice educated with B16-F10 and B16-F1 exosomes or control particles for 28 d (n = 5 mice per group; error bars represent s.e.m.; NS = not significant by ANOVA).
We found an increase in the size and number of metastases at both typical (i.e., lungs and ipsilateral lymph nodes) and atypical sites (i.e., contralateral lymph nodes, mesentery, and brain) at this stage of tumorigenesis (Supplementary Table 4). BM-educated mice had a 3—fold higher metastatic burden and larger metastatic lesions in the lung and 2—fold elevation of BMDCs, as compared to BM-controls (Fig. 3d). Interestingly, BMDCs were present in metastatic and non-metastatic regions, although in tumor areas the number of BMDCs per area was increased (Supplementary Fig. 3b). Furthermore, BM cell education with B16-F10 exosomes increased primary tumor growth and metastatic burden by 10—fold in Lewis lung carcinoma (Supplementary Fig. 4), demonstrating a crucial role for BM cell education by tumor exosomes in regulating tumor metastasis.
To further define the mechanisms of exosome education of BMDC, we analyzed the effect of B16-F10 and B16-F1 exosomes on BM progenitor cells. Twenty-eight days of exosome treatment led to a 2—fold increase in the frequency of pro-angiogenic c-Kit+Tie2+ cells in BM, while c-Kit+Sca1+ hematopoietic stem cells were not affected (Fig. 3e). Exosome education did not affect other BM-derived propulations, including CD11b+, CD11b+Gr1+, F4/80+, or VEGFR1+ cells (data not shown), which are commonly mobilized by growth factors and chemokines18,21,37.
Horizontal transfer of exosomal Met to BM progenitors
Exosomes are known to promote horizontal transfer of molecules to recipient cells 8,13,40. Given the differential effects of B16-F1 versus B16-F10 exosomes on metastatic potential41, we identified proteins that were highly expressed in the B16-F10 exosomes but present at much lower levels in the B16-F1 exosomes by proteomic profiling (Supplementary Table 5). Some candidates with known roles in tumorigenesis and metastasis include the Met oncoprotein, CD44, Hsp70 and annexin A6 24,42. Given the importance of MET signaling in migration, invasion, angiogenesis and BM cell mobilization24,25,43,44, we focused our functional analyses on this proto-oncogene. We hypothesized that exosomes could horizontally transfer MET from melanoma to BM progenitor cells, and could represent a new mechanism promoting metastatic progression. We verified that Met and phospho-Met levels were increased in both B16-F10 exosomes and cells compared to B16-F1 (Fig. 4a). After 28 d of exosome education, Met expression was increased in BM progenitor cells from mice injected with B16-F10 exosomes (Fig. 4b) but not in the B16-F1 group. Notably, Cd44 levels were not affected by treatment with either B16-F10 or B16-F1 exosomes (Fig. 4b).
Figure 4. Met analysis in tumor and BM cells.
(a) Western blot analysis of Met and phospho-Met in B16-F1 and B16-F10 exosomes and cells (b) QRT-PCR analysis of Met and Cd44 in lineage-negative BM cells after B16-F10 or B16-F1 exosome education; error bars represent s.e.m.; NS = not significant by ANOVA. (c) Flow cytometric analysis of c-Kit and Met expression on BM cells after overnight incubation with fluorescently labeled exosomes (20 μg ml−1, PKH26+). FL2 fluorescence indicates exosome uptake (right panel). (d) Flow cytometric analysis of Met expression in BM c-Kit+Tie2+ cells (upper panel) and Lin–c-Kit+Tie2+ circulating blood cells (lower panel) of mice educated with B16-F10 and B16-F1 exosomes (red area=percentage of Met+ cells). Error bars represent s.e.m.; NS = not significant by ANOVA. (e) Analysis of metastasis in mice educated with 5μg of B16-F10, B16-F10shMet and control. Scale bar, 200 μm. Quantification of total photon flux 21d post B16-F10-luciferase cell injection (lower panel). Error bars represent s.e.m.; P value by ANOVA. (f) Multiplex protein analysis of MET and phospho-MET (Tyr1349) in the circulating exosomes from a retrospective series of frozen plasma derived from melanoma subjects and controls. Controls (n = 7); Stage III (n = 24); Stage IV (n = 15). P values by ANOVA. (g) Flow cytometric analysis depicting the percentage of MET+ BM progenitor cells in the blood of individuals with melanoma. Controls (n = 7); stage I–III (n = 10); stage IV (n = 9). Error bars represent s.e.m; P values by ANOVA.
To determine the functional consequences of Met expression within exosomes, we reduced Met levels by 40% and phospho-Met levels by 80% using Met shRNAs (B16-F10shMet, Supplementary Fig. 5a). Using fluorescently-labeled exosomes isolated from B16-F10, B16-F10shMet and B16-F1 cells, we demonstrated a 6—fold increase in c-Kit+Met+ BM progenitors after treatment with B16-F10 exosomes as compared to treatment with B16-F10-shMet, B16-F1 and control exosomes (Fig. 4c and Supplementary Fig. 5b). Notably, exosome uptake was similar regardless of their origin and Met expression (Fig. 4c, right inset). In order to determine whether B16-F10 exosomes contribute to MET pathway activation, we analyzed the phosphorylation of downstream mediators of HGF/MET signaling, such as phospho S6-kinase (mTOR pathway) and phospho-ERK (MAPK pathway), known effectors of MET-mediated BM progenitor cell mobilization43,44. Pre-treatment of BM cells with B16-F10 exosomes resulted in HGF-induced S6 and ERK phosphorylation compared to non-treated or B16-F1 exosome pre-treated BM (Supplementary Fig. 5c). Additionally, we observed that a Met inhibitor (Crizotinib) prevented HGF-induced phosphorylation of S6-kinase and ERK after B16-F10 exosome treatment, demonstrating the specificity of this pathway (data not shown).
