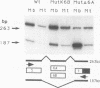
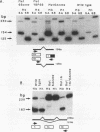
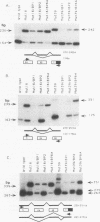
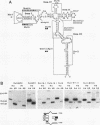
Abstract
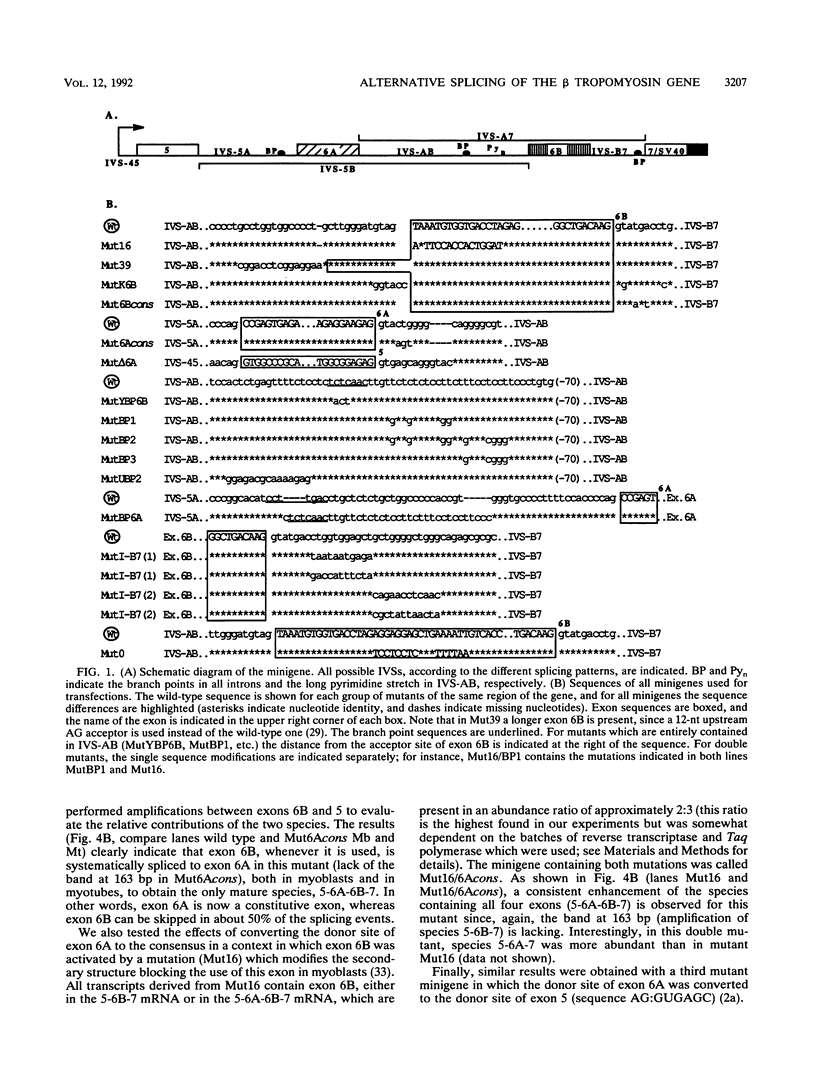
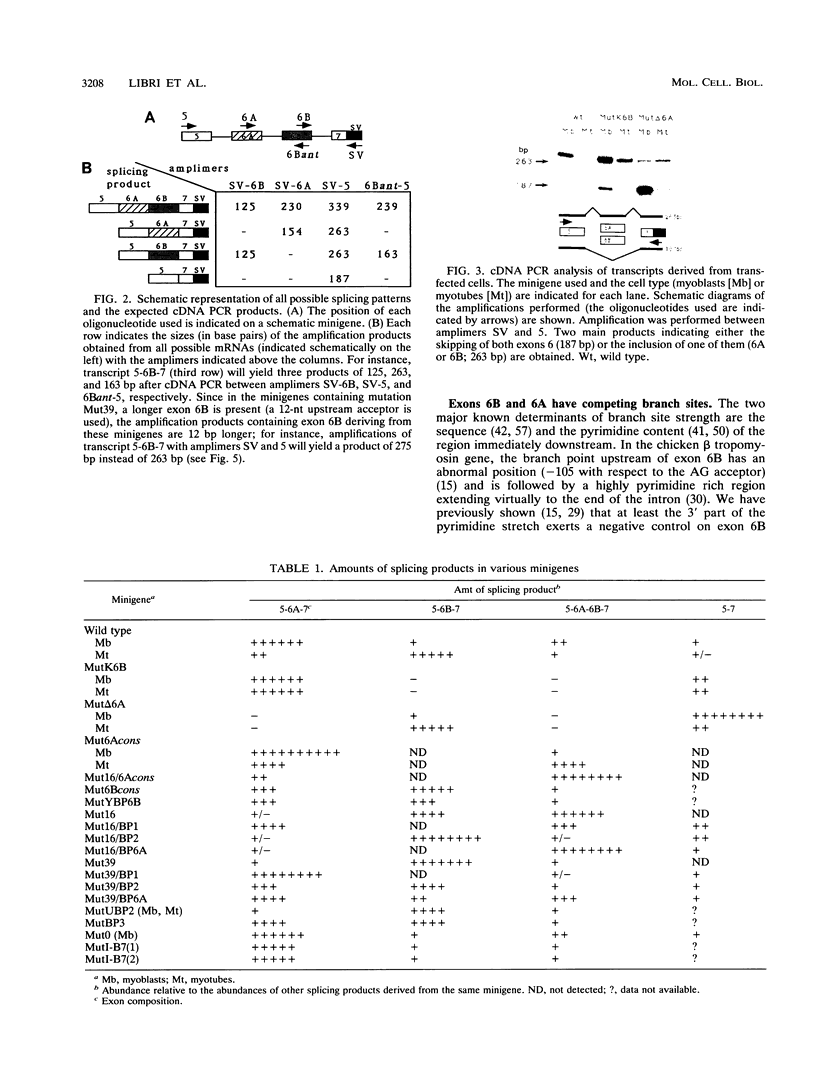
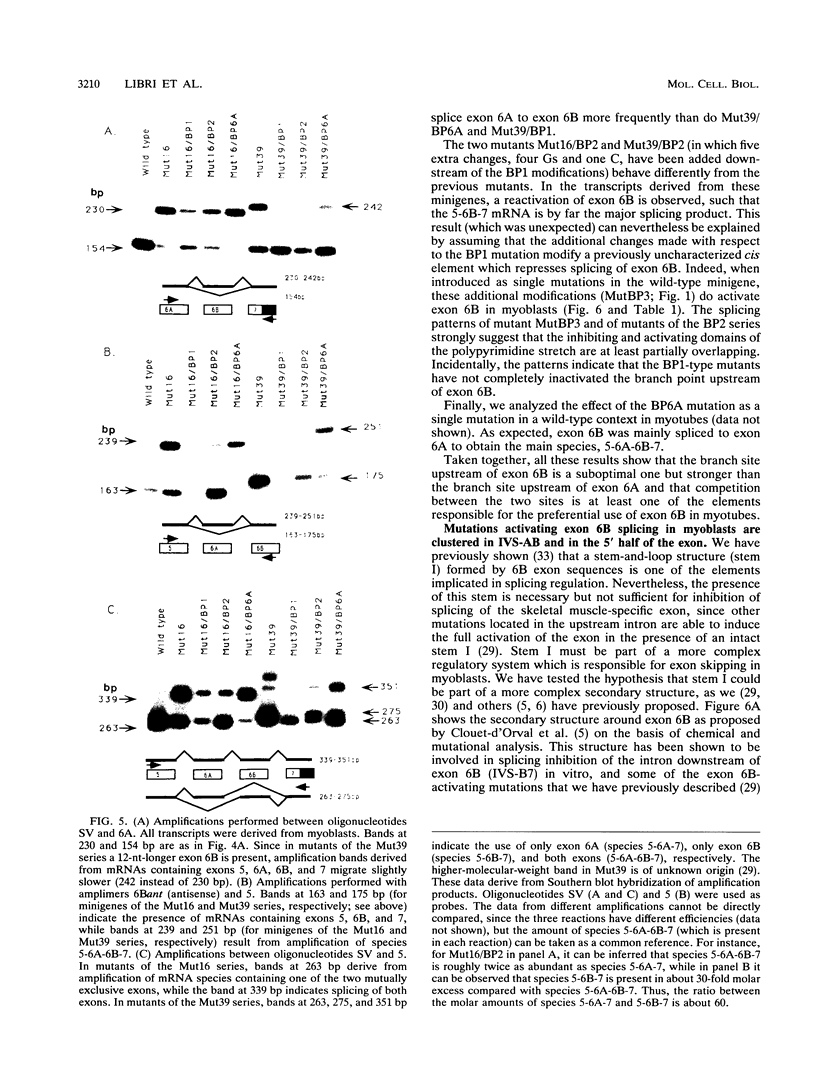
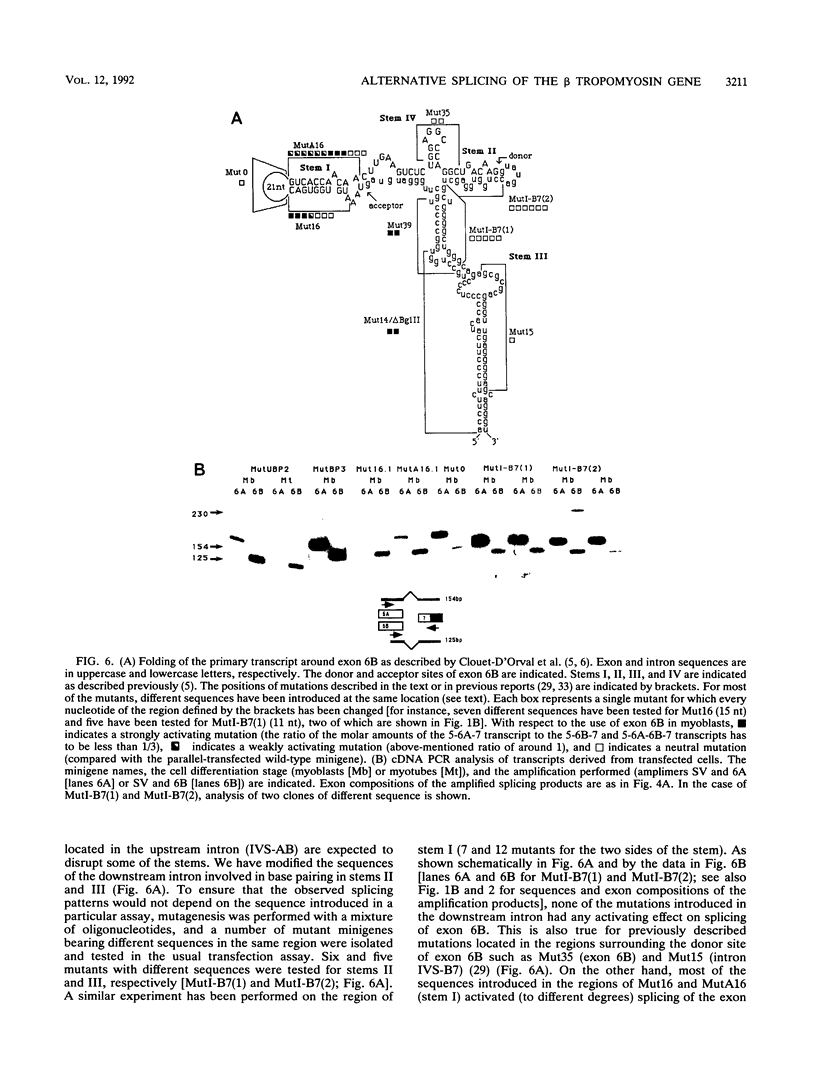
The chicken beta tropomyosin gene contains two sets of alternatively spliced, mutually exclusive exons whose utilization is developmentally regulated. Exons 6A and 6B are used in nonmuscle cells (or undifferentiated muscle cells) and skeletal muscle cells, respectively. A complex arrangement of cis-acting sequence elements is involved in alternative splicing regulation. We have performed an extensive mutational analysis on the sequence spanning the region from exon 6A to the constitutive exon 7. A large number of mutant minigenes have been tested in transfection assays of cultured myogenic cells, and the splicing products have been analyzed by cDNA polymerase chain reaction. We demonstrate that in undifferentiated myoblasts, exon 6B is skipped as a result of a negative control on its selection, while exon 6A is spliced as a default choice. We provide evidence that the focal point of such a regulation is localized in the intron upstream of exon 6B and probably involves the blockage of its associated branch point. In differentiated myotubes, in contrast, both exons are accessible to the splicing machinery. We show that the preferential choice of exon 6B in this splicing environment depends on the existence of a competition between the two exons for the flanking constitutive splice sites. We demonstrate that both the donors and the branch points of the two exons are involved in this competition.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Chou T. B., Mims I., Zachar Z. On/off regulation of gene expression at the level of splicing. Trends Genet. 1988 May;4(5):134–138. doi: 10.1016/0168-9525(88)90136-9. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton-Carafa Y., Brody J. M., Brody E. Determination of an RNA structure involved in splicing inhibition of a muscle-specific exon. J Mol Biol. 1991 Oct 5;221(3):837–856. doi: 10.1016/0022-2836(91)80179-x. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J. L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991 Jul 26;66(2):373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gelfand M. S. Statistical analysis of mammalian pre-mRNA splicing sites. Nucleic Acids Res. 1989 Aug 11;17(15):6369–6382. doi: 10.1093/nar/17.15.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Gil A., Sharp P. A., Jamison S. F., Garcia-Blanco M. A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991 Jul;5(7):1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Cheley S., Kuismanen E., Finn L. A., Yamawaki-Kataoka Y. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol Cell Biol. 1986 Nov;6(11):3582–3595. doi: 10.1128/mcb.6.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., Weber K. S. Identification of two distinct intron elements involved in alternative splicing of beta-tropomyosin pre-mRNA. Genes Dev. 1990 Jan;4(1):98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Drosophila sex determination: a cascade of regulated splicing. Cell. 1989 Mar 24;56(6):905–906. doi: 10.1016/0092-8674(89)90619-3. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Sakamoto H., Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990 Mar 29;344(6265):461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991 Jul 26;66(2):383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Lemonnier M., Balvay L., Mouly V., Libri D., Fiszman M. Y. The chicken gene encoding the alpha isoform of tropomyosin of fast-twitch muscle fibers: organization, expression and identification of the major proteins synthesized. Gene. 1991 Nov 15;107(2):229–240. doi: 10.1016/0378-1119(91)90323-4. [DOI] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Lemonnier M., Meinnel T., Fiszman M. Y. A single gene codes for the beta subunits of smooth and skeletal muscle tropomyosin in the chicken. J Biol Chem. 1989 Feb 15;264(5):2935–2944. [PubMed] [Google Scholar]
- Libri D., Marie J., Brody E., Fiszman M. Y. A subfragment of the beta tropomyosin gene is alternatively spliced when transfected into differentiating muscle cells. Nucleic Acids Res. 1989 Aug 25;17(16):6449–6462. doi: 10.1093/nar/17.16.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Mouly V., Lemonnier M., Fiszman M. Y. A nonmuscle tropomyosin is encoded by the smooth/skeletal beta-tropomyosin gene and its RNA is transcribed from an internal promoter. J Biol Chem. 1990 Feb 25;265(6):3471–3473. [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lindquester G. J., Flach J. E., Fleenor D. E., Hickman K. H., Devlin R. B. Avian tropomyosin gene expression. Nucleic Acids Res. 1989 Mar 11;17(5):2099–2118. doi: 10.1093/nar/17.5.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Fiszman M. Y. A new muscle phenotype is expressed by subcultured quail myoblasts isolated from future fast and slow muscles. J Biol Chem. 1983 Mar 25;258(6):3883–3888. [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Patton J. G., Mayer S. A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991 Jul;5(7):1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Opazo N., Nadal-Ginard B. Alpha-tropomyosin gene organization. Alternative splicing of duplicated isotype-specific exons accounts for the production of smooth and striated muscle isoforms. J Biol Chem. 1987 Apr 5;262(10):4755–4765. [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S. Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 1991 Nov;5(11):2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Stolow D. T., Berget S. M. Identification of nuclear proteins that specifically bind to RNAs containing 5' splice sites. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):320–324. doi: 10.1073/pnas.88.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Wieczorek D. F., Smith C. W., Nadal-Ginard B. The rat alpha-tropomyosin gene generates a minimum of six different mRNAs coding for striated, smooth, and nonmuscle isoforms by alternative splicing. Mol Cell Biol. 1988 Feb;8(2):679–694. doi: 10.1128/mcb.8.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991 Jan;10(1):207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Rose S. D., Berget S. M. U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Mol Cell Biol. 1987 Aug;7(8):2877–2883. doi: 10.1128/mcb.7.8.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]