Abstract
BACKGROUND
The loss of E-cadherin (ECAD) protein expression has been linked to aggressive head and neck squamous cell carcinoma (HNSCC). Promoter hypermethylation of the cadherin 1, type 1 (CDH1) gene (encoding ECAD) is 1 mechanism by which this protein can be inactivated, although this epigenetic alteration of the gene has not been linked conclusively to poorer patient outcome and, in fact, may be associated with better patient prognosis.
METHODS
The authors investigated the prevalence of CDH1 promoter hypermethylation in a population-based case series of 340 primary HNSCC tumors using methylation-specific polymerase chain reaction. They also studied the association between CDH1 hypermethylation and patient demographic characteristics using multivariate analysis and examined the impact of CDH1 hypermethylation on patient survival using both univariate and multivariate methods.
RESULTS
Hypermethylation of CDH1 was significantly more prevalent (P <.03) among individuals with a low smoking history independent of whether they were seropositive for human papillomavirus type 16 (HPV-16). Patients who had tumors with CDH1 hypermethylation had significantly better overall survival compared with patients who had tumors without hypermethylation (P <.02; log-rank test). This effect was independent of HPV-16 status and demonstrated a significant hazard ratio of 0.5 (95% confidence interval, 0.3–0.9) in a model that controlled for HPV-16 serology, age, sex, and tumor stage.
CONCLUSIONS
The current results suggested that hypermethylation of CDH1 occurs more commonly in patients with HNSCC who are low smokers, suggesting that an additional factor may be driving this epigenetic alteration. Clinically, CDH1 hypermethylation may hold powerful prognostic potential in addition to that observed with HPV serology, and the authors concluded that it should be pursued in additional studies.
Keywords: hypermethylation, epigenetic, head and neck squamous cell carcinoma, survival, E-cadherin
Head and neck squamous cell carcinoma (HNSCC) is the ninth most common cancer in men in the United States; and, in 2003, approximately 390,000 new cases of oral cancer and 160,000 cases of laryngeal cancer were diagnosed worldwide among both sexes.1 In the United States, it is estimated that, in 2008, there will be approximately 30,000 new diagnoses of oral and pharyngeal cancer (excluding cancers of the lip) and approximately 10,000 new diagnoses of laryngeal cancer; approximately 11,000 deaths will result from these diseases.2 Greater than 95% of all oral cancers are squamous cell carcinoma (SCC).
It is well accepted today that alcohol and tobacco synergistically enhance the risk of oral cancer. 3–9 In addition to these factors, Loning et al10 first suggested a correlation between human papillomavirus (HPV) and HNSCC in 1985, and evidence of a possible association has been increasing in Europe and North America.11–13 In a previous study, we observed significant relative risks of 1.5 and 6.0 for SCC of the oral cavity and pharynx, respectively, associated with seropositivity for HPV type 16 (HPV-16), and we demonstrated that patients with HPV-16 seropositivity have significantly better overall survival. 14 The mechanism for this survival advantage related to viral presence remains unclear.
The hypermethylation of tumor suppressor gene promoters is a well characterized epigenetic alteration in many tumor types, including HNSCC, and it has been established that it leads to the transcriptional silencing of the downstream gene. Loss of expression of the E-cadherin (ECAD) protein (encoded by the cadherin 1, type 1 [CDH1] gene) has been implicated in increased metastatic potential and poorer prognosis in HNSCC,15,16 and this has been attributed to the epithelial-to-mesenchymal transition (EMT) associated with ECAD loss.17 It is noteworthy that several studies have demonstrated a lack of association between CDH1 methylation, loss of protein expression, and patient prognosis15,18,19; and the studies that demonstrated associations often used cultured cells and not human tissue samples.20 In fact, in a relatively large study of laryngeal and hypopharyngeal tumors, the methylation of CDH1 had a nonsignificant association with better survival. 21 Those studies suggested that the hypermethylation of CDH1 may represent a prognostic marker distinct from ECAD protein expression. Thus, for the current study, we comprehensively examined the promoter hypermethylation of CDH1 in a population-based case series of HNSCC and studied the correlation of this alteration with risk factors associated with head and neck carcinogenesis and as well as its correlation with patient survival.