We then investigated the functional consequences of exosome-mediated MET signaling in bone marrow progenitor cells. Education with B16-F10, but not B16-F1 exosomes, increased Met expression in vasculogenic (c-Kit+Tie2+) and hematopoietic (c-Kit+-Sca1+) progenitors in the circulation but not in the BM (Fig. 4d; Supplementary Fig. 5d), suggesting that Met expression enhanced BM cell mobilization. To determine the effects of exosome-dependent MET signaling on primary tumor growth and metastasis, we implanted B16-F10 tumor cells in mice educated with exosomes from B16-F10, B16-F10shMet and controls. While no differences were observed in primary tumor growth (data not shown), mice educated with B16-F10shMet exosomes had reduced lung and bone metastasis compared to mice educated with B16-F10 exosomes (Fig. 4e). Moreover, vasculogenic and hematopoietic precursors were decreased in the BM and peripheral blood of B16-F10shMet exosome-educated mice bearing B16-F10 tumors (data not shown).
To determine the clinical relevance of our findings, we analyzed MET and phospho-MET levels in circulating exosomes isolated from Stage III and IV melanoma subjects (same cohort as in Fig. 1). We found higher levels of total and phospho-MET in both Stage III and IV melanoma individuals compared to control subjects (Fig. 4f). Moreover, MET expression was increased in both BM progenitor cells (CD45−C-KITlow/+) and pro-vasculogenic cells (CD45−C-KIT+/lowTIE2+) isolated from the blood of subjects with Stage IV disease compared to Stage I–III and control subjects (Fig. 4g and Supplementary Fig. 5e). Our data suggest that MET expression in circulating exosomes and BM progenitor cells could be new predictors or early markers of metastatic disease.
Rab27a regulates exosome production and metastasis
As Ras-related RAB proteins control exosome biogenesis 30, we analyzed the expression of RAB genes (RAB1A, RAB5A, RAB5B, RAB7, RAB27A, and RAB27B) using a panel of 39 human-derived melanoma cell lines. Interestingly, those cell lines producing high concentrations of exosomes also expressed high levels of RAB27A, RAB5B and RAB7 mRNA relative to those cell lines expressing lower levels of exosomes (Fig. 5a, b). Given the importance of RAB27 in exosome release, we reasoned that inactivating this gene would reduce exosome production. The Rab27a isoform was selected for knockdown as it was selectively expressed in melanoma-derived cell lines (Fig. 5a and data not shown). We effectively reduced the levels of Rab27a in B16-F10 and SK-Mel28 cells (Fig. 5c) which resulted in a 50% decrease in exosome secretion (Fig. 5d and Supplementary Fig. 6a), demonstrating a role for Rab27a in exosome production by melanoma cells. Although no significant differences in total protein, protein content, exosome-specific markers or Met/phospho-Met levels were seen between control and RAb27a-knockdown cells, we did observe a decrease in the secretion of PlGF-2, PDGF-AA and osteopontin as a consequence of reducing Rab27a levels (Supplementary Fig. 6b, c and Fig. 5e). Importantly, Rab27a knockdown in B16-F10 and SK-Mel 28 cells reduced primary tumor growth, lung colonization and metastasis (Fig. 5f), which were associated with a decrease in BMDCs recruited to these sites (Fig. 5g and Supplementary Fig. 6d). The levels of circulating tumor-derived exosomes in Rab27a knockdown tumor bearing mice were minimal and therefore insufficient to educate and mobilize BMDCs.
Figure 5. Disrupting Rab27a expression reduces exosome release, tumor growth, and metastasis.
(a) QRT-PCR analysis of RAB genes in melanoma (SK-Mel-#), breast cancer (MCF7, MDA-MB-231, SkBr3) and pancreatic adenocarcinoma (AsPc1) human cell lines. Red denotes high (≥ 2—fold), white intermediate (< 2—fold and > 1.5—fold), and blue low (≤ 1.5—fold) RAB expression in melanoma relative to breast cancer and pancreatic cell lines. (b) Measurement of the total protein in the exosomes secreted per million human melanoma cells in culture. (c) QRT-PCR analysis of Rab27a expression after shRNA knockdown of Rab27a in B16-F10 and SK-Mel-28 cell lines. (d) Measurement of exosome protein per million cells after shRNA knockdown of Rab27a in B16-F10 and SK-Mel-28 cell lines. Control scramble shRNA and parental cells were used as a reference. (e) Characterization and densitometric analysis of conditioned medium derived from B16-F10-shScramble and -shRab27a cell lines. (f) Analysis of primary tumor growth and metastasis in shScramble, sh-Rab27a- B16-F10 and SK-Mel28 cell lines subcutaneously injected into the flank of C57BL/6 and NOD SCID mice, respectively. Metastases were macroscopically counted (B16-F10) or quantified by QRT-PCR for mCherry (SK-Mel-28), n = 5 mice per group; error bars represent s.e.m.; ***P < 0.001 by ANOVA. (g) Analysis of BMDCs (GFP+-green) and tumor cells (mCherry-red) in B16-F10-shScramble and -shRab27a primary tumors (upper panel, scale bar, 200 μm) and lungs (lower panels, scale bar, 50 μm). Quantification of the metastatic area and total BMDCs is shown on the right, n = 5 mice per group; Error bars represent s.e.m; P value by ANOVA.
By injecting increasing amounts of exosomes isolated from B16-F10shScramble and B16-F10-shRab27a cell lines in mice followed by flank injection of tumor cells we observed a dose-dependent increase in metastatic burden (Supplementary Fig. 7a, b). These data suggest that quantitative differences in exosome production can alter metastatic potential. Since overexpression of Rab27a in B16-F1, LLC and B16-F10 cell lines led to cell death, we were unable to determine the sufficiency of Rab27a in exosome production and modulating metastasis (data not shown). Collectively, these data suggest that Rab27a knockdown in melanoma models reduced exosome production and circulating exosome levels, preventing the recruitment of BMDCs necessary for metastatic progression.
