Summary
We have developed a robust in vivo small molecule screen that modulates heart size and cardiomyocyte generation in zebrafish. Three structurally-related compounds (Cardionogen-1 to -3) identified from our screen enlarge the size of the developing heart via myocardial hyperplasia. Increased cardiomyocyte number in Cardionogen-treated embryos is due to expansion of cardiac progenitor cells. In zebrafish embryos and murine embryonic stem (ES) cells, Cardionogen treatment promotes cardiogenesis during and after gastrulation, whereas inhibits heart formation before gastrulation. Cardionogen-induced effects can be antagonized by increasing Wnt/β-catenin signaling activity. We demonstrate that Cardionogen inhibits Wnt/β-catenin-dependent transcription in murine ES cells and zebrafish embryos. Cardionogen can rescue Wnt8-induced cardiomyocyte deficiency and heart-specific phenotypes during development. These findings demonstrate that in vivo small molecule screens targeted on heart size can discover compounds with cardiomyogenic effects and identify underlying target pathways.
Introduction
Dysregulation of heart development and growth is a hallmark of most cardiovascular diseases (Olson, 2004). Screening for small molecules that regulate cardiomyocyte generation will further our understanding of cardiac developmental mechanisms and aid in discovering novel therapeutics for heart diseases. Zebrafish has emerged as a model organism used in multiple steps of the drug discovery process through their use in in vivo phenotypic screens (Lehar et al., 2008; Murphey and Zon, 2006). Whole embryo screens offer considerable advantages in drug discovery by evaluating target cell populations and organs as well as other pleiotropic effects. Established heart-specific zebrafish transgenic lines permit visualization of green fluorescent proteins in developing hearts. These features make zebrafish particularly well-suited for discovering small molecule regulators of cardiac development and growth.
Despite advances in modern medicine, management of myocardial infarction and heart failure remains a major challenge. Developing therapies that can stimulate cardiomyocyte regeneration in areas of infarction would have an enormous medical and economic impact. Both embryonic and adult stem cells have received considerable attention as donor cells for therapeutic applications. Use of pluripotent embryonic stem (ES) cells is largely limited by ethical issues and concerns of their tumorigenic potential (Behfar et al., 2002), while recent trials featuring adult donor stem cells have demonstrated only modest clinical benefits (Lunde et al., 2006; Schachinger et al., 2006). These findings demonstrate a limited capacity of donor stem cells to differentiate into cardiomyocytes and highlight the need to develop small molecules that induce differentiation of exogenous and endogenous stem cells towards cardiac cell lineages.
Zebrafish cardiac development begins during early stages of embryogenesis. Generation of the required number of cardiomyocytes involves both production of cardiac progenitor cells and proliferation of embryonic cardiomyocytes (Stainier 2001). The size of the embryonic heart primarily reflects cardiac cell number and cell size (Jia et al., 2007). Several signaling pathways, including bone morphogenic protein (BMP), Wnt, fibroblast growth factor (FGF), Notch and retinoic acid, are implicated in the initial selection of myocardial progenitors from a multipotential stem cell population (Keegan et al., 2005; Marques et al., 2008; Reiter et al., 2001; Rones et al., 2000). Among them, the Wnt signaling pathway has received considerable attention for its roles in development, stem cell formation, regeneration and cancer progression(Logan and Nusse, 2004; Moon et al., 2004). Canonical Wnt signaling is mediated by binding of secreted (Wnt) proteins to specific Frizzled receptor complexes and results in inactivation of GSK-3β leading to dephosphorylation and stabilization of cytoplasmic β-catenin. β-catenin then translocates into the nucleus and activates T cell factor (Tcf)/Lymphoid-enhancing factor (Lef)-mediated transcription (Logan and Nusse, 2004; Moon et al., 2004). During zebrafish heart development, Wnt/β-catenin signaling regulates heart development in a temporally biphasic fashion. It induces cardiac specification before gastrulation but inhibits heart formation during and after gastrulation (Ueno et al., 2007). Although core components of the Wnt signaling pathway are clearly defined and highly conserved, tissue-specific modifiers of the pathway remain a mystery (Logan and Nusse, 2004; Moon et al., 2004).
In this study, we screened a small molecule library for compounds using an in vivo cardiac development assay. A novel small molecule family containing three structurally-related compounds (Cardionogen-1, 2, 3) was identified based on their ability to selectively enlarge the size of the embryonic heart. We show that Cardionogen is a biphasic modulator of cardiogenesis, either promoting or inhibiting heart formation depending on the stage of treatment. Cardionogen treatment also promotes murine ES cells to differentiate into beating cardiomyocytes, demonstrating that the bioactivity of this small molecule family is functionally conserved in mammalian cells. We indicate that Cardionogen inhibits Wnt/β-catenin-dependent transcriptional activity in murine ES cells (EC50 of ~23 nM) and zebrafish embryos. Furthermore, Cardionogen can rescue cardiac cell and chamber deficiency induced by Wnt8 after gastrulation and reverse cardiac cell expansion caused by Wnt8 overexpression before gastrulation. These findings indicate that Cardionogen interferes with Wnt signaling during cardiac development and growth.
Results
In vivo chemical screens for small molecule modulators of heart development
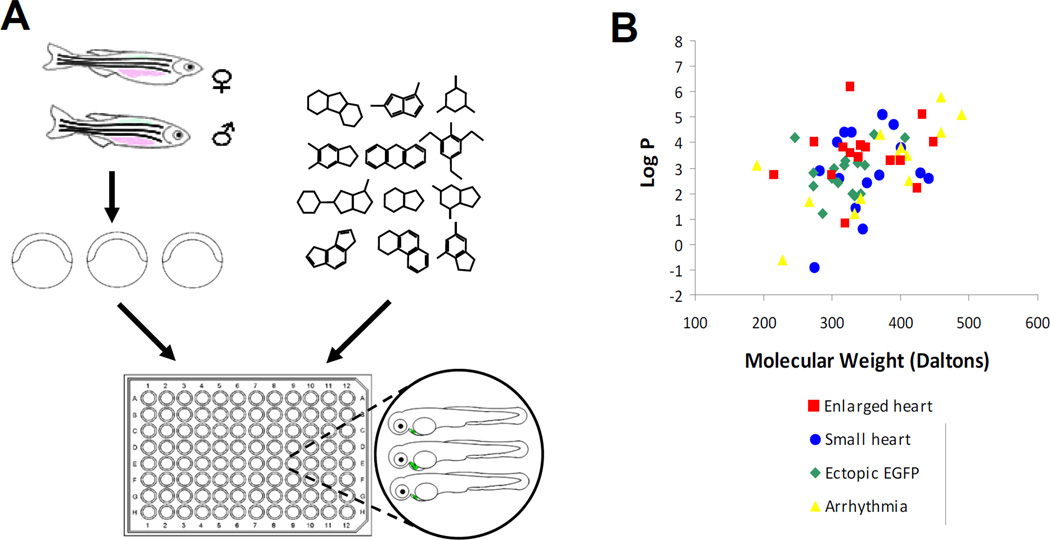
The size of the zebrafish embryonic heart primarily reflects the number and size of cardiomyocytes during development (Jia et al., 2007). We hypothesized that screening for small molecules that increase heart size in zebrafish embryos may identify compounds that induce cardiomyocyte differentiation and proliferation without causing pleiotropic effects. To test this hypothesis, we conducted an in vivo cardiac development screen using transgenic zebrafish embryos [Tg(cmlc2:EGFP)] in which expression of enhanced green fluorescent protein (EGFP) is under the control of the cardiac myosin light chain 2 (cmlc2) promoter (Burns et al., 2005). The primary focus of our screen was heart size, which was assessed by visual inspection using fluorescent microscopy. We adopted and modified a small molecule screening procedure using zebrafish embryos (Peterson et al., 2004; Stern et al., 2005). Briefly, aliquots of test compounds were delivered into individual wells of 96-well tissue culture plates (Fig. 1A). cmlc2-EGFP transgenic embryos were harvested from crosses of transgenic fish and added to test wells at 5 hour post fertilization (hpf), the onset of gastrulation when cardiac progenitor cells begin to form (Stainier 2001). We examined embryonic heart size, cardiac morphology and contractility of treated embryos at 24, 48 and 72 hour post fertilization (hpf). Additionally, overall morphologies of embryos (e.g., dorsal-ventral and anterior-posterior axis) and other organs, including the brain, eyes, notochord and somites, were carefully examined to determine whether development of these organs was affected, providing a preliminary assessment of compound selectivity and toxicity.
Fig. 1. In vivo small molecule screen identified bioactive compounds that affect heart development.