MATERIALS AND METHODS
Study Population
The study population was described previously.22,23 Briefly, incident cases of HNSCC were identified from 9 medical facilities in the Boston, Massachusetts metropolitan area, with histologic classification of malignancy based on pathology reports from the participating hospitals and confirmed by an independent study pathologist. All patients who were enrolled in the study provided written, informed consent as approved by the institutional review boards of the participating institutions. Archived pathology specimens were used for analysis of promoter hypermethylation, and a total of 350 FFPE tumor samples were available with adequate tissue for analysis, from which 340 samples were analyzed successfully for promoter hypermethylation of CDH1. Data on HPV-16 serology from the parent case-control study14 were reported previously. Table 1 highlights the demographic characteristics of the population studied.
TABLE 1.
Demographics of the Patients With Head and Neck Squamous Cell Carcinoma Examined for CDH1 Hypermethylation (n=340)
| Characteristic | No. | Prevalence |
|---|---|---|
| Age, y | ||
| <50 | 68 | 20 |
| 50–59 | 115 | 33.8 |
| 60–69 | 85 | 25 |
| >70 | 72 | 21.2 |
| Sex | ||
| Women | 89 | 26.2 |
| Men | 251 | 73.8 |
| Pack-years smoked* | ||
| 0–5 | 82 | 24.9 |
| 5–34 | 99 | 31.1 |
| >34 | 148 | 45 |
| Average alcoholic drinks per wk† | ||
| 0–2.5 | 49 | 14.7 |
| 2.5–6 | 50 | 15 |
| 6–14 | 57 | 17 |
| >14 | 178 | 53.3 |
| HPV-16 seropositivity | ||
| Negative | 253 | 74.4 |
| Positive | 87 | 25.6 |
| CDH1 hypermethylation | ||
| Negative | 227 | 66.8 |
| Positive | 113 | 33.2 |
| Tumor stage‡ | ||
| 1 | 25 | 16.5 |
| 2 | 31 | 20.4 |
| 3 | 37 | 24.3 |
| 4 | 58 | 38.8 |
| Tumor site§ | ||
| Oral cavity and tongue | 194 | 57.4 |
| Pharynx | 86 | 25.4 |
| Larynx | 58 | 17.2 |
HPV-16 indicates human papillomavirus type 16; the CDH1, cadherin 1, type 1/E-cadherin gene.
Data on pack-years of smoking were missing for 11 individuals.
Data on alcohol consumption were missing for 6 individuals.
The components used to derive tumor stage were not fully available for 188 patients.
Tumor site was not available for 2 patients.
DNA Extraction and Sodium Bisulfite Modification
Tumor sections with the greatest proportion of malignant tissue were selected for use by the study pathologist. Three 20-μM sections were cut from each FFPE tumor sample, and the sections were transferred into microcentrifuge tubes. The paraffin was dissolved using Histochoice Clearing Agent (Sigma-Aldrich, St. Louis, Mo) followed by 2 washes with 100% ethanol and 1 wash with phosphate-buffered saline. Then, the samples were incubated in sodium dodecyl sulfate (SDS)-lysis solution (50 mM Tris-HCl, pH 8.1; 10 mM ethylene diamine tetracetic acid; 1% SDS) with proteinase K (Qiagen, Valencia, Calif) overnight at 55 °C. Decrosslinking was performed by adding NaCl (final concentration, 0.7 M) and incubating at 65 °C for 4 hours. DNA was recovered using the Wizard DNA clean-up kit (Promega, Madison, Wis) according to the manufacturer’s protocols. Sodium bisulfite modification of the DNA was performed using the EZ DNA Methylation Kit (Zymo Research, Orange, Calif) according to manufacturer’s protocol, with the addition of a 5-minute initial incubation at 95 °C before addition of the denaturation reagent. The decrosslinking steps in the extraction and in the 95 °C incubation ensure more complete melting of the DNA and, thus, more complete sodium bisulfite conversion for these highly cross-linked, formalin-fixed specimens.