DISCUSSION
Accumulating evidence indicates that exosomes are mediators of metastasis3,11,13,15,22,23,45,46. Multiple cell types, including fibroblasts, endothelial cells, and BM progenitor cells, contribute to the generation of metastatic microenvironments,39,47 and may be influenced by horizontal transfer of molecules (i.e. proteins and miRNAs) by exosomes8,40,48,49. However, the molecular and cellular mechanisms underlying this association have not been determined. Here, we show that exosomes play a role in tumor progression and metastatic niche formation that is distinct from that of tumor-derived growth factors, extracellular proteins and chemokines 18,20,21,37,50,51. Tumor-derived exosomes promote the “education” and mobilization of BMDCs that support tumor vasculogenesis, invasion and metastasis. Importantly, tumor-derived exosomes recruit BMDCs through up-regulation of pro-inflammatory molecules at pre-metastatic sites. Specifically, pre-conditioning the BM with exosomes from a highly metastatic cancer (B16-F10) can increase the metastatic tumor burden and distribution of target tissues, even for tumors of different origins with low metastatic capacity. Our work in tumor-derived exosome biology complements Paget and Ewing’s theories of metastatic spread, offering an explanation of organotropic involvement in certain metastatic diseases 52,53.
By comparing the protein content of exosomes from highly metastatic and poorly metastatic melanoma cells, we identified MET signaling as a principal mediator of BM progenitor cell “education”. The MET oncogene mediates cellular transformation, tumor cell proliferation, survival, motility, invasion and metastasis 26,28,29,44,54,55. Here, we propose a novel role for exosome-packaged MET in metastatic melanoma. The unique cargo of exosomes from other tumor types may “educate” distinct bone marrow populations, thus directly impacting metastatic propensity and outcome. Interestingly, the BM of mice “educated” with the low-metastatic B16-F1 exosomes, lacking the MET receptor, reduced (though not reaching statistically significance) metastatic burden and organotropism of highly metastatic B16-F10 primary tumors, suggesting that non-metastatic exosomes may educate bone marrow to prevent metastatic disease.
The molecular mechanisms of exosome biogenesis and secretion are poorly described. We found that, among 39 melanoma-derived cell lines studied, specific RAB genes involved in exosome secretion (RAB1A, RAB5B, RAB7, RAB27A) were highly expressed in cells lines secreting high exosome levels. Reducing the expression of Rab27a, a regulator of protein trafficking and melanoma proliferation 31,56, decreased exosome production and pro-angiogenic factor (PlGF-2, osteopontin and PDGF-AA) release from tumor cells, hindering BMDC mobilization, tumor growth and metastasis. Our data, together with other studies showing that overexpression of Rab27a correlates with breast cancer invasiveness and metastasis57,58, suggest that Rab27a should be considered as a potential therapeutic target in cancer.
Our human studies identified a specific “melanoma signature” comprised of TYRP2, VLA-4, HSP70, an HSP90 isoform, and MET in circulating exosomes from subjects with advanced melanoma, which could be used as an indicator of metastatic disease and tumor burden. We found that the co-expression of TYRP2 and MET in exosomes, as well as increased protein per exosome, predicted disease progression. Finally, we demonstrated that MET expression is elevated in circulating pro-vasculogenic progenitors in metastatic subjects with melanoma. Thus, our work identifies quantitative and qualitative signatures, along with specific BM progenitor cell populations mobilized as representative hallmarks of metastatic disease.
Although membrane vesicles of cancer cells have been suggested to contribute to the horizontal propagation of oncoproteins and genetic material (RNA)5,8,13,14,40,48, our study is the first to demonstrate that transfer of the MET oncoprotein from tumor-derived exosomes to BM progenitor cells promote the metastatic process in vivo. Importantly, we demonstrate that exosomes can alter the BM in a durable manner; these results suggest that genetic or epigenetic changes could be involved in this phenomenon as BM cells retain the “educated” phenotype following engraftment into a new host.
Here, we propose a novel mechanism that controls metastatic progression through the crosstalk between tumor-derived exosomes and BM progenitor cells. Collectively, our data identified exosome-mediated transfer of the oncoprotein MET as a key regulator of BM “education”, mobilization, and metastatic progression.
Microarray Data
Microarray raw data tables have been deposited in the Gene Expression Omnibus (accession number GSE36584).
METHODS
Exosome purification and tracking analysis
Cells were cultured in media supplemented with 10% exosome-depleted fetal bovine serum (FBS, Hyclone). FBS was depleted of bovine exosomes by ultracentrifugation at 100,000 × g for 70 min. Supernatant fractions collected from 48–72 h cell cultures were pelleted by centrifugation at 500 × g for 10 min. The supernatant was centrifuged at 20,000 × g for 20 min. Exosomes were then harvested by centrifugation at 100,000 × g for 70 min. The exosome pellet was resuspended in 20 ml of PBS and collected by ultracentrifugation at 100,000 × g for 70 min (Sorvall Surespin 630 rotor). Circulating exosomes were isolated from mouse and human plasma as above with an additional filtration through 1.2 μm nylon filters (GE) before the last step of ultracentrifugation. For centrifugation onto sucrose cushions, the samples were diluted 1:10 in PBS after centrifugation at 20,000 × g and then collected by ultracentrifugation (100,000 × g for 70 min) on a 40% sucrose cushion. The floating exosome fraction was collected again by ultracentrifugation as above, and the final pellet was resuspended in PBS. For retrospective studies using frozen plasma, 2 ml of cell-free plasma were centrifuged at 500 × g for 10 minutes, then the supernatant was centrifuged at 20,000 × g for 20 min. Exosomes were then harvested by centrifugation at 100,000 × g for 70 min. The exosome pellet was resuspended in PBS and collected by ultracentrifugation at 100,000 × g for 70 min (Sorvall S100AT5 rotor). The LM10 nanoparticle characterization system (NanoSight) equipped with a blue laser (405 nm) was used for real-time characterization of the vesicles.