(A) Schematic diagram displaying the screening process for compounds that affect heart development. Three Tg(cmlc2-EGFP) embryos were transferred to each well in E3 buffer at the concentration of test compounds to 10 µM. (B) Log P values (partition coefficients between octanol and water) of all active compounds identified in the screen plotted against their respective molecular weights. The bioactive compounds have molecular weights ranging from 200 to 500 Daltons and log P values ranging from −1 to +7. Positive log P value=hydrophobic. Negative log P value=hydrophilic. Bioactive compounds were subclassified by cardiac phenotypes observed with treatment (10 µM).
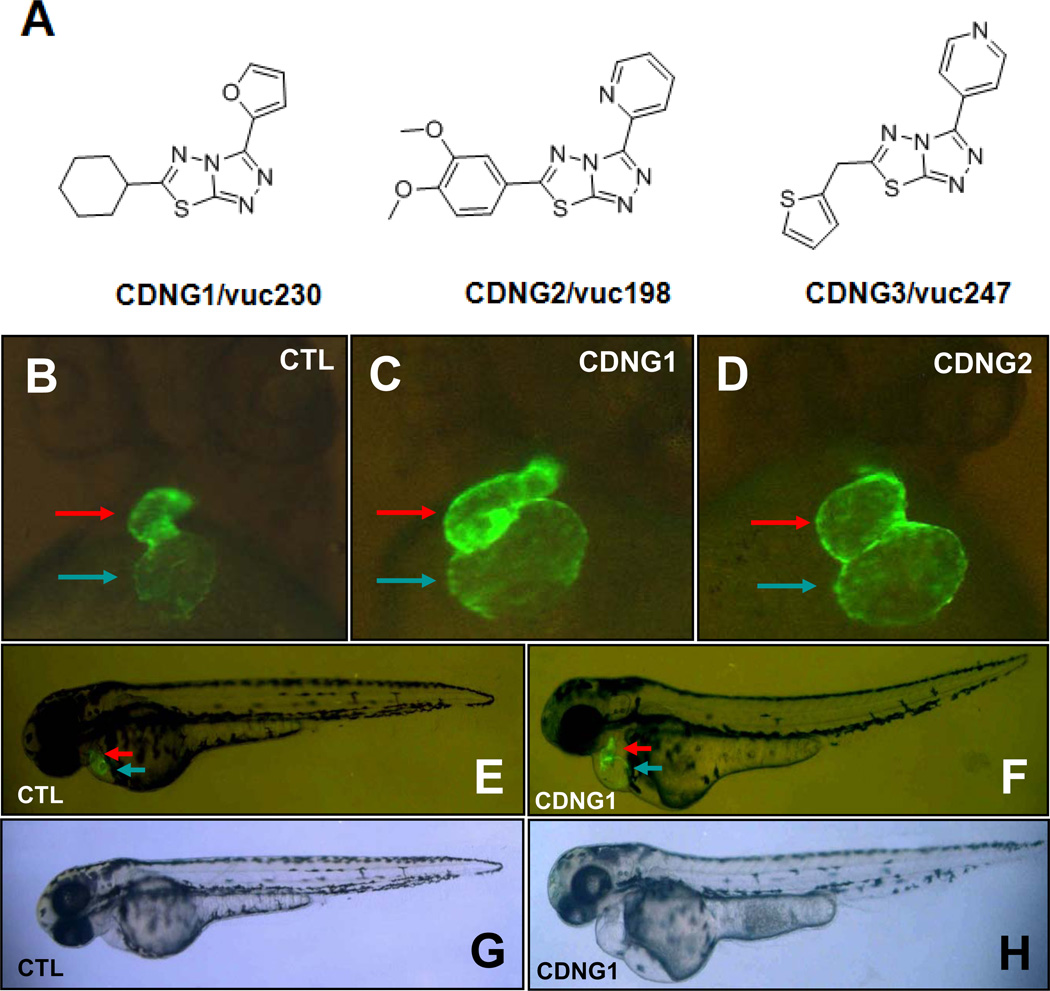
A subset of the small molecule library (4,000 compounds with diverse structures) provided by the Vanderbilt Institute of Chemical Biology was screened. Sixty-one compounds were identified as having significant effects on heart size, morphology and contractility. Of these compounds, 15 caused an enlarged heart phenotype, 17 induced ectopic EGFP expression, 13 caused arrhythmias, and 16 delayed development at the heart tube stage. Notably, almost all bioactive compounds were hydrophobic as indicated by positive log P values (the partition coefficient between octanol and water), suggesting that hydrophobic compounds effectively penetrate zebrafish embryos (Fig. 1B). This observation is consistent with the results of previous small molecule screens in zebrafish (Sachidanandan et al., 2008). vuc230, vuc198 and vuc247, named as Cardionogen (CDNG) -1, -2 and -3, were among the most potent inducers of a large heart phenotype identified in our screen. Notably, these small molecules are structurally-related and contain the same core motif, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazole (Fig. 2A). Treatment with Cardionogen-1/vuc230 [IUPAC: 6-cyclohexyl-3-furan-2-yl-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole] or Cardionogen-2/vuc198 [IUPAC: 6-(3,4-dimethoxyphenyl)-3-pyridin-2-yl-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole] significantly enlarged both the atrium and ventricle (Fig. 2B,C,D). Cardionogen-3/vuc247 [IUPAC: 3-pyridin-4-yl-6-(thiophen-2-ylmethyl)-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole] affected heart size to a lesser extent than Cardionogen-1 and -2 (data not shown). Cardionogen treatment did not cause apparent defects of overall embryonic morphology or other organ development (Fig. 2E–H), suggesting that this class of small molecules has selective activity on heart development and growth.
Fig. 2. Cardionogen increases heart size during zebrafish development.
(A) Chemical structures of Cardionogen family, including CDNG1/vuc230, CDNG2/vuc198 and CDNG3/vuc247. Fluorescent optics displaying (B) Untreated control heart (CTL), (C) CDNG1-treated heart, (D) CDNG2-treated heart, (E) untreated control embryo (CTL) and (F) CDNG1-treated embryo, in Tg(cmlc2:EGFP) embryos at 60 hpf. Light optics showing (G) control embryos (CTL), (H) CDNG1-treated embryos at 60 hpf. Ventral view (B–D). Lateral view (E–H). Red arrow: ventricle. Blue arrow: atrium. CDNG1 treatment (30 µM ; 5 to 60 hpf).
Cardionogen induces cardiac hyperplasia in a biphasic manner
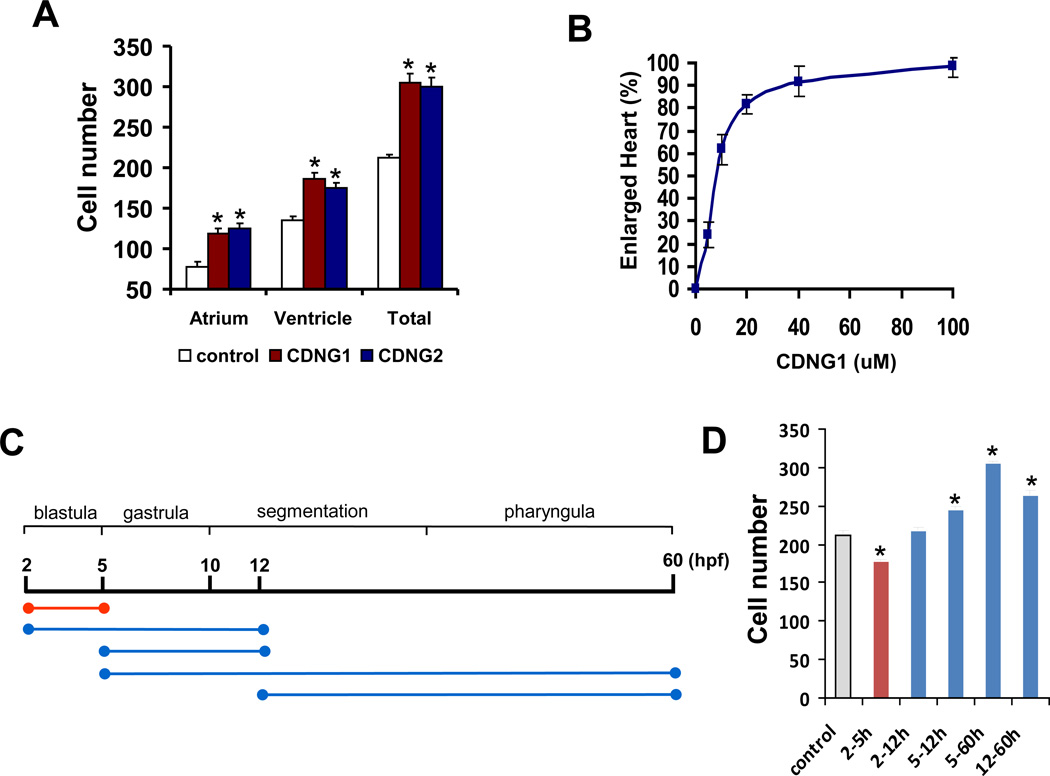
To assess whether heart enlargement is due to hyperplastic and/or hypertrophic growth, we evaluated cardiac cell number, cell size, and proliferative rates following treatment with both Cardionogen-1 and -2. We quantified total cardiomyocyte number in Tg(cmlc2:DsRed-nuc) embryos, in which a gene encoding red fluorescent protein fused to a nuclear localization signal (DsRed-nuc) is under the control of the cmlc2 promoter (Mably et al., 2003). In these transgenic embryos, individual cardiomyocyte nuclei are marked by red fluorescence, permitting quantitative assessment of cardiomyocyte number using confocal microscopy analysis (Fig. S1A). To this end, we compared Cardionogen-treated embryos versus controls in a series of flat-mount confocal sections. Cardionogen treatment caused an increase of cardiac cell numbers in both the atrium and ventricle (Fig. 3A). To test whether hypertrophy of cardiomyocytes also contributed to increases in heart size, we measured cell size in Tg(cmlc2:EGFP) embryos using confocal microscopy. No alterations in cell size were observed with Cardionogen treatment (Fig. S1B). Whole-mount immunohistochemistry assays (n=10 embryos) using antibodies recognizing the mitosis marker phosphohistone 3 (H3P) revealed that Cardionogen-1 did not increase cardiomyocyte proliferation in comparison to control embryos (data not shown). Dosage-response analyses revealed that Cardionogen exponentially reached its optimum activity at 40 µM and maintained its peak activity at higher concentration ranges (Fig. 3B). No differences in survivorship or general morphology were noted at any of the test concentration. Taken together, our results indicate that Cardionogen enlarges heart size via cardiac hyperplasia, which is not attributed to increases in the proliferative growth of cardiomyocytes.