Methylation-specific Polymerase Chain Reaction
We specifically choose to use traditional methylation- specific polymerase chain reaction (MSP) for the analysis of promoter hypermethylation in these studies, because we performed a matched analysis between fresh-frozen and formalin-fixed, paraffin-embedded (FFPE) samples and observed the greatest concordance (>95%) for methylation detection using this method (data not shown). We also previously examined potential biases in the sensitivity of using this assay against the relative-quantitative Taqman-based methods24 and observed no evidence for potential bias based on tumor quantity or tumor stage in the samples analyzed. Finally, this method allows for the detection of large numbers of genes from the limited DNA samples available on many of these tumors, whereas the quantitative assays require larger DNA quantities for multiple amplifications of specific genes and reference genes.
Sodium bisulfite-modified DNA was used as the template for MSP, as described previously,25 using primers specific for the methylated promoters of CDH1.26 All MSP analyses are optimized to detect >5% methylated substrate in each sample. To control for the presence of modified DNA, primers were used that were specific to a modified region of the actin beta gene ACTB that contained no CpG sites.27 Modified circulating blood lymphocyte DNA (obtained from a control patient) and the same lymphocyte DNA that was completely methylated using SssI DNA methylase and modified by treatment with sodium bisulfite were used as the negative and positive controls, respectively, in each run.
Statistical Methods
Data were analyzed using SAS software, and all P values represent 2-sided statistical tests. Unconditional logistic regression was used to examine the association between demographics and/or risk factors for HNSCC and CDH1 hypermethylation, including all factors listed in Table 1. Smoking initially was categorized into quartiles of pack-years smoked in controls, with nonsmokers in the lowest quartile. Because the effect estimates for all categories greater than the lowest were similar, we examined whether never smokers differed from those in the lowest category of smokers and, thus, created the following tertiles of smoking: never (0 pack-years), low (from >0 to 10 pack-years), and high (>10 pack-years). Similar categorization was performed for average alcoholic drinks per week; and, because no significant correlation was observed, this variable was removed from the final model. HPV-16 titer was dichotomized as positive versus negative, with positive representing all individuals who had an HPV-16 antibody titer greater than the limit of detection, as reported previously. 14 To create a parsimonious model, all variables initially were included, and those that were identified as nonsignificant were removed as long as their removal did not alter the effect estimates of the other covariates by greater than 15% (ie, if the nonsignificant variable was not a confounder). Sex and age (categorically by decade) also were included in the final model to control for residual confounding.
Patient overall survival was examined first using Kaplan-Meier survival probability curves, and differences between strata were tested using the log-rank test. To control for additional variables related to patient survival, Cox proportional hazards modeling was used. These survival probability models included variables that represented CDH1 hypermethylation and HPV-16 seropositivity and were controlled for patient age (in decades), sex, and tumor stage (I/II vs III/VI).