Human studies
Human peripheral blood samples were obtained from stage I–IV melanoma subjects at MSKCC, with histologically confirmed melanoma. For retrospective studies of circulating exosomes in frozen plasma, Stage III subjects were followed up for 1 to 4 years. Stage IV subjects were followed up over 42 months. All individuals provided informed consent for blood donation on an MSKCC IRB-approved protocol.
Exosome labeling and treatment
5–10 μg of total exosome protein were injected via tail vein in a total volume of 100–200 μl PBS. Synthetic unilamellar 100 nm liposomes (Encapsula Nanoscience) or PBS were used as controls in all studies (data not shown). For exosome-tracking experiments, purified exosomes were fluorescently labeled using PKH67 membrane dye (Sigma). Labeled exosomes were washed in 20 ml of PBS, collected by ultracentrifugation as above and resuspended in PBS. We verified no dye contamination occurred in PKH67-labeled exosome preparations, by ultracentrifugation on 40% sucrose cushions as describe above (data not shown).
Metastasis and lung colonization studies
To analyze the role of exosomes in tumor metastasis, 8–10 week-old C57Bl/6 female mice were injected in the flank with 1 × 106 B16-F10mCherry. Seven days later, 10 μg of B16-F10 exosomes were injected three times a week for 3 weeks. Metastasis was evaluated by mCherry expression at days 19 and 28. Alternatively, C57Bl/6 female mice were injected three times a week with the indicated dose of exosomes for 28 d. After exosome injection, mice received 1 × 106 B16-F10mluciferase subcutaneous flank injections. Controls included PBS and synthetic unilamellar liposome injections. Luciferase imaging was performed using the IVIS Spectrum system (Caliper, Xenogen). Tumor bearing mice were anesthetized (isoflurane/O2) and D-luciferin (50 mg Kg−1 in 100 μl PBS) was administered. Five minutes later mice were euthanized and organs were analyzed for luciferase expression. Data were quantified with Living Imaging software 4.2. For analysis of Rab27a knockdown tumor cell growth, 1 × 106 B16-F10 cells (in 100 μl of PBS) or 2 × 106 SK-Mel28mCherry cells (in a mix of 1:1 serum-free RPMI:growth factor reduced Matrigel) were injected in the flank of C57Bl/6 or NOD SCID female mice, respectively. For lung colonization experiments, 1 × 105 B16-F10 cells or 5 × 105 SK-Mel-28 cells (in 100 μl of PBS) were injected via tail vein in C57Bl/6 female mice.
GFP BM education
eGFP-transgenic mice males (C57Bl/6-TgN(ActbEGFP)1Osb/J, Jackson Laboratory) were injected three times a week with 5–10 μg of B16-F10 exosomes over 28 d. BM was harvested by flushing femurs and tibias of eGFP donor animals and 5 × 106 eGFP total BM cells were transplanted into lethally irradiated (950 rads) C57Bl/6 female mice via retro-orbital injection. After 4 weeks, the eGFP BM-reconstituted C57Bl/6 mice were injected subcutaneously in the flank with 1 × 106 B16-F10mCherry or LLCmCherry cells. Mice were sacrificed after 28 or 35 d. Tissues were dissected and fixed in 4% paraformaldehyde (PFA) or in a mix of 2% PFA-20% sucrose solution overnight, embedded in Tissue-tek O.C.T. (Electron Microscopy Sciences) and blocks were frozen in a dry ice/ethanol bath. Additionally, tissues were snap-frozen in liquid nitrogen for RNA/protein extraction. Tissues used to evaluate metastasis in BM-educated mice and controls (Supplementary Table 4) were paraffin-embedded, stained with hematoxilin/eosin and metastases were evaluated by microscopy. All mouse work was performed in accordance with institutional, IACCUC and AAALAS guidelines. All animals were monitored for abnormal tissue growth or ill effects according to AAALAS guidelines and sacrificed if excessive deterioration of animal health was observed.
Mouse flow cytometry and antibodies
Lungs and tissues were prepared for flow cytometry by mincing followed by digestion at 37 °C for 45–60 min with an enzyme cocktail (Collagenase A and DNaseI, Roche). Single cell suspensions were filtered through a 70 μm strainer. Peripheral blood was obtained by retro-orbital bleeding directly into anti-coagulant tubes (EDTA) and red blood cells were lysed using ACK lysis buffer (Gibco). Cell suspensions were blocked with Fc-block (CD16/CD32, BD), washed in PBS-1% BSA and incubated with the following primary antibodies diluted in PBS-1% BSA: CD11b-FITC (M1/70), VEGFR2-PE (Avas12α1), lineage negative cell detection kit (biotinylated B220, CD3ε, TER119, CD11B, GR-1), followed by streptavidin-PE-CY7, c-Kit-APC (2B8), Sca1-PE (D7), Gr1-PE (RB6-8C5), F4/80-APC (BM8), Tie2-PE (TEK4), CD29-APC (HMb1), CD105-PE (MJ7/18) and Met-FITC (ebioclone4). Fluorochrome-conjugated antibodies were purchased from BD or eBioscience and used at pre-determined saturating concentrations. Data acquired on a BD FACS Calibur™cytometer with CellQuest software (BD) was analyzed using FlowJo™ software (TreeStar).