Fig. 3. Cardionogen induces cardiac hyperplasia in a biphasic manner.
(A) Bar chart showing increased cardiac cell number in the atrium and ventricle in CDNG1- and CDNG2-treated embryos at 30 µM (5–60 hpf). (B) Dose-response curve for heart enlargement with CDNG1 treatment. Tg(cmlc2:EGFP) (n=30) embryos were treated at 5 hpf using CDNG1 compound at various concentrations (0–100 µM). Heart sizes were scored at 60 hpf. No differences in survivorship or general morphology were noted at any of the test concentration. (C) Schematic representation showing pulse treatment intervals used to define Cardionogen bioactivity period in cardiomyogenesis. (D) Graph showing cardiomyocyte number in embryos treated by CDNG1 pulse during different stages. Three to five embryos were subjected to confocal imaging under each condition with CDNG1 treatment (30 µM), and cardiomyocyte numbers were counted using confocal microscopy analysis. All cell counts were conducted at 60hpf. Error bars indicate standard deviation in three independent measurements (A, B; D). Asterisks indicate statistical significance between treated and control embryos (*p<0.01) (A; D). (see also Figure S1)
The results presented above suggest that Cardionogen may affect earlier stages of cardiac cell differentiation. To assess the temporal activity of Cardionogen, we quantified cardiomyocyte number in transgenic embryos [Tg(cmlc2:DsRed-nuc) ] following pulse treatments of Cardionogen-1. In the first set of pulse experiments, Cardionogen-1 was added at 2 hpf (the beginning of blastula) and washed away at progressively later stages of development, including 5 hpf (the onset of gastrula) and 12 hpf (the 5-somite stage) when nkx2.5 starts to express (Fig. 3C). Cardiac cell counts were conducted at 60 hpf. Cardionogen treatment had an inhibitory effect on cardiomyocyte generation with treatment from 2 hpf to 5 hpf (178 ± 3) in comparison with controls (212 ± 5) (Fig. 3D). Extension of Cardionogen treatment to 12 hpf recovered the loss of cells (219 ± 4) (Fig. 3C, D). In a second set of pulse experiments, Cardionogen-1 was added at 5 hpf and washed away at 12 hpf or 60 hpf (Fig. 3C). We quantified cell number at 60 hpf and observed increased cardiac cell numbers with treatment from 5 hpf to 12 hpf (245 ± 5) and from 5 hpf to 60 hpf (305 ± 4), compared to controls (212 ± 5)(Fig. 3D). When treating embryos from 12 hpf to 60 hpf, cardiomyocyte number was reduced from the peak but still had a marked increase compared to controls (265 ± 5 versus 212 ± 5) (Fig. 3C, D). Together, these results suggest that Cardionogen has a temporally biphasic pattern of cardiomyogenic activity during development. Specifically, Cardionogen treatment promotes cardiogenesis during and after gastrulation, whereas treatment prior to gastrulation inhibits cardiomyocyte formation.
Cardionogen causes expansion of cardiac progenitor cells
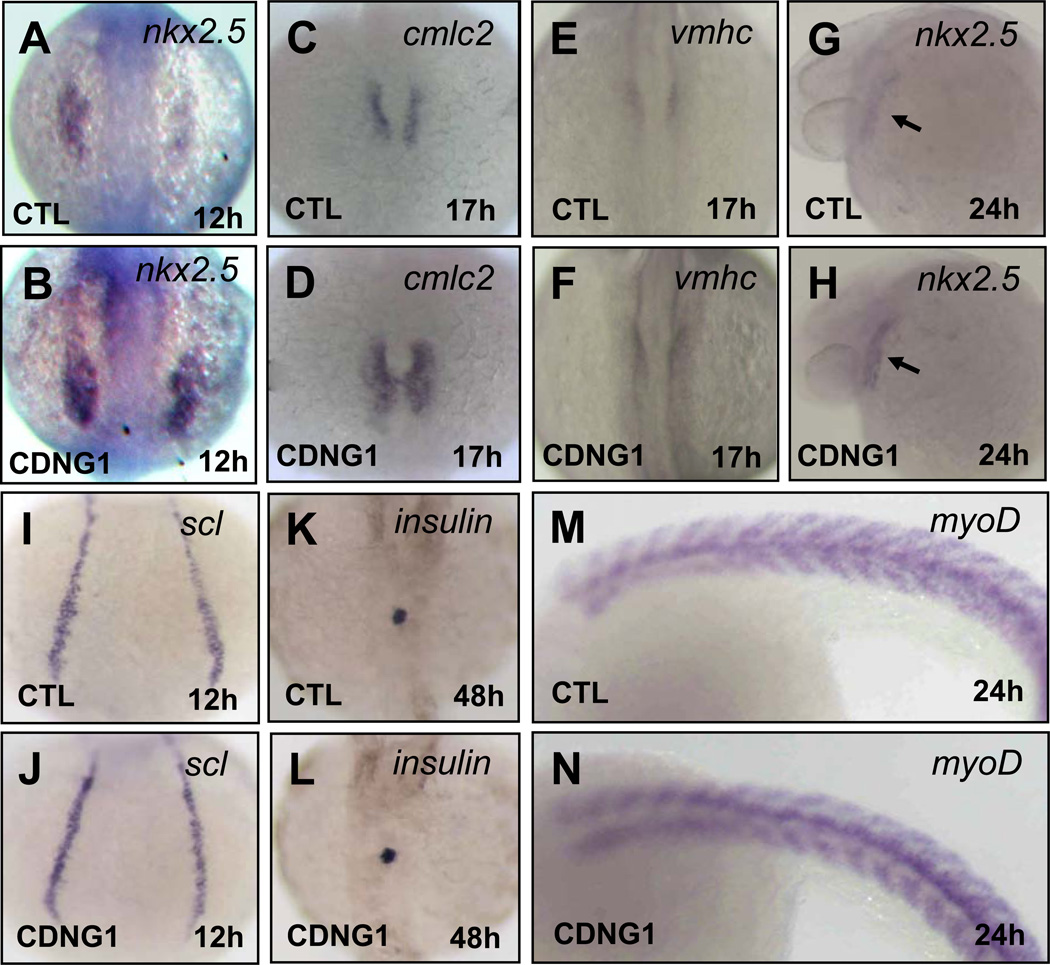
To understand how Cardionogen treatment regulates cardiogenesis, we performed in situ hybridization analyses with the cardiac progenitor marker nkx2.5 and cardiac differentiation markers cmlc2 and ventricular myosin heavy chain (vmhc)(Yelon et al., 1999). In Cardionogen-1 treated embryos, marked increases in nkx2.5 expression were observed in the anterior lateral plate mesoderm, the definitive heart field (the heart forming region) (Schoenebeck et al., 2007) at 12 hpf, as well as the heart tube stage at 24 hpf (Fig. 4A,B; G,H). Cardiomyocyte populations of Cardionogen-treated embryos were also expanded at the onset of myocardial differentiation, as evidenced by increased expression of cmlc2 and vmhc at 17 hpf (Fig 4C,D,E,F). In contrast, expression of scl, a hematopoietic progenitor marker in the head and the lateral mesoderm, was not affected in treated embryos compared to controls (Fig. 4I,J). Similarly, expression of insulin, a pancreas marker, and myoD, a skeletal myoblast marker, did not appear to be affected in Cardionogen-treated embryos (Fig. 4K,L; M,N). These observations indicate that Cardionogen promotes cardiac hyperplasia via expanding the cardiac progenitor cell population.