RESULTS AND DISCUSSION
In our population-based case series of primary HNSCC (1 of the largest assembled to date), hypermethylation of the CDH1 gene (encoding ECAD) was detected in 113 of 340 HNSCC tumors (33%) by using MSP. This prevalence is consistent with that reported previously in an independent pilot sample of patients with HNSCC28 and in other large studies of HNSCC.21,29
To gain a better understanding of the etiology of this epigenetic alteration, next, we examined the association between CDH1 hypermethylation and risk factors for HNSCC using multivariate logistic regression analysis, thereby allowing for the controlling of confounders while examining the direction, strength, and significance of the associations between CDH1 hypermethylation and these demographic characteristics. We observed no significant association between hypermethylation of CDH1 and patient age at diagnosis, patient sex (Table 2), alcohol use, disease stage, or tumor location (data not shown). A significant association was observed between CDH1 hypermethylation and patient smoking, in which we observed that patients who smoked <10 pack-years had an increased odds of hypermethylated CDH1 (odds ratio, 2.4; P <.03) (Table 2) when the analysis was controlled for age, sex, and HPV-16 seropositivity. Never smokers had a slightly elevated, although nonsignificant, odds of CDH1 methylation. Because these low-tertile smokers were more likely to have disease-related HPV infection,30 we also included HPV-16 serology in the model but observed no correlation with HPV-16 seropositivity (Table 2) or with HPV-16 DNA in the tumors (data not shown). Together, these data suggest that CDH1 methylation occurs more often in low-tertile smokers, but not in those with HPV positivity, suggesting that a different factor may be selecting for this epigenetic alteration, which others have suggested may occur early in HNSCC carcinogenesis.18,19
TABLE 2.
Multivariate Analysis of Association Between Patient Demographics and CDH1 Hypermethylation*
| Characteristic |
CDH1 Methylation Status: No. of Patients (%)
|
OR [95% CI] | |
|---|---|---|---|
| Negative | Positive | ||
| Age, y | |||
| <50 | 43 (63) | 25 (37) | Referent |
| 50–59 | 76 (66) | 39 (34) | 0.9 [0.5–1.7] |
| 60–69 | 57 (67) | 28 (33) | 0.9 [0.4–1.7] |
| >70 | 51 (71) | 21 (29) | 0.7 [0.3–1.4] |
| Sex | |||
| Women | 54 (61) | 35 (39) | Referent |
| Men | 173 (69) | 78 (31) | 0.7 [0.4–1.2] |
| Pack-years smoked | |||
| Never | 38 (61) | 24 (39) | 1.4 [0.8–2.5] |
| >0–10 | 16 (50) | 16 (50) | 2.4 [1.1–5]† |
| >10 | 164 (70) | 71 (30) | Referent |
| HPV-16 seropositivity | |||
| Negative | 168 (66) | 85 (34) | Referent |
| Positive | 59 (68) | 28 (32) | 0.9 [0.5–1.6] |
CDH1 indicates the cadherin 1, type 1/E-cadherin; OR, odds ratio; CI, confidence interval; HPV-16, human papillomavirus type 16.
Note: The model was controlled for all variables in the table. Eleven patients were missing data on smoking history; they were coded as missing and were included in the model.
P <.03.
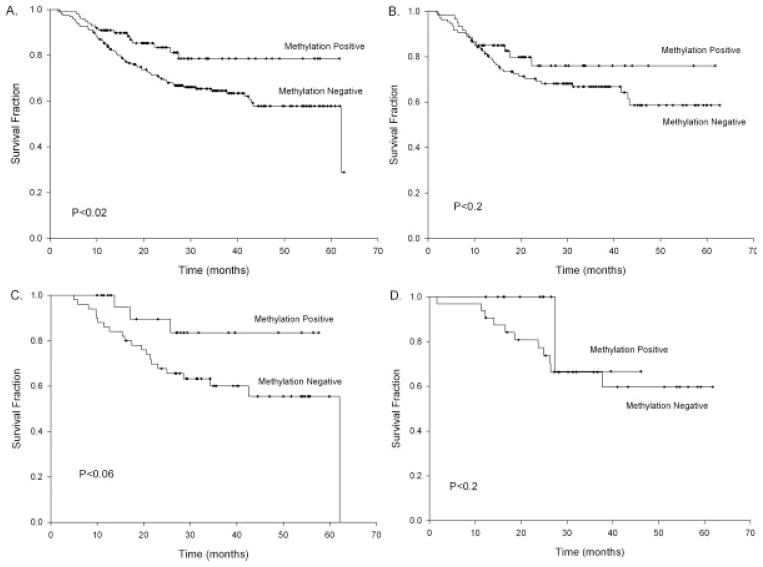
Because this alteration is occurring more often in patients with low tobacco exposure, and because these patients tend to have a different and often better clinical course than patients with heavy tobacco and alcohol use, we examined the impact of CDH1 hypermethylation on patient survival. For our initial univariate analyses, Kaplan-Meier survival probability analysis was used, and the results are depicted in Figure 1. Overall, patients who had CDH1 hypermethylation had increased survival compared with patients who did not have CDH1 hypermethylation (Fig. 1A) (log-rank P <.02). Next, we performed analyses stratified by the location of the tumor and observed the greatest