Flow cytometry for human samples
Peripheral blood obtained from human subjects was centrifuged at 500 × g for 10 min at 4 °C. Plasma was used for exosome isolation and analysis. Peripheral blood cells were isolated using Ficoll-Paque gradient (GE Healthcare). The buffy coat was separated and cells were washed in PBS-1% BSA and collected by centrifugation at 400 × g for 5 min at 4 °C. Residual red blood cells were lysed for 4 min at 4 °C using ACK lysis buffer (Gibco). Cells were incubated with flurochrome-conjugated antibodies diluted in PBS-1% BSA and washed in PBS-1% BSA. Flow cytometric analysis was performed using antibodies to: CD45-eFluor450 (2D1), C-KIT/CD117-pe (104D2), CD34-Alexa700 (581), TIE2-Alexa647 (CD202B) (33.1), MET-FITC (eBioclone 97). Antibody-fluorochrome conjugates were purchased from BD Biosciences, eBioscience or Biolegend and used as suggested by manufacturer. Data acquired on a BD FACS Canto™ was analyzed using FlowJo™ (Tree Star) software.
Cell lines and cell culture
B16-F10, B16-F1, LLC, MCF-7, As-Pc1, and MDA-MB-231 cells were cultured in DMEM. SkBr3, SW480, SW620 and human melanoma cells (SK-Mel-#) were cultured in RPMI supplemented with penicillin (100 U ml−1) and streptomycin (100μg ml−1) and 10% exosome-depleted FBS. Cells were obtained from ATCC. Human melanoma cell lines were obtained from MSKCC.
Electron microscopy
Exosomes purified as described above were fixed in 2% PFA (w/v) in 200 mM phosphate buffer (pH 7.4). Fixed exosomes were dropped onto a formvar-carbon coated grid and left to dry at room temperature for 20 min. After washing in PBS, the exosomes were fixed in 1% glutaraldehyde for 5 min, washed in water, and stained with saturated aqueous uranyl oxalate for 5 min. Samples were then embedded in 0.4% w/v uranyl acetate, 1.8% w/v methylcellulose and incubated on ice for 10 min. The excess liquid was removed. The grid was dried at room temperature for 10 min and viewed at 20,000 and 50,000 magnification using an electron microscope (model 910, Carl Zeiss.).
Lung leakiness experiments
Ten micrograms of total exosome protein were injected via tail vein. Conditioned media (CM) was prepared by filtering supernatant fractions of cultured B16-F10 cells through a 0.22 μm filter and ultracentifuging it at 100,000 × g to deplete exosomes. One hundred microliters of CM were injected via tail vein injection. As controls, mice were injected with melan-a exosomes, PBS or synthetic 100 μm unilamellar liposomes. Twenty hours after exosome treatment, mice were injected with two miligrams of Texas Red®-lysine fixable dextran 70,000 MW (Invitrogen) via retro-orbital injection. One hour after dextran injection, mice were sacrificed and perfused with 40 ml of PBS. Lungs were dissected and fixed in a mix of 2% PFA and 20% sucrose overnight.
Immunofluorescence
For immunofluorescence, 12 μm O.C.T. tissue sections were stained with antibodies to CD31 (1:100) and VCAM-1 (1:100) (BD). For vasculature analysis, each animal was injected with 50 μg Alexa Fluor 594-conjugated Isolectin GS-IB4 (Molecular Probes) 10 min before sacrifice. GFP and mCherry+ cells were detected by their intrinsic signal. Fluorescent images were obtained using a Nikon confocal microscope (Eclipse TE2000U) and analyzed using Nikon software (EZ-C1 3.6). To determine the vessel density in metastatic lesions, digital images of mCherry/GFP stained sections were analyzed with ImageJ Software (NIH) by determining the pixel intensity per square micrometer.
BM progenitor cell enrichment, in vitro treatment and response to HGF
Mice were sacrificed and BM was isolated by flushing the femur and tibia with PBS using a 25G1/2″ needle (BD). Cell suspensions were filtered through 70 μm cell strainers (BD) prior to centrifugation. To enrich for progenitors, lineage positive (Lin+, ie: CD5, CD45R, CD19, CD11b, Ly6G/C, 7-4, TER119) BM cells were immunomagnetically depleted using the EasySep® Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stem Cell Technologies) as per manufacturer’s instructions. For in vitro BM treatment with exosomes, BM cells were flushed as above and cultured in Stem Span (Stem Cell Technologies) for 16 hours in the presence or absence of 20 μg ml−1 exosomes as indicated. For HGF analysis BM cells were then stimulated with 5 ng ml−1 HGF (Peprotech) for 4 hours and collected for Western blot analysis in RIPA buffer with phosphatase inhibitors (1 mM sodium orthovanadate and 5mM β-glycerol phosphate). For Met signaling inhibition, cells were incubated with 20 nM Crizotinib (Selleck Bio) 1 h before HGF stimulation.