Fig. 4. Cardionogen promotes expansion of the cardiac progenitor cell population.
Expression of nkx2.5 (A, B), cmlc2 (C, D), vmhc (E, F), nkx2.5 (G, H), scl (I, J), insulin (K, L) and myoD (M, N) in CDNG1-treated and control embryos were examined using in situ hybridization. Expression of nkx2.5 (39 out of 43 embryos in A, B), cmlc2 (42 out of 47 embryos), vmhc (41 out of 45 embryos) and nkx2.5 (38 out of 41 embryos in G, H) are increased. Expression of scl (41 out of 43 embryos), insulin (42 out of 45 embryos) and myoD (39 out of 42 embryos) are not altered. 12 hpf (A,B; I,J); 17 hpf (C–F); 24 hpf (G,H;M,N); 48 hpf (K,L). Dorsal view (A–F;I–L); Lateral view (G,H;M,N). CTL: control. Black arrow: heart tube. CDNG1: 30 µM.
Cardionogen promotes mouse ES cells to differentiate into beating cardiomyocytes
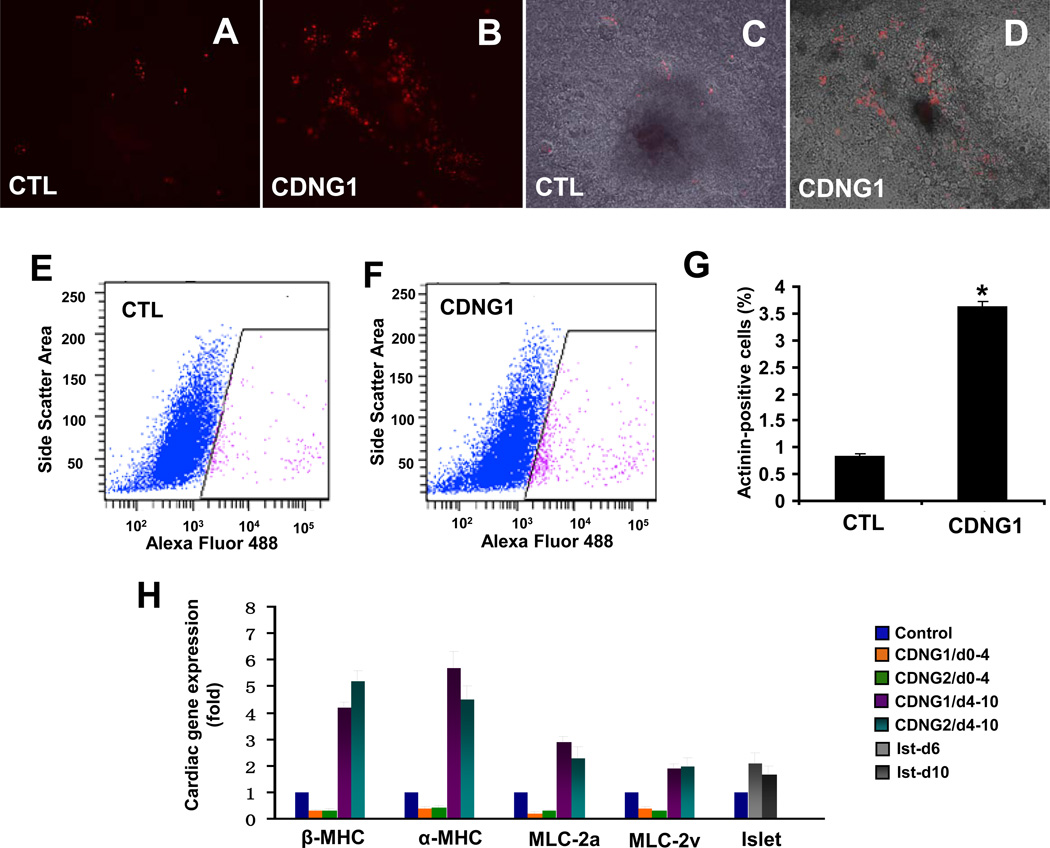
To examine Cardionogen activity in a mammalian model, we employed a murine ES cell line [Tg(αMHC:DsRed-Nuc)] that was stably transfected with a DsRed-Nuc construct driven by the promoter of α–myosin heavy chain (αMHC) (Hao et al., 2008; Subramaniam et al., 1991). In this transgenic line, differentiating cardiomyocytes are marked by prominent nuclear red fluorescence (Hao et al., 2008). Embryoid bodies (EBs) were initiated (marked as day 0) in cultured ES cells. Pulse treatment of Cardionogen-1 during the initial ES cell differentiation from day 0 to day 4 failed to induce cardiomyocyte differentiation when examined at day 12 (data not shown). Cardionogen-1 treatment from day 4 to day 10 significantly promoted ES cell differentiation into cardiomyocytes that express DsRed (Fig. 5A–D). We have found that Cardionogen-1 at 1 µM and 5 µM but not at 0.1 µM induces ES cell cardiac differentiation (Fig. 5A–D; data not shown). This treatment period coincides with the late differentiation of three germ layers in the EBs. Furthermore, cardiomyocytes generated from treated EBs formed foci that spontaneously and rhythmically contracted (Movie S1; Movie S2). Finally, we determined the fraction of ES cell culture expressing aMHC using flow cytometry. Cardionogen treatment from day 4 to day 10 increased the cardiac cell percentage by 4.36 fold (Fig. 5E, F, G). The overall cardiac differentiation percentages are consistent with previous studies using Wnt3a or GSK3β inhibitor BIO (Naito et al., 2006; Ueno et al., 2007).
Fig. 5. Cardionogen induces murine ES cells to differentiate into cardiomyocytes.
(A, B) Fluorescent optics revealing myocardial differentiation (red areas) in 0.1% DMSO-treated (CTL) and 1 µM CDNG1-treated CGR8-ES cells [Tg(αMHC:DsRed-nuc)]. (C, D) Bright-field pictures merged with fluorescent images of control (A) and CDNG1-treated cells (B). (E, F) Flow-cytometry analyses revealing the fraction of ES cells expressing αMHC:DsRed-nuc in 0.1% DMSO-treated and CDNG1-treated ES cells. X-axis: intensity of Alexa Flour488 immunostaining. Y-axis: side scatters area. (G) Analysis of sarcomeric α–Actinin by flow cytometry indicates that CDNG1-treatment enhances cardiomyocyte content by 4.36 fold (from 0.83 ± 0.06% to 3.62 ± 0.09%). GCR8-ES cells were treated with 0.1% DMSO and 1 µM CDNG1 from day 4 to day 10 and analyzed at day 12. (H) Bar chart depicting relative expression folds of βMHC, αMHC, MLC-2a, MLC-2v, islet in 1 µM CDNG1- and CDNG2-treated ES cells, compared to 0.1% DMSO-treated controls. βMHC, αMHC, MLC-2a, MLC-2v were examined at day 12; islet was examined at day 6 and day10. GAPDH was used as internal controls for normalization. CTL values were arbitrarily set to 1. Graphs (G, H) show mean ± s. d., performed in triplicate; *p<0.01 compared with control. (see also Movie S1 and S2)
To further assess the induction of cardiac cell differentiation, we examined expression of cardiac differentiation markers by real-time PCR. Treating ES cells from day 4 to day 10 with Cardionogen-1 and -2 significantly increased expression of cardiac sarcomere genes, including βMHC, αMHC, myosin light chain-2a (MLC-2a) and myosin light chain-2v (MLC2v) (Fig. 5H); while pulse treatment of ES cells with Cardionogen from day 0 to day 4 down-regulated expressions of these four cardiac markers in RNA samples prepared at day 12 (Fig. 5H). Furthermore, Cardionogen treatment of ES cells starting at day 4 induced the expression of cardiac progenitor cell marker islet (Fig. 5H). Hence, Cardionogen treatment after the appearance of cells representing the different germ layers promoted cardiomyocyte differentiation, whereas treatment before this stage led to a decrease in cardiogenesis. These studies reveal a biphasic pattern of cardiomyogenic activity of Cardionogen during murine ES cell differentiation, which closely mimics results obtained from our studies in zebrafish.