survival advantage in patients with pharyngeal cancer (Fig. 1C) (log-rank P <.06), whereas the patients with cancers of oral or laryngeal origin (Fig. 1B,D, respectively) had similar survival irrespective of their CDH1 methylation status. To determine whether this effect on survival was independent of the effects of HPV-16 positivity, which reportedly leads to improved patient survival,12,14 and whether it was independent of tumor stage, we used Cox proportional hazards modeling to control for these possible confounders. That model (Table 3) suggested that, in all patients with HNSCC, CDH1 hypermethylation led to a 50% reduced instantaneous risk (hazard ratio [HR], 0.5; 95% confidence interval [CI], 0.3–0.9) and that this was independent of HPV-16 seropositivity, tumor stage, age, and sex, all of which were controlled for in the model. In a stratified model of the patients with pharyngeal cancer, a lower but not statistically significant HR of 0.3 (95% CI, 0.1–1.2) was observed and, again, was controlled for HPV-16 seropositivity (which was significant: HR, 0.3; 95% CI, 0.1–0.8), age, sex, and tumor stage. This lack of statistical significance for CDH1 methylation in this stratified model was probably caused by the lack of statistical power in this analysis of a smaller subset of patients. These results suggest that CDH1 hypermethylation is an independent prognostic marker in HNSCC and may hold the greatest promise in pharyngeal tumors, although additional larger studies will be needed to confirm these findings.
FIGURE 1.
These Kaplan-Meier survival probability plots compare cadherin 1, type 1/E-cadherin (CDH1) hypermethylation-positive tumors and hypermethylation-negative tumors in all patients with head and neck squamous cell carcinoma (n = 340) (A), only in patients with oral cancers (n = 194) (B), only in patients with pharyngeal cancers (n = 86) (C), and only in patients with laryngeal cancers (n = 58) (D). Tick marks represent censored values, and P values shown on graphs are the results of log-rank tests.
TABLE 3.
Proportional Hazards Model of Survival in Patients With Head and Neck Squamous Cell Carcinoma*
| Covariate | HR (95% CI) |
|---|---|
| CDH1 hypermethylation | |
| Negative | Referent |
| Positive | 0.5 (0.3–0.9) |
| HPV-16 seropositivity | |
| Negative | Referent |
| Positive | 0.4 (0.2–0.8) |
HR indicates hazard ratio; CI, confidence interval; CDH1, the cadherin 1, type 1/E-cadherin; HPV-16, human papillomavirus type 16.
Note: The model included all variables in the table and was controlled for age, sex, and tumor stage.
ECAD is involved in several cellular pathways: It plays a variety of roles related to cell-cell interactions, and it has a critical role in the EMT.17 It is believed that the EMT is a process critical to tumor invasion and metastasis; thus, loss of ECAD protein often is linked to a more aggressive phenotype.16,31 At the same time, hypermethylation of CDH1, which encodes the ECAD protein, has not been linked to this phenotype and, in fact, as demonstrated in our work, may play a role in early and less aggressive disease. 18,19,21 These findings suggest that the loss of ECAD protein observed in a large number of later stage or metastatic HNSCC may be tied to a different mechanism of inactivation, possibly through cellular inhibitors, such as SNAIL,32 or through posttranslational modification of the ECAD protein.33 Thus, an early loss of ECAD through hypermethylation of CDH1 may lead to a tumor phenotype that is more susceptible to treatment or is less invasive or metastatic, thereby providing the survival advantage observed in the current study. Conversely, retention of ECAD and loss later through other mechanisms may provide for a more aggressive tumor phenotype.