Quantitative real-time PCR and short-hairpin RNA interference studies
Frozen tissues or cell lines were analyzed for specific gene expression using pre-designed TaqMan® assays (specific assay numbers: mouse Met,: Mm01156972_m1, mouse Cd44: Mm01277163, human RAB27A: Hs00608302_m1, mouse Rab27a: Mm00469997_m1, S100a8: Mm00496696-g1, S100a9: Mm00656925_m1) or GFP or mCherry-specific primers18 using SybrGreen PCR reagents (Applied Biosystems). Briefly, total RNA was extracted from tissues or cells using the RNeasy kit (Qiagen), and reverse-transcribed using Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR (QRT-PCR) was performed on a 7500 Fast Real Time PCR System (Applied Biosystems), using TaqMan Universal PCR Master Mix (Applied Biosystems). Relative expression was normalized to β-actin levels. For shRNA-mediated knockdown of Rab27a, lentiviral vectors encoding shRNAs were purchased from Thermo Fisher. Human Rab27a shRNA sense sequences were: V3LHS_300918: 5′-CCCAGTGTACTTTACCAATATA-3′, V3LHS_300917: 5′-CAGGGAAGACCAGTGTACTTTA-3′. Rab27a knockdown efficiency was similar for both human shRNAs but V3LHS_300918 is shown in figures. The sense sequence for mouse Rab27a shRNA was V3LHS_300916: 5′-ACAGGAGAGGTTTCGTAGCTTA-3′ and the scramble sense sequence used was: 5′-ATCTCGCTTGGGCGAGAGTAAG-3′. Mouse Met shRNA sense sequences were: V3LMM_456078: 5′-CCAGACTTTTCATACAAGAATA-3′, V2LMM_30812: 5′-CCCTATGTAGA-TCCTGTAATAA-3′. Met knockdown efficiency was similar for both shRNAs (routinely exceeding 90% as determined by GFP expression) but V3LMM_456078 is shown in figures.
Proteomic analysis
Identification of exosomal proteins was performed using reverse phase high-pressure liquid chromatography mass spectrometry (HPLC-MS). Samples were denatured at 90 °C, reduced with 10 mM DTT at 51 °C for 1 h and alkylated with 50 mM iodoacetamide at 25 °C for 45 min. Proteins were digested with trypsin (Promega) overnight at 25 °C. Tryptic peptides were concentrated by vacuum centrifugation and desalted using C18STAGE Tips prior to mass spectrometric analysis. Samples were loaded by an Eksigent AS2 autosampler onto a 75 μm fused silica capillary column packed with 11 cm of C18 reverse phase resin (5 μm particles, 200 A pore size; Magic C18; Michrom BioResources). Peptides were resolved on a 180 min 1–100% buffer B gradient (buffer A = 0.1 mol L−1 acetic acid, buffer B = 70% acetonitrile in 0.1 mol L−1 acetic acid) at a flow rate of 200 nl min−1 (1200 series, Agilent,). The HPLC was coupled to a mass spectrometer (LTQ-Orbitrap, ThermoFisher Scientific) with a resolution of 30,000 for full MS followed by seven data-dependent MS/MS analyses. Collision-induced dissociation (CID) was used for peptide fragmentation. Each sample was analyzed at least two times. All MS data were analyzed with Proteome Discoverer software (version 1.2,Thermo Fisher Scientific) using the SEQUEST algorithm to search against human and mouse UniProt databases. The peptides were constrained to be tryptic and up to 2 missed cleavages were allowed. Carbamidomethylation of cysteine was specified as a fixed, and oxidation of methionine as a variable modification. The precursor ion tolerance was set to 25 ppm, and fragment ion mass tolerance to 0.8 Da. Search results were analyzed individually and data for replicates were combined and evaluated. For validation of proteomic analysis we performed western blot analysis for selected proteins (i.e Met, CD44, Annexin A2, Annexin A6; Fig. 4a and data not shown) using equal amounts of exosomes derived from B16-F1 and B6-F10 cells.
Microarray preparation and analysis
For analyzing the genes modified in lungs by B16-exosomes, a total of 10 μg of exosome protein were injected by tail vein, and 24 or 48 h later mice were sacrificed and lungs were dissected and stored in RNA-later solution (Ambion) until RNA extraction. A total of 3 mice were used per time point and the experiment was performed in triplicate. Total RNA was isolated using the RNeasy Mini Kit (Qiagen). The Affymetrix One-round in vitro transcription (IVT) RNA Amplification Kit was used to amplify 1.5 μg of total RNA. The cDNA was synthesized with a primer containing oligo(dT) and T7 RNA polymerase promoter sequences. Double-stranded cDNA was then purified and used as a template to generate biotinylated cRNA. The quantity and quality of the amplified cRNA was assessed using NanoDrop ND-1000 Spectrophotometer (Thermo Scientific) and Agilent Bioanalyzer. The biotinylated cRNA was fragmented and hybridized to Affymetrix Mouse Genome 430 Plus 2.0 GeneChip arrays containing 39,000 transcripts per array. After hybridization, GeneChip arrays were washed, stained, and scanned using a GeneChip Scanner 3000 7G according to Affymetrix Expression Analysis instructions. Affymetrix GeneChip Operating Software was used for image acquisition. The target signal intensity from each chip was scaled to 500. Triplicate samples from each exosome-treated and control lung samples were analyzed to identify differentially expressed genes. For statistical analysis, we selected genes whose expression differed between groups by a factor of at least 2. A hierarchical clustering method was applied to group the genes and samples on the basis of the similarities in gene expression, and the unsupervised analyses were visualized using the SOTA and TreeView software assuming euclidean distances between genes (http://bioinfo.cnio.es/cgi-bin/tools/clustering/sotarray). We tested the null hypothesis of equal means between the two groups using t-test, computing p-values using a permutation test. Fold expression was calculated from the expression ratio in the exosome-treated conditions with respect to control samples.