Cardionogen inhibits Wnt/β-catenin-mediated transcription
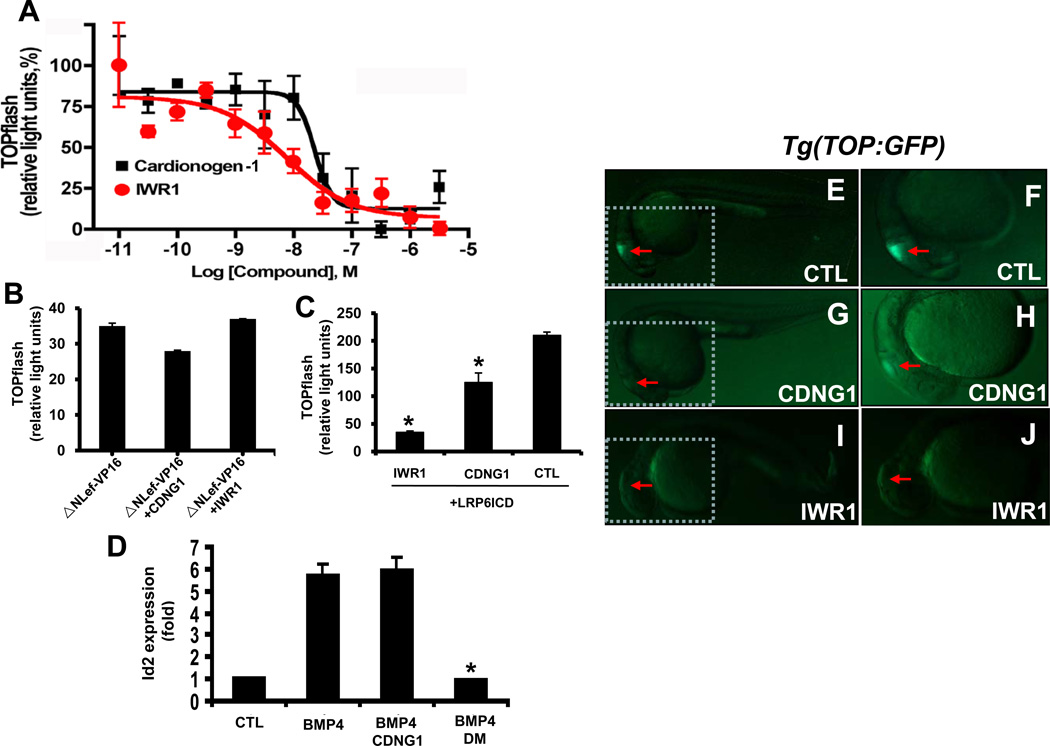
Cardionogen promotes cardiomyocyte generation following gastrulation and inhibits heart formation prior to gastrulation. Inversely, Wnt/β-catenin signaling during and after gastrulation inhibits cardiogenesis, whereas signaling before gastrulation induces heart formation (Naito et al., 2006; Ueno et al., 2007). These apparently opposing activities of Cardionogen and Wnt/β-catenin signaling led us to hypothesize that Cardionogen modulates heart development by antagonizing Wnt//β-catenin activity. The canonical Wnt/β-catenin pathway is a highly conserved signaling pathway, whereby activation of Wnt signaling causes β-catenin to translocate into the nucleus and bind to Tcf and Lef transcription factors, resulting in activation of downstream gene expression (Logan and Nusse, 2004; Moon et al., 2004). To determine whether Cardionogen opposes Wnt/β-catenin activity, we performed β-catenin/Tcf-mediated transcription assays. We employed a murine CGR8 embryonic stem cell line that was stably transfected with a TOPflash construct containing six copies of the Tcf/Lef binding site upstream of the thymidine kinase minimal promoter and luciferase cDNA (Ishitani et al., 1999). In this assay, Wnt3a was used to activate TOPflash luciferase activity. Cardionogen-1 inhibited Wnt3a/β-catenin-mediated luciferase activity in a dose-dependent manner. An effector Cardionogen concentration for half-maximal response (EC50) and maximal response is 23 nM and 100 nM, respectively (Fig. 6A). IWR1, a known Wnt inhibitor (Chen et al., 2009), reduced TOPflash activity, as a positive control (Fig. 6A). In contrast, Cardionogen-1 failed to inhibit TOPflash activity induced by ΔNLef-VP16 (Fig. 6B). ΔNLef-VP16 can activate TOPflash activity independently of catenin, in which ΔNLef lacks β-catenin binding site and fused with transactivation domain VP16 (Aoki, 1999). As a control, IWR1 also failed to reduce ΔNLef-VP1- induced TOPflash activity (Fig. 6B). Together, these findings suggest that Cardionogen inhibits β-catenin-dependent Wnt signaling. We next examined whether Cardionogen disrupted Wnt/β-catenin signaling within responding cells. LRP6ICD, a Wnt coreceptor, can constitutively activate Wnt signaling within cells (Tahinci E, 2007; Tamai, 2004). GCR8-ES cells were transfected with LRP6ICD. Cardionogen-1 treatment resulted in a reduction of LRP6ICD-induced TOPflash activity, while IWR1 causes loss of the same luciferase activity (Fig. 6C). Thus, Cardionogen blocks Wnt/β-catenin signaling within responding cells, rather than disrupting ligand-cell interaction or ligand production. We finally determined whether Cardionogen treatment altered other signaling activities. Cardionogen-1 did not reduce BMP4-induced Id2 expression, while Dorsomorphin (DM), a known BMP4 inhibitor, reduced the Id2 expression (Fig. 6D). Furthermore, Cardionogen-1 failed to alter Notch- or SRF/MAPK-induced transcription in ES cells (Fig. S2A, B). These findings demonstrate the Cardionogen specificity for Wnt signaling.
Fig. 6. Cardionogen inhibits Wnt3a/β-catenin-mediated transcription.
(A) Cardionogen-1 inhibits Wnt3a-induced TOPflash activity in ES cells. CGR8-ES cells were treated with Wnt3a (100 ng/ml)-conditioned media plus CDNG1 or IWR1 compounds in a series concentrations. Dose-response curves represent TOPflash activities normalized to cell number (mean ± s.d.; performed in quadruplicate). The calculated EC50 for Cardionogen-1 and IWR1 are 23 nM and 7.5 nM, respectively. Graphs were made in Prism 4 (GraphPad Software, Inc.) with nonlinear regression fit to a sigmoidal dose-response curve. (B) Cardionogen does not inhibit Lef/Tcf transcription that is independent of β-catenin activity. CGR8-ES cells were transfected with ΔNLef-VP16 and treated with 1 µM Cardionogen-1 or 1 µM IWR1. Graph represents TOPflash activities (mean ± s.d.; performed in triplicate). (C) Cardionogen inhibits LRP6-mediated Wnt signaling. CGR8-ES cells were transfected with a constitutively active LRP6ICD and treated with 1 µM Cardionogen-1, 1 µM IWR1 or 0.1% DMSO (CTL). TOPflash activity is graphed (mean ± s.d; performed in triplicate; *p<0.01). (D) Bar chart showing relative expression fold of BMP4-induced Id2 expression (Hua et al., 2006; Nakahiro et al., 2010) in the presence of CDNG1, BMP4, BMP4+CDNG1, BMP4+DM, compared to 0.1% DMSO (CTL). Bmp4: 30 ng/ml; CDNG1:1 µM; DM: 1 µM. Id2 expression normalized to GAPDH is graphed (mean ± s.d; performed in triplicate; *p<0.01). CTL values were arbitrarily set to 1. (E) Fluorescent optics revealing GFP fluorescence in the midbrain in Tg(TOP:GFP) embryos. (F) GFP fluorescence in the enlargement of brain region (insert in E). (G) CDNG1 treatment (30 µM; 5–24 hpf) reduces GFP fluorescence in the midbrain of Tg(TOP:GFP) embryos. (H) Reduction of GFP fluorescence in the enlargement of brain region (insert in G). (I) IWR1 treatment (30 µM; 5–24 hpf) eliminates GFP fluorescence in the midbrain of Tg(TOP:GFP) embryos. (J) Loss of GFP fluorescence in the enlargement of the brain region (insert in I). Red arrow: GFP expression. 24 hpf (E–J). (see also Figure S2 and S3)
To test whether Cardionogen inihibits Wnt signaling in developing embryos, we examined effects of Cardionogen on GFP expression in Tg(TOP:GFP) embryos, in which GFP transgene expression is under the control of four consensus Lef binding sites and a minimal cFos promoter (Dorsky et al., 2002). In Tg(TOP:GFP) embryos, GFP fluorescence is only observed in the midbrain using fluorescent microscopy analysis (Fig. 6E, F; (Dorsky et al., 2002). We were unable to observe GFP fluorescence in other embryo regions, which is consistent with previous studies (Dorsky et al., 2002). Nevertheless, treating Tg(TOP:GFP) embryos with Cardionogen-1 reduced GFP fluorescence in the midbrain (Fig. 6G, H). As a control, IWR1 treatment caused loss of GFP fluorescence in the same region (Fig. 6I, J). These data indicate that Cardionogen reduces Wnt/β-catenin signaling activity in zebrafish embryos, and further suggest that IWR1 is more potent Wnt pathway antagonist than Cardionogen, which are consistent with EC50 of Cardionogen-1 and IWR1 using TOPflash reporter assays in murine ES cells (23 nM versus 7.5 nM). Notably, IWR1 treatment resulted in defects in the anterior-posterior body development and loss of tail structure posterior to the yolk extension (Fig. 6I; Fig. S3A, B). We next examined whether IWR1 affects heart development. At early stages, IWR1 treatment caused an increased expression of cardiac progenitor cell marker nkx2.5 (Fig. S3C, D) and cardiac differentiation marker cmlc2, compared to controls (Fig. S3E, F). However, at late stages, IWR1-treated embryos displayed defects in cardiac chamber formation. Wild-type embryos form clearly two cardiac chambers (atrium and ventricle) (Fig. S3G; I). In contrast, IWR1-treated embryos develop only one cardiac chamber (Fig. S3H; J). We determined the chamber identity using a ventricular marker vmhc and an atrial marker atrial myosin heavy chain (amhc). vmhc expression analyses indicated that the single chamber formed in IWR1-treated embryos possessed ventricular identity compared to controls (Fig. S3K, L). amhc expression analyses revealed atrial myocytes aligned bilaterally in IWR1-treated embryos, resulting in failure to form the atrium, compared to controls (Fig. S3M, N).
