SUMMARY
Partitioning of chromosomes into euchromatic and heterochromatic domains requires mechanisms that specify boundaries. The S. pombe JmjC family protein Epe1 prevents the ectopic spread of heterochromatin and is itself concentrated at boundaries. Paradoxically, Epe1 is recruited to heterochromatin by HP1 silencing factors that are distributed throughout heterochromatin. We demonstrate here that the selective enrichment of Epe1 at boundaries requires its regulation by the conserved Cul4-Ddb1Cdt2 ubiquitin ligase, which directly recognizes Epe1 and promotes its polyubiquitylation and degradation. Strikingly, in cells lacking the ligase, Epe1 persists in the body of heterochromatin thereby inducing a defect in gene silencing. Epe1 is the sole target of the Cul4-Ddb1Cdt2 complex whose destruction is necessary for the preservation of heterochromatin. This mechanism acts parallel with phosphorylation of HP1/Swi6 by CK2 to restrict Epe1. We conclude that the ubiquitin-dependent sculpting of the chromosomal distribution of an antisilencing factor is critical for heterochromatin boundaries to form correctly.
INTRODUCTION
Eukaryotic genomes are organized into active and inactive domains referred to as euchromatin and heterochromatin. This functional organization plays an important role in chromosome segregation, telomere maintenance, and genome stability. Given the repressive nature of heterochromatin, the regulation of its assembly is critical for genome homeostasis. The fission yeast Schizosaccharomyces pombe has become a powerful model system for dissecting mechanisms of eukaryotic heterochromatin control (for an extensive review, see Grewal, 2010). Its genome contains three distinct major heterochromatic domains: pericentromeric otr repeats, subtelomeric regions, and the silent mating type locus mat2/mat3. As in metazoans and plants, a key step in heterochromatin assembly is the recruitment of a histone H3 lysine 9 (H3K9) methyltransferase (Clr4 in S. pombe) to chromatin. The methylation of H3K9 by Clr4 is required for the recruitment of the HP1 family proteins Swi6 and Chp2, which then appear to spread along the DNA fiber. It is thought that nucleation of heterochromatin is guided by RNA interference (RNAi)-dependent and -independent mechanisms. In the RNAi-dependent pathway, heterochromatic sequences are transcribed by RNA polymerase II (Pol II) during S phase, and siRNAs are subsequently generated by the RNAi machinery. Whereas pericentromeric heterochromatin requires the RNAi-dependent pathway for its establishment, both pathways act redundantly at the telomeres and the silent mating type locus.
Initial H3K9 methylation and subsequent binding of HP1 proteins lead to the spreading of this repressive modification, but the phenomenon of heterochromatin spread is still poorly understood (reviewed in Talbert and Henikoff, 2006). HP1 proteins, whose chromatin association depends on H3K9 methylation, seem to be involved in recruiting the upstream H3K9 methyltransferase. This has been suggested to result in methylation of neighboring nucleosomes, thereby creating a positive feedback loop in assembly and spreading of heterochromatin over large distances in cis. Spreading is a stochastic process that can result in metastable but heritable silencing of neighboring euchromatic genes, a phenomenon known as position effect variegation (PEV) (Gaszner and Felsenfeld, 2006). The invasion of heterochromatin into adjacent euchromatic regions is prevented by boundary elements, which terminate the chain of events involved in spreading. The mechanisms by which boundaries are formed are complex, and a number of models have been proposed that include tethering of boundary elements to subnuclear regions and recruiting silencing-opposing activities (Gaszner and Felsenfeld, 2006). In S. pombe, the euchromatin-heterochromatin borders are characterized by sharp transitions of euchromatic and heterochromatic histone modifications (Cam et al., 2005). Specific boundary elements composed of inverted repeat (IR) sequences are found at the boundaries flanking the silent mating type locus and the pericentromeric regions of chromosomes 1 and 3 (Cam et al., 2005; Noma et al., 2006). The left and right boundary elements at the silent mating type locus (IR-R/L) contain recruitment sites for transcription factor TFIIIC, which has been suggested to delineate heterochromatic domains by sequestering the boundary elements to the nuclear periphery (Noma et al., 2006). Conversely, the pericentromeric inverted repeat (IRC) elements are associated with Pol II-dependent transcription and show an enrichment of euchromatic marks (Cam et al., 2005; Noma et al., 2006). Other boundaries at pericentromeric regions are characterized by tRNA gene clusters, which seem to be critical for barrier function (Scott et al., 2006).