Western blot, antibodies and multiplex analysis
Exosomes or cells were lysed with RIPA buffer containing a complete protease inhibitor tablet (Roche). Lysates were cleared by centrifugation at 14,000 × g for 20 min. Supernatant fractions were used for Western blot. Protein extracts were resolved by SDS-PAGE and probed with the indicated antibodies. For Western Blot or immunofluorescence analysis we used antibodies to: Hsp90 (1:1000, Stressgen #ADI-SPA-830), Hsp70 (1:1000, Stressgen #SPA-810-F), Hsc70 (1:500, Stressgen #ALX-804-067), Rab-family sampler kit (Rab4, 5, 7, 9, 11, (1:500, Cell Signaling: #9385), Rab27a (1:500, Abnova # H0005873-M02), TYRP-2 (1:500, PEP-8h), VLA4 (1:500, Chemicon clone PS2), Melan-A (1:1000, Santa Cruz #28871), Alix (1:1000, Cell Signaling #2171), TSG101 (1:500, Santa Cruz #sc-7964), Met/Phospho Met (1:1000, Cell signaling #3127 and #3077), -phospho-S6 kinase (1:1000 Cell Signaling #9802), phospho ERK (1:1000, Cell signaling #4370). Antibodies to GAPDH or β-actin (Santa Cruz, 1:1000 #sc-25778 and 1:5000 #sc-130656) were used as loading controls. The intensities of the immunoreactive bands were quantified by densitometry using ImageJ software (NIH). For analysis of protein expression profiles in conditioned media of cells we used the mouse angiogenesis antibody array (R&D) using 200 μg of cell extracts or 450 μl of 24 h CM. The MET/PhosphoMET analysis kits were purchased from MesoScale Discovery (MSD). All reagents were provided with the MSD kit and the experiment was performed according to the manufacturer’s instructions using 10 μg of protein extract. MSD plates were read on the MSD Sector Imager 2400 plate reader (MSD). The raw data was measured as electrochemiluminescence signal (light) detected by photodetectors and analysed using the Discovery Workbench 3.0 software (MSD). The graphs depict mean photon intensity.
Statistical Analysis
Error bars in graphical data represent means ± s.e.m. Mouse experiments were performed in duplicate, using at least 5 mice per treatment group. All in vitro experiments were performed at least in duplicate. Statistical significance was determined using a two-tailed Student’s t test, and p-values of P < 0.05 were considered statistically significant by ANOVA. For tumor growth analyses, we performed two-way ANOVA statistical analysis using GraphPad Prism software.
Supplementary Material
Suppl. Table 1. Melanoma exosome signature. List of main proteins found in exosomes derived from 5 melanoma cell lines (B16-F10, SK-MEL-28, SK-MEL-202, SK-MEL-35 and SK-MEL-265) by mass-spectrometry.
Suppl. Table 2. Tissue distribution of tail vein-injected B16-F10-labelled exosomes in different organs after 24 or 48 hours analyzed by confocal microscopy.
Suppl. Table 3. Differentially expressed genes in mice lungs after 24 and 48 hours of B16-F10-exosome tail vein injection (red = increased expression (> 2 fold), blue= decreased expression (> 2 fold)
Suppl. Table 4. Analysis of B16 metastasis by H&E staining in sections derived from different tissues of mice with B16-F10 flank tumors after transplantation with B16-F10 exosome-educated bone marrow (BM-educated) or BM-treated with control exosome particles (BM-control)
Suppl Table 5. Specific proteins shed in B16-F10 exosomes compared to B16-F1 by Mass Spectrometry
Acknowledgments
We thank C. Ghajar, M. Bissell, A. Cano, J. Wels and S.R. Granitto for critical reading of this paper and their suggestions. We also thank the members of our laboratories for helpful discussions and WCMC electron microscopy and microarray core facilities for their support. We thank Dr. Hearing, (NIH/NCI) for providing the TYRP2 antibody and Dr. Dorothy C Bennett, (St. George’s University of London) for providing the melan-a cell line. Our work is supported by grants from the Children’s Cancer and Blood Foundation (HP, DL), The Hartwell Foundation (DL), The Manning Foundation (DL, BCS), Pediatric Oncology Experimental Therapeutics Investigator’s Consortium (HP, DL), Stavros S. Niarchos Foundation (DL), Champalimaud Foundation (HP, DL), The Nancy C. and Daniel P. Paduano Foundation (HP, DL), The Mary Kay Foundation (ANH, DL), AHEPA 5th District (DL), The Malcolm Hewitt Wiener Foundation (DL), NCI (DL, NCI-R01CA 098234-01), National Foundation for Cancer Research (DL), Susan G. Komen for the Cure (HP, DL), NCI-U54-CA143836 training grant (DL), FICYT foundation (GGS), University of Oviedo Foundation (GGS), Sussman Family Fund (JB), Charles and Marjorie Holloway Foundation (JB), Manhassat Breast Cancer Fund (JB), CA87637-06 (JB), Fundação de Amparo a Pesquisa do Estado de São Paulo, FAPESP (VRM and BCS), NIH (Y.K, R01-CA134519 and R01-CA141062), National Science Foundation Grant CBET-0941143 and an American Society for Mass Spectrometry research award (BAG).
Footnotes
Author Contributions
H.P. developed the hypothesis, designed the experimental approach, performed the experimental work, analyzed the data, coordinated the project and wrote the manuscript. M.A. performed mass spectrometry studies. S.L. conducted experimental work, I.M. performed flow cytometry studies and analysis, B.C.S. conducted mouse work and proteomic characterization of exosomes. G.M.B. conducted gene expression studies and analysis of microarray data. M.H.R. conducted mouse work and BM education studies, C.W. conducted mouse work and human studies, G.G.S developed BM education assays, A.N.H. quantified exosomes in human plasma, B.A.G. ran mass spectrometry samples and assisted with data interpretation. C.H. obtained human blood specimens. M.K.C. and J.Y. contributed to the characterization of human plasma exosomes. J.S., R.K., V.M., M.S.B, J.D.W. P.C. and Y.K discussed the hypothesis and contributed to data interpretation and experimental design. J.B. coordinated the project, interpreted data and wrote the manuscript. D.C.L conceived the hypothesis, led the project, interpreted the data, and wrote the manuscript.