Cardionogen reverses Wnt-induced cardiac phenotypes
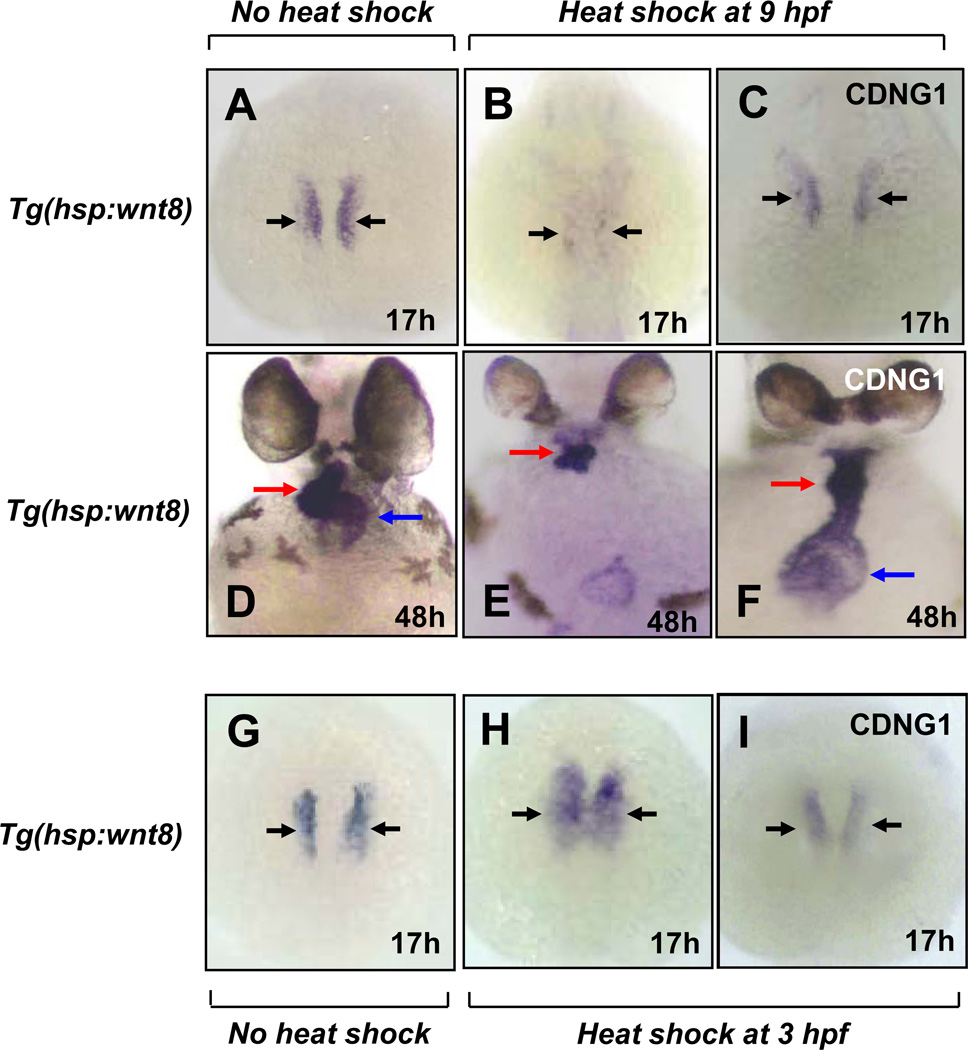
Since Cardionogen represses β-catenin/Tcf-mediated transcription in murine cells and zebrafish embryos, we evaluated whether Cardionogen can rescue Wnt-induced cardiac inhibitory phenotypes in zebrafish embryos. To test this possibility, we used transgenic zebrafish embryos [Tg(hsp:wnt8-EGFP)], in which wnt8 fused to EGFP is under the control of a heat shock promoter (Ueno et al., 2007). We heat shocked transgenic embryos to elevate wnt8 expression at the end stage of gastrulation (9 hpf), then treated these embryos with Cardionogen-1. While cardiomyocyte formation and cmlc2 expression are normally inhibited in Tg(hsp:wnt8-EGFP) embryos following heat shock (Fig. 7A, B), Cardionogen-1 treatment largely rescued cardiomyocyte deficiency labeled by cmlc2 expression (Fig. 7B, C). The same cardiac rescue effects were also observed by monitoring nkx2.5 expression at the 8-somite stages (data not shown). Remarkably, the rescue effects persisted through late stages of heart development. While wnt8-induced embryos failed to form the atrium and a large part of the ventricle at 48 hpf (Fig. 7D, E), Cardionogen-1 treatment completely restored the atrium and ventricle, but looping defects persisted (Fig. 7E, F). Notably, the small eye size induced by wnt8 overexpression was not rescued in embryos with Cardionogen treatment (Fig. 7D, E, F), suggesting that Cardionogen may block Wnt signaling in the cardiac mesoderm but not other tissues. In a second set of experiments, we examined whether Cardionogen could inhibit expansion of cardiac cell domains induced by wnt8 overexpression before gastrulation. Transgenic embryos [Tg(hsp:wnt8-EGFP)] were heat shocked to induce wnt8 expression at 3 hpf before gastrulation, followed by treatment with Cardionogen-1. While cmlc2 expression was expanded in the lateral plate mesoderm of heat shocked control embryos (Fig. 7G, H), Cardionogen-1 treatment reduced the expansion of the cmlc2 domain (Fig. 7H, I). Together, these findings demonstrate that Cardionogen reverses Wnt8-induced heart-specific phenotypes.
Fig. 7. Cardionogen rescues Wnt8-induced cardiac phenotypes.
(A–C) cmlc2 expression in non-heat-shocked embryos (A), heat-shocked embryos (B) and CDNG1-treated, heat-shocked embryos (C). 23 out of 25 embryos were rescued. (D–F) Rescue of heart formation labeled by cmlc2 expression in CDNG1-treated, heat-shocked embryos (F) compared to heat-shocked embryos (E) and non-heat-shocked embryos (D). 25 out of 26 embryos were rescued. (G–I) Inhibition of cardiac expansion labeled by cmlc2 expression in CDNG1-treated, heat- shocked embryos (I), compared to heat-shocked embryos (H) and non-heat-shocked embryos (G). 21 out of 24 embryos were inhibited. Heat shock: 38.5° C for 15 min at 3 hpf (H, I) and 9 hpf (B,C;E,F). Black arrow: cardiomyocytes. Red arrow: ventricle. Blue arrow: atrium.