The current results, suggesting that CDH1 hypermethylation is a significant prognostic marker of better survival in HNSCC, may hold clinical importance. This study is one of the largest to date examining CDH1 and is unique in its population-based nature. Thus, the results from this study may be easier to generalize to other populations compared with the results from studies that used convenience series designs. This study’s epidemiologic nature and use of questionnaire-based exposure assessment also allowed us to examine the etiologic contributors to these molecular alterations, which is often not possible in hospital-based convenience series, in which the measures of exposure often are derived from chart review and, thus, often are incomplete or misclassified. The size of this study also allowed for a more complete examination of the correlation between epigenetic alteration of CDH1 and survival, which has been difficult in previous studies.21 Further studies should characterize this alteration and should determine more specifically whether patients with this alteration have differential responses to the treatment regimens commonly used in HNSCC. Larger prospective studies also are needed to define better the clinical utility of this biomarker.
Acknowledgments
Supported by National Institutes of Health grants R01CA078609 and R01CA100679, by the Friends of Dana-Farber, and by the Flight Attendants Medical Research Institute (C.J.M.).
We thank the collaborating clinicians and research staff involved in this study.
References
- 1.IARC. Cancer Incidence, Mortality and Prevalence Worldwide (2002 Estimates) Lyon, France: IARC; 2004. GLOBOCAN 2002. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Blot WJ, McLaughlin JK, Winn DM, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 4.Rothman K, Keller A. The effect of joint exposure to alcohol and tobacco on risk of cancer of the mouth and pharynx. J Chronic Dis. 1972;25:711–716. doi: 10.1016/0021-9681(72)90006-9. [DOI] [PubMed] [Google Scholar]
- 5.Elwood JM, Pearson JC, Skippen DH, Jackson SM. Alcohol, smoking, social and occupational factors in the aetiology of cancer of the oral cavity, pharynx and larynx. Int J Cancer. 1984;34:603–612. doi: 10.1002/ijc.2910340504. [DOI] [PubMed] [Google Scholar]
- 6.Olsen J, Sabreo S, Fasting U. Interaction of alcohol and tobacco as risk factors in cancer of the laryngeal region. J Epidemiol Commun Health. 1985;39:165–168. doi: 10.1136/jech.39.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen J, Sabroe S, Ipsen J. Effect of combined alcohol and tobacco exposure on risk of cancer of the hypopharynx. J Epidemiol Commun Health. 1985;39:304–307. doi: 10.1136/jech.39.4.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franco EL, Kowalski LP, Oliveira BV, et al. Risk factors for oral cancer in Brazil: a case-control study. Int J Cancer. 1989;43:992–1000. doi: 10.1002/ijc.2910430607. [DOI] [PubMed] [Google Scholar]
- 9.Zeka A, Gore R, Kriebel D. Effects of alcohol and tobacco on aerodigestive cancer risks: a meta-regression analysis. Cancer Causes Control. 2003;14:897–906. doi: 10.1023/b:caco.0000003854.34221.a8. [DOI] [PubMed] [Google Scholar]
- 10.Loning T, Ikenberg H, Becker J, Gissmann L, Hoepfer I, zur Hausen H. Analysis of oral papillomas, leukoplakias, and invasive carcinomas for human papillomavirus type related DNA. J Invest Dermatol. 1985;84:417–420. doi: 10.1111/1523-1747.ep12265517. [DOI] [PubMed] [Google Scholar]
- 11.Hammarstedt L, Dahlstrand H, Lindquist D, et al. The incidence of tonsillar cancer in Sweden is increasing. Acta Otolaryngol. 2007;127:988–992. doi: 10.1080/00016480601110170. [DOI] [PubMed] [Google Scholar]
- 12.Gillison ML, Shah KV. Human papillomavirus-associated head and neck squamous cell carcinoma: mounting evidence for an etiologic role for human papillomavirus in a subset of head and neck cancers. Curr Opin Oncol. 2001;13:183–188. doi: 10.1097/00001622-200105000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Ringstrom E, Peters E, Hasegawa M, Posner M, Liu M, Kelsey KT. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clin Cancer Res. 2002;8:3187–3192. [PubMed] [Google Scholar]
- 14.Furniss CS, McClean MD, Smith JF, et al. Human papillomavirus 16 and head and neck squamous cell carcinoma. Int J Cancer. 2007;120:2386–2392. doi: 10.1002/ijc.22633. [DOI] [PubMed] [Google Scholar]
- 15.Pyo SW, Hashimoto M, Kim YS, et al. Expression of E-cadherin, P-cadherin and N-cadherin in oral squamous cell carcinoma: correlation with the clinicopathologic features and patient outcome. J Craniomaxillofac Surg. 2007;35:1–9. doi: 10.1016/j.jcms.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Diniz-Freitas M, Garcia-Caballero T, Antunez-Lopez J, Gandara-Rey JM, Garcia-Garcia A. Reduced E-cadherin expression is an indicator of unfavourable prognosis in oral squamous cell carcinoma. Oral Oncol. 2006;42:190–200. doi: 10.1016/j.oraloncology.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 18.Yeh KT, Shih MC, Lin TH, et al. The correlation between CpG methylation on promoter and protein expression of E-cadherin in oral squamous cell carcinoma. Anticancer Res. 2002;22(6C):3971–3975. [PubMed] [Google Scholar]
- 19.de Moraes RV, Oliveira DT, Landman G, et al. E-cadherin abnormalities resulting from CPG methylation promoter in metastatic and nonmetastatic oral cancer. Head Neck. 2007;30:85–92. doi: 10.1002/hed.20666. [DOI] [PubMed] [Google Scholar]
- 20.Mese H, Sasaki A, Nakayama S, et al. Analysis of cellular sensitization with cisplatin-induced apoptosis by glucose-starved stress in cisplatin-sensitive and -resistant A431 cell line. Anticancer Res. 2001;21(2A):1029–1033. [PubMed] [Google Scholar]
- 21.Dikshit RP, Gillio-Tos A, Brennan P, et al. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110:1745–1751. doi: 10.1002/cncr.22975. [DOI] [PubMed] [Google Scholar]
- 22.Ting Hsiung D, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 23.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119:1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 24.Marsit CJ, Karagas MR, Andrew A, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in nonsmall cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 27.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32–E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–4236. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 29.Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer. 2003;105:41–46. doi: 10.1002/ijc.11028. [DOI] [PubMed] [Google Scholar]
- 30.Applebaum KM, Furniss CS, Zeka A, et al. Lack of association of alcohol and tobacco with HPV16-associated head and neck cancer. J Natl Cancer Inst. 2007;99:1801–1810. doi: 10.1093/jnci/djm233. [DOI] [PubMed] [Google Scholar]
- 31.Chang HW, Chow V, Lam KY, Wei WI, Yuen A. Loss of E-cadherin expression resulting from promoter hypermethylation in oral tongue carcinoma and its prognostic significance. Cancer. 2002;94:386–392. doi: 10.1002/cncr.10211. [DOI] [PubMed] [Google Scholar]
- 32.Becker KF, Rosivatz E, Blechschmidt K, Kremmer E, Sarbia M, Hofler H. Analysis of the E-cadherin repressor Snail in primary human cancers. Cells Tissues Organs. 2007;185(1–3):204–212. doi: 10.1159/000101321. [DOI] [PubMed] [Google Scholar]
- 33.Masterson J, O’Dea S. Posttranslational truncation of E-cadherin and significance for tumour progression. Cells Tissues Organs. 2007;185(1–3):175–179. doi: 10.1159/000101318. [DOI] [PubMed] [Google Scholar]