References
- 1.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 2.Iero M, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 3.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 4.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 7.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Ratajczak J, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 9.Nazarenko I, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70:1668–1678. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 10.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, et al. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–2499. doi: 10.2353/ajpath.2010.090777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang X, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 14.Hao S, et al. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp Oncol. 2006;28:126–131. [PubMed] [Google Scholar]
- 15.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao D, et al. Bone marrow-derived endothelial progenitor cells contribute to the angiogenic switch in tumor growth and metastatic progression. Biochim Biophys Acta. 2009;1796:33–40. doi: 10.1016/j.bbcan.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 22.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 23.Jung T, et al. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia. 2009;11:1093–1105. doi: 10.1593/neo.09822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 25.Stella GM, Benvenuti S, Comoglio PM. Targeting the MET oncogene in cancer and metastases. Expert Opin Investig Drugs. 2010;19:1381–1394. doi: 10.1517/13543784.2010.522988. [DOI] [PubMed] [Google Scholar]
- 26.Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 27.Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer. 2010;46:1260–1270. doi: 10.1016/j.ejca.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006;12:3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 29.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 30.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 31.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2009;12:19–30. 11–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 32.Braeuer RR, Zigler M, Villares GJ, Dobroff AS, Bar-Eli M. Transcriptional control of melanoma metastasis: the importance of the tumor microenvironment. Semin Cancer Biol. 2011;21:83–88. doi: 10.1016/j.semcancer.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fidler IJ. Critical determinants of melanoma metastasis. J Investig Dermatol Symp Proc. 1996;1:203–208. [PubMed] [Google Scholar]
- 34.Grammatikakis N, et al. The role of Hsp90N, a new member of the Hsp90 family, in signal transduction and neoplastic transformation. J Biol Chem. 2002;277:8312–8320. doi: 10.1074/jbc.M109200200. [DOI] [PubMed] [Google Scholar]
- 35.Fidler IJ, Nicolson GL. Organ selectivity for implantation survival and growth of B16 melanoma variant tumor lines. J Natl Cancer Inst. 1976;57:1199–1202. doi: 10.1093/jnci/57.5.1199. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, et al. Pulmonary vascular destabilization in the premetastatic phase facilitates lung metastasis. Cancer Res. 2009;69:7529–7537. doi: 10.1158/0008-5472.CAN-08-4382. [DOI] [PubMed] [Google Scholar]
- 37.Hiratsuka S, et al. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10:1349–1355. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- 38.Lucas R, Verin AD, Black SM, Catravas JD. Regulators of endothelial and epithelial barrier integrity and function in acute lung injury. Biochem Pharmacol. 2009;77:1763–1772. doi: 10.1016/j.bcp.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor DD, Taylor CG, Jiang CG, Black PH. Characterization of plasma membrane shedding from murine melanoma cells. Int J Cancer. 1988;41:629–635. doi: 10.1002/ijc.2910410425. [DOI] [PubMed] [Google Scholar]
- 42.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 43.Jalili A, Shirvaikar N, Marquez-Curtis LA, Turner AR, Janowska-Wieczorek A. The HGF/c-Met axis synergizes with G-CSF in the mobilization of hematopoietic stem/progenitor cells. Stem Cells Dev. 2010;19:1143–1151. doi: 10.1089/scd.2009.0376. [DOI] [PubMed] [Google Scholar]
- 44.Tesio M, et al. Enhanced c-Met activity promotes G-CSF-induced mobilization of hematopoietic progenitor cells via ROS signaling. Blood. 2011;117:419–428. doi: 10.1182/blood-2009-06-230359. [DOI] [PubMed] [Google Scholar]
- 45.Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009;283:168–175. doi: 10.1016/j.canlet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 46.Logozzi M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guise T. Examining the metastatic niche: targeting the microenvironment. Semin Oncol. 2010;37 (Suppl 2):S2–14. doi: 10.1053/j.seminoncol.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Aliotta JM, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–245. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aliotta JM, et al. Alteration of marrow cell gene expression, protein production, and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells. 2007;25:2245–2256. doi: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Psaila B, Kaplan RN, Port ER, Lyden D. Priming the ‘soil’ for breast cancer metastasis: the pre-metastatic niche. Breast Dis. 2006;26:65–74. doi: 10.3233/bd-2007-26106. [DOI] [PubMed] [Google Scholar]
- 52.Ewing J. W. B. Saunders Co; Philadelphia: 1928. [Google Scholar]
- 53.Paget G. The distribution of secondary growths in cancer of the breast. Lancet. 1889;133:571–573. [PubMed] [Google Scholar]
- 54.Scott KL, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 56.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang JS, Wang FB, Zhang QG, Shen ZZ, Shao ZM. Enhanced expression of Rab27A gene by breast cancer cells promoting invasiveness and the metastasis potential by secretion of insulin-like growth factor-II. Mol Cancer Res. 2008;6:372–382. doi: 10.1158/1541-7786.MCR-07-0162. [DOI] [PubMed] [Google Scholar]
- 58.Hendrix A, et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J Natl Cancer Inst. 2010;102:866–880. doi: 10.1093/jnci/djq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table 1. Melanoma exosome signature. List of main proteins found in exosomes derived from 5 melanoma cell lines (B16-F10, SK-MEL-28, SK-MEL-202, SK-MEL-35 and SK-MEL-265) by mass-spectrometry.
Suppl. Table 2. Tissue distribution of tail vein-injected B16-F10-labelled exosomes in different organs after 24 or 48 hours analyzed by confocal microscopy.
Suppl. Table 3. Differentially expressed genes in mice lungs after 24 and 48 hours of B16-F10-exosome tail vein injection (red = increased expression (> 2 fold), blue= decreased expression (> 2 fold)
Suppl. Table 4. Analysis of B16 metastasis by H&E staining in sections derived from different tissues of mice with B16-F10 flank tumors after transplantation with B16-F10 exosome-educated bone marrow (BM-educated) or BM-treated with control exosome particles (BM-control)
Suppl Table 5. Specific proteins shed in B16-F10 exosomes compared to B16-F1 by Mass Spectrometry