Discussion
In this study, we describe an in vivo small molecule screen in zebrafish capable of identifying chemical modulators of cardiac development. We have identified a family of novel small molecules (named Cardionogen) that enlarges the size of the embryonic heart by promoting cardiomyocyte formation. Cardionogen either induces or inhibits the expansion of cardiac progenitor cells, depending on the timing and stages of treatment. Importantly, we have linked Cardionogen to the Wnt signaling pathway. Cardionogen inhibits Wnt/β-catenin signaling activity in murine ES cells (EC50 of ~23 nM) and zebrafish embryos. Cardionogen can rescue Wnt8-induced cardiomyocyte deficiency and heart-specific phenotypes during development. These findings have demonstrated that a complex but sensitive development screen targeting organ size can identify both active small molecules and their target pathways.
Embryonic heart size primarily reflects cardiac cell number and size. However, dysregulation of myocardial patterning and morphology may also cause alterations in overall heart size. As such, failure of concentric myocardial growth in heart of glass, santa and valentine mutants leads to an enlarged heart phenotype without changing cardiomyocyte number (Mably et al., 2006; Mably et al., 2003). Therefore, it is critical to determine whether bioactive small molecules enlarge heart size by modulating cardiomyocyte number, cell size, and/or myocardial patterning. Notably, only a few zebrafish genetic mutants have been identified to affect cardiac cell number, suggesting that certain limitations (e.g., functional redundancy) may prevent identification of relevant genes and pathways in cardiomyocyte generation through mutagenesis studies. Small molecule screens may overcome these obstacles by identifying compounds that interact with these important pathways. We observed that Cardionogen induces murine ES cells to differentiate into cardiomyocytes at 1 µM and 5 µM but not at 0.1 µM, indicating that a higher concentration is necessary to promote cardiogenic differentiation in EBs than the active concentration (0.1 µM) to completely block canonical Wnt signaling in undifferentiated ES cells. This might be due to the fact that these two assays are conducted at two distinct points of ES cell differentiation, each marked with stage-specific expression of various Wnt ligands (Schulz et al., 2009). From the TOPflash dose-response analyses, IWR1 is a more potent Wnt inhibitor than Cardionogen in murine ES cells. These findings are consistent with activities of Cardionogen and IWR1 in zebrafish Tg(TOP:GFP) embryos, in which GFP fluorescence in the midbrain is reduced in CDNG1-treated embryos but eliminated in IWR1-treated embryos. It is noted that Cardionogen dose-response curve is steep compared to IWR1 dose curve (Fig. 6A), suggesting cooperative effects of Cardionogen in inhibiting TOPflash activity. Multiple Cardionogen molecules may bind to one target protein to reduce TOPflash activity quickly, resulting in a rise in the slope of the dose-response curve. However, to demonstrate this effect will require detail molecular interaction studies of Cardionogen with its potential binding proteins in the Wnt pathway.
Wnt signaling is a key regulator of a variety of developmental processes, including primitive streak formation, mesoderm and endoderm induction and patterning, the anterior-posterior (AP) axis development, neural differentiation and heart formation (Kimelman, 2006; Logan and Nusse, 2004; Moon et al., 2004). Blocking Wnt signaling in zebrafish development after gastrulation with dickkopf homolog 1 (dkk1) induces cardiac progenitor cell formation but truncates the posterior axis at late stages (Ueno et al., 2007). Similarly, we observed that IWR1 treatment increased the expression of cardiac progenitor cell marker nkx2.5 but caused AP axis defects and disruption of the atrium formation. The failure to form the atrium might be due to overall embryonic defects (i.e., AP axis abnormality in combination with other mesoderm defects) in IWR1-treated embryos. Notably, Cardionogen treatment induces cardiac progenitor cell formation without causing tail truncation and atrium disruption. In addition, Cardionogen rescued the Wnt8-induced heart defects but not the Wnt8-induced small eye phenotypes. This demonstrates the benefits of a whole organism-based chemical screen. We believe that the phenotypic differences between zebrafish treated with IWR1 and Cardionogen are more likely due to their differences in mechanism of Wnt inhibition rather than “off target” effects, although this could be a possibility. Cardionogen might affect only a select population of Wnt-responding cells in embryos, considering that Cardionogen does not cause overall Wnt-dependent embryonic phenotypes but affecting Wnt signaling in the heart and the midbrain. The fact that IWR1 inhibits Wnt signaling in both HEK and ES cells, and can cause typically Wnt-dependent embryonic phenotypes suggest that IWR1 antagonizes Wnt signaling in broader cell types and tissues than Cardionogen. Importantly, we have not observed any phenotype independent of Wnt signaling defects in embryos treated with IWR1, suggesting the “off target” effect is unlikely or is minimal if it has. We wondered whether Cardionogen reduces Tcf/Lef-mediated luciferase activities in human embryonic kidney (HEK) cells. Notably, Cardionogen-1 does not inhibit Wnt3a-induced Topflash activity in HEK cells while IWR1 does (Fig. S2C). Thus, we propose that Cardionogen selectively reduces Wnt/β-catenin signaling activity in certain cell-types and tissues (i.e., heart and others) during development. We speculate that Cardionogen may inhibit tissue (heart)-specific modifiers that activate Wnt signaling. Alternatively, Cardionogen might interfere with interactions between Wnt pathway components and tissue (heart)-specific transcriptional factors.
The core components in the Wnt signaling pathway have been clearly defined and studied. However, tissue-specific modifiers of the pathway remain unknown (Logan and Nusse, 2004; Moon et al., 2004). Identifying in vivo binding factors of Cardionogen may provide novel insights into the mechanisms related to tissue-specific modifiers of Wnt signaling pathway. Recent studies have established that inhibition of the canonical Wnt pathway after the germ layer cell formation is necessary to promote cardiomyocyte differentiation from human cardiovascular stem cells (Yang et al., 2008). Our findings further support these human ES cell studies, as treatment of murine ES cells with Cardionogen after the germ layer cell formation promotes cardiac differentiation. Thus, evaluating the potential of Cardionogen on human adult and ES cells, along with other known cardiogenic small molecules (Hao et al., 2008; Saderk et al., 2008; Wu et al., 2004), is the next logical step in defining therapeutic regimens to enhance repopulation of infarcted myocardium and restore function in diseased hearts.
Experimental Procedures
Zebrafish strains and maintenance
Zebrafish strains used in this study were raised according to standard procedures(Westerfield, 2000). Transgenic lines Tg(cmlc2:EGFP), Tg(cmlc2:DsRed-nuc) and Tg(hsp:wnt8-EGFP) have been previously described(Burns et al., 2005; Ueno et al., 2007) ((Mably et al., 2003).
Cardiac phenotype-based small molecule screen
Experimental compounds were obtained from Vanderbilt Institute of Chemical Biology. Aliquots (1µL) of the diluted chemical compounds (1mM) were delivered into each of the wells using a Labcyte Echo 550. Immediately following aliquot addition, E3 buffer (69µL) was delivered into each well. Well12A was served as a negative control (1% DMSO). Embryos in were generated by crossing homozygous Tg(cmlc2:EGFP) transgenic fish. Harvested embryos were placed in E3 and incubated at 28.5°C. When embryos reached 5 hpf (50% epiboly), 3 embryos were transferred to each well in a 30µL aliquot of E3 buffer, bringing the final compound concentration to 10µM. Plates were incubated at 28.5°C and screened using fluorescent microscopy at 24, 48 and 72 hpf.
Partition coefficient analysis
Log P values and molecular weights of bioactive compounds were obtained from the National Center for Biotechnical Information (http://pubchem.ncbi.nih.gov)
Cardionogen synthesis
Synthesis of Cardionogen series (vuc198 and vuc230) was designed and conducted by Vanderbilt Chemical Synthesis Core. Compound identities were confirmed by 1H NMR.
Analyses of cardiac cell number, in situ hybridization and immunohistochemistry
The transgenic zebrafish lines Tg(cmlc2:DsRed-nuc) and Tg(cmlc2:EGFP) were employed to assess changes in cardiomyocyte number and size by confocal microscopy (Jia et al., 2007; Mably et al., 2003). Whole mount in situ hybridizations were carried out as described (Zhong et al., 2001) using antisense ribonucleotide probes for nkx2.5, cmlc2, vmhc, scl, myoD and insulin. Immunofluorescence was performed as described (Jia et al., 2007) using primary anti-phosphorylated histone H3 antibody (1:100; Santa Cruz Biotechnology) and secondary antibody Alexa Fluor 555 donkey anti-rabbit conjugate (1:200; Molecular Probes).