Epe1 (enhancer of position effect) was previously identified in a screen for mutants in S. pombe that display propagation of heterochromatin beyond its natural borders. Mutants of epe1+ show enhanced PEV at the silent mating type locus and pericentromeric regions (Ayoub et al., 2003). Epe1 is critical for the boundary function of the pericentromeric IRC elements and mediates their Pol II-dependent transcription (Zofall and Grewal, 2006). The mechanism by which Epe1 antagonizes heterochromatin spread is unknown. Epe1 contains a JmjC domain that is present in many histone demethylases but lacks a conserved residue predicted to be involved in binding of a catalytic iron atom. Furthermore, no histone demethylase activity has been detected for Epe1 in vitro (Tsukada et al., 2006). Despite being an antisilencing factor, Epe1 interacts with the HP1 proteins Swi6 and Chp2 in vivo and in vitro and is itself recruited to heterochromatin in an HP1-dependent manner (Sadaie et al., 2008; Zofall and Grewal, 2006). In particular, Epe1 facilitates the recruitment of Pol II to heterochromatic regions (Zofall and Grewal, 2006). Perhaps due to this role in Pol II-dependent transcription, mutants of epe1+ have perturbed levels of heterochromatic siRNAs and are affected in the stability of heterochromatic domains (Trewick et al., 2007). In addition, Epe1 appears to compete for binding to heterochromatin with the HDAC effector complex SHREC (Shimada et al., 2009). These findings raised the important question of how heterochromatin is protected from the silencing-antagonizing activity of Epe1 that it recruits.
Histones have long been known to be substrates for the ubiquitin system. Conjugation involves the transfer of ubiquitin to a lysine residue within the substrate by an enzymatic cascade comprising an activating enzyme (E1), a conjugating enzyme (E2), and an ubiquitin ligase (E3), the latter determining substrate specificity of ubiquitylation. Ubiquitylation plays a crucial role in the regulation of chromatin. For instance, monoubiquitylation of histone H2A is associated with silencing of the mammalian Hox gene cluster (Wang et al., 2004), whereas ubiquitylation of histone H2B is a prerequisite for methylation of H3K4 and H3K79 (Nakanishi et al., 2009; Sun and Allis, 2002).
Methylation of H3K9 in S. pombe requires a multisubunit E3 that associates with the H3K9 methyltransferase Clr4 in the CLRC complex and is necessary for chromatin recruitment of Clr4 (Hong et al., 2005). This E3 enzyme, Cul4-Rik1Dos1/Dos2, is related to the cullin-RING finger family of ubiquitin ligases (CRLs), in particular the conserved Cul4-Ddb1DCAF complexes. Common to this family is a modular architecture that employs a cullin family scaffold, a RING finger protein that recruits the ubiquitin-conjugating E2 enzyme, and a substrate recognition factor (Jackson and Xiong, 2009). Ddb1 is a specific adaptor protein of Cul4 RING finger ligases (Cul4-Ddb1) and recruits the substrate recognition factor that confers specificity to the ubiquitylation reaction. Most of the identified substrate recognition factors (DCAFs, Ddb1/Cul4 associated factors) contain WD40 repeats (Lee and Zhou, 2007). However, in the Cul4-Rik1Dos1/Dos2 complex, the conserved Ddb1 adaptor is replaced by Rik1 and the substrate recognition DCAF subunit is replaced by Dos1/Dos2. As the Cul4-Rik1Dos1/Dos2 E3 seems to function particularly in silencing, it appears to be a specialized paralog of the conserved Cul4 CRLs. Despite its requirement for heterochromatin formation, the corresponding substrate has not been identified.
Here, we report the identification of a regulatory mechanism essential for proper boundary formation and heterochromatic silencing in S. pombe, which unexpectedly requires the action of the canonical CRL Cul4-Ddb1Cdt2. We demonstrate that the Cul4-Ddb1Cdt2 complex directly recognizes and promotes ubiquitylation and degradation of the boundary factor Epe1. Strikingly, this pathway controls the distribution of this antisilencing factor within heterochromatic domains and restricts Epe1 to the heterochromatic boundaries. We show that this heterochromatin-sculpting function of Cul4-Ddb1Cdt2 is sufficient to explain its requirement for silencing. Our studies define a ubiquitin-dependent degradation event necessary for heterochromatin formation and demonstrate that it functions to shape heterochromatin.