Murine ES Differentiation Assay
Murine CGR8-ES cells were stably transfected with the α-MHC-DsRed-Nuc plasmid, maintained, and differentiated as described (Hao et al., 2008). Cells were cultured on gelatin-coated culture plates with Glasgow Minimum Essential Medium (GMEM; Sigma) containing 10% heat inactivated FBS, 20 µM L-Glutamine, 50 µM β-Mercaptoethanol, and 100 units/ml Leukemia Inhibitory Factor (ESGRO-Chemicon). CGR8-ES cells were differentiated in Iscove’s Modified Dulbecco’s Medium (IMDM; GIBCO) containing 20% heat inactivated FBS, 16 µM L-Glutamine, non-essential amino acids, and 80 µM β-Mercaptoethanol. EBs were initiated (day 0) and grown in hanging drops for two days. Each EB initially consisted of 500 cells in 20 µl of IMDM differentiation medium. Formed EBs were washed down into an uncoated Petri dish and suspended in IMDM differentiation medium for two more days. On day 4, EBs were transferred to gelatin-coated plates, allowed to attach, and incubated in differentiation medium until analyses on day 12. The medium was replaced every 48 hours, and differentiating cell cultures were microscopically examined for the presence of contractile cardiomyocytes marked by red fluorescence. Cells were harvested at day 12 and RNAs were isolated using RNeasy kit (Qiagen) for real-time RT-PCR (see Supplement Experimental Procedure).
Fluorescence Activated Cell Sorting (FACS)
Embryoid Bodies created with CGR8-ES cell line were washed in 1X PBS and dissociated with collagenase (20mg/ml) in 1X PBS. These cells were centrifuged for 3.5 minutes at 3500 rpm. Supernatant was removed and cells were washed with FACS buffer (1X PBS and 5% Fetal bovine serum). Washed cells were fixed in 2%PFA/PBS for 10 mins, and probed using the first α-Actinin monoclonal antibody (Sigma; 1:2,000 dilution) then the secondary antibody goat anti-mouse IgG Alexa Fluor 488 (Invitrogen; 1:200 dilution). Cells were sorted on LSR-II (BD Biosciences) and analyzed using FACS Diva v6.1.3 software.
Reporter Assay
Murine CGR8-ES cells were transfected with constructs of pTOPflash reporter (Upstate Biotechnology Inc., now Millipore), RBP-Jk reporter or SRF/MAPK reporter (SABiosciences Inc) using Lipofectamine 2000 transfection reagent (Invitrogen Inc). These transfected cells were grown in feeder-free conditions as monolayers in GMEM medium supplemented with 10% FBS, 100 units/ml LIF, 2 mM L-glutamine, and 50 µM β-mercaptoethanol, in a humidified 5% CO2 atmosphere at 37°C as described (Meyer et al., 2000). HEK293 cells were maintained in DMEM, 10% FBS and antibiotics. Cells were then lysed with 1X Passive Lysis Buffer (Promega). Luciferase activity was measured by Steady-Glo Luciferase Assay (Promega) on a Monolight 2010 Luminometer (Analytical Luminescence Laboratory) and normalized to viable cell number using the CellTiter-Glo Assay (Promega). Graphs were made in Prism 4 (GraphPad Software, Inc.) with nonlinear regression fit to a sigmoidal dose-response curve.
Wnt8-induced cardiac phenotype assays
Tg(hsp:wnt8-EGFP) fish were out-crossed to wild type AB line. Progeny embryos were all heterozygous for the transgene. Embryos at 3 hpf or 9 hpf were placed in 15 ml Falcon tubes with E3 buffer. The tubes were then submerged in 38.5 °C water bath for 15 minutes for heat shock, while non-heat shock control embryos were maintained at 25 °C. After heat shock, embryos were washed with E3 at room temperature. Heat shocked embryos were either treated with CDNG1 (90 µM) or returned to E3 buffer as controls. All embryos were incubated at 25 °C until fixation at 17hpf or 48hpf for in situ hybridization analyses.
Highlights.
in vivo compound screen in zebrafish identifies modulators of cardiac development
Identified chemical family (Cardionogen) in regulating cardiomyocyte number
Functionality of Cardionogen is conserved in mouse embryonic stem cells
Chemical genetics indicates Cardionogen acts by inhibiting Wnt/β-catenin signaling
Supplementary Material
Acknowledgments
We are indebted to John Guan for his invaluable assistance in fish care and Alex Waterson for their efforts in chemical synthesis. We thank Geoffrey Burns for cmlc2-EGFP and cmlc2-DsRed fish, Randall Moon for hsp-wnt8 fish, G J. Robbins for providing Myh6 promoter plasmid and P. ten Dijke for BRE2-Luc construct. We are grateful to Bruce Appel, Scott Baldwin, Wenbiao Chen, Josh Gamse, Daqing Jin and members of our laboratories for comments on the manuscript and helpful discussions. This research is supported in part by grants NIH-NS064852 (TPZ), HL083958 (AKH), Vanderbilt-VICTR104 (TPZ) and Fudan-EZH1322001(TPZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc Natl Acad Sci USA. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, Puceat M. Stem cell differentiation requires a paracrine pathway in the heart. Faseb J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nature Chemical Biology. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/β-Catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241 doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, Peterson RT, Hatzopoulos AK, Hong CC. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS ONE. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Zhang YQ, Dabernat S, Kritzik M, Dietz D, Sterling L, Sarvetnick N. BMP4 regulates pancreatic cell expansion through Id2. J Biol Chem. 2006;281:13574–13580. doi: 10.1074/jbc.M600526200. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Jia H, King IN, Chopra SS, Wan H, Ni TT, Jiang C, Guan X, Wells S, Srivastava D, Zhong TP. Vertebrate heart growth is regulated by functional antagonism between Gridlock and Gata5. Proc Natl Acad Sci U S A. 2007;104:14008–14013. doi: 10.1073/pnas.0702240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- Kimelman D. Mesoderm induction: from caps to chips. Nat Rev Genet. 2006;7:360–372. doi: 10.1038/nrg1837. [DOI] [PubMed] [Google Scholar]
- Lehar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Lunde K, Solheim S, Aakhus S, Arnesen H, Abdelnoor M, Egeland T, Endresen K, Ilebekk A, Mangschau A, Fjeld JG, et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction. N Engl J Med. 2006;355:1199–1209. doi: 10.1056/NEJMoa055706. [DOI] [PubMed] [Google Scholar]
- Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Current Biology. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Jaconi M, Landopoulou A, Fort P, Puceat M. A fluorescent reporter gene as a marker for ventricular specification in ES-derived cardiac cells. FEBS Lett. 2000;478:151–158. doi: 10.1016/s0014-5793(00)01839-1. [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006;39:255–261. doi: 10.1016/j.ymeth.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahiro T, Kurooka H, Mori K, Sano K, Yokota Y. Identification of BMP-responsive elements in the mouse Id2 gene. Bochem Biophys Res Commun. 2010;399:416–421. doi: 10.1016/j.bbrc.2010.07.090. [DOI] [PubMed] [Google Scholar]
- Olson EN. A decade of discoveries in cardiac biology. Nature Medicine. 2004;10:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nature Biotechnology. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Developmental Biology. 2001;234:330–338. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- Sachidanandan C, Yeh JR, Peterson QP, Peterson RT. Identification of a novel retinoid by small molecule screening with zebrafish embryos. PLoS ONE. 2008;3:e1947. doi: 10.1371/journal.pone.0001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saderk H, Hannack B, Choe E, Wang J, Latif S, Garry MG, Garry DJ, Longggod J, Frantz DE, E.N O, et al. Cardiogenic small molecules that enhance stem cell function in myocardial repair. Proc Natl Acad Sci U S A. 2008;105:6063. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachinger V, Erbs S, Elsasser A, Haberbosch W, Hambrecht R, Holschermann H, Yu J, Corti R, Mathey DG, Hamm CW, et al. Intracoronary bone marrow-derived progenitor cells in acute myocardial infarction. N Engl J Med. 2006;355:1210–1221. doi: 10.1056/NEJMoa060186. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H, Kolde R, Adler P, Aksoy I, Asastassiadis K, Mader M, Hatzopoulos A. The FunGene ES database: a genomic resource for mouse stem cell differentiation. PloS One. 2009;4:6804. doi: 10.1371/journal.pone.0006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nature Reviews. 2001;2:39–47. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1:366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- Tahinci E TC, Franklin JL, Salic A, Christian KM, Lee LA, Coffey RJ, Lee E. LRP6 is required for convergent extension during Xenopus gastrulation. Development. 2007;134:4095–4106. doi: 10.1242/dev.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. J Am Chem Soc. 2004;126:1590–1591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Developmental Biology. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Childs S, Leu JP, Fishman MC. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.