RESULTS
A Targeted Knockout Screen Identifies Factors Required for Pericentromeric Silencing
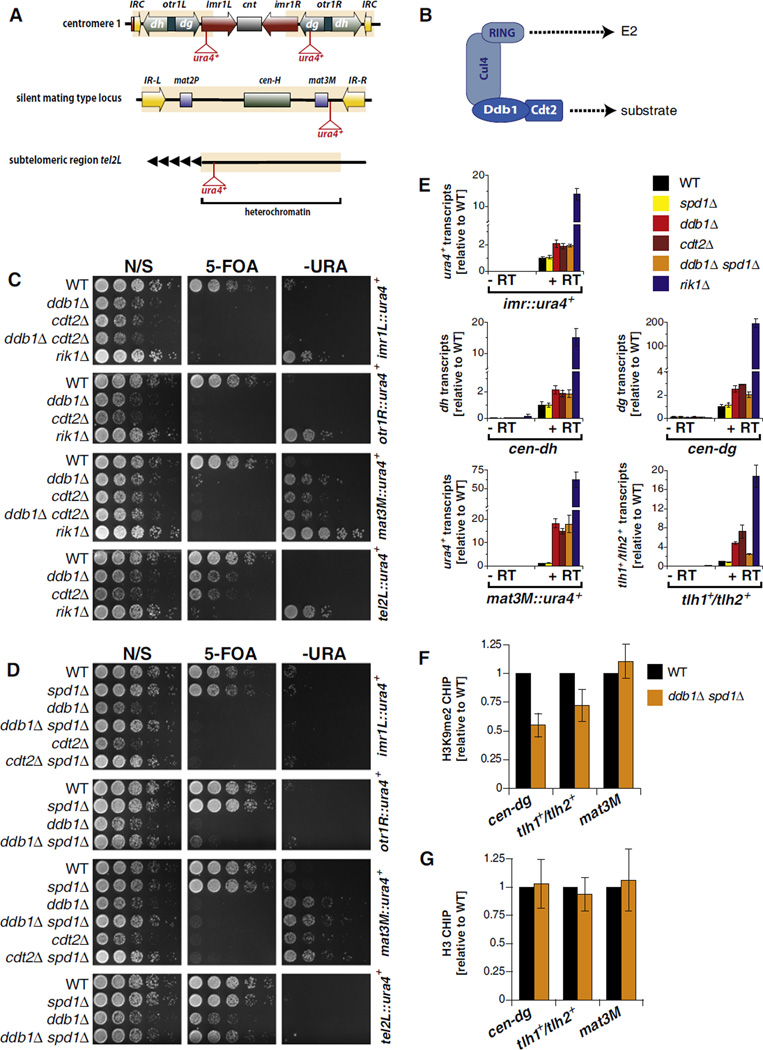
To identify factors required for heterochromatin formation, we disrupted candidate genes in fission yeast harboring a pericentromeric ura4+ reporter gene whose silencing can be assayed using the drug 5-FOA that counterselects for ura4+-expressing cells (Ekwall et al., 1999). In S. pombe, heterochromatin marked by the HP1 protein Swi6 colocalizes with the spindle pole body (SPB) during interphase (Appelgren et al., 2003). In fact, many other heterochromatic proteins display a similar SPB-like localization or dot-like staining within the nucleus (Matsuyama et al., 2006). A high-throughput study reported that 346 S. pombe proteins display such a localization pattern when fused to yellow fluorescent protein (YFP) and expressed from an inducible promoter (Matsuyama et al., 2006). We successfully deleted 166 of these genes in an imrL::ura4+ reporter strain. In addition, we deleted 23 other genes that display sequence motifs suggestive of a potential role in chromatin biology plus a few control genes encoding known silencing factors (Figure S1H and Table S1 available online).
We screened this collection of 195 deletion mutants on 5-FOA media and isolated 12 mutants with a previously undescribed loss-of-silencing phenotype (Figure S1A). Among those mutants were 11 genes that encode SBP/nuclear dot proteins and 1 factor with a nucleoplasmic localization, Ddb1, the well-studied adaptor component of the canonical Cul4 CRLs. The ddb1Δ mutant showed a 5-FOA silencing phenotype comparable to cells lacking the histone H3K9 methyltransferase Clr4, which is essential for heterochromatin formation (Figure S1A, left panel). Among the SPB/nuclear dot candidates, deletion of SPCC1393.05 also resulted in a strong silencing defect, and we have described an initial analysis of this gene, ers1+, elsewhere (Rougemaille et al., 2008). The remaining SBP/nuclear dot mutants exhibited weaker phenotypes, both in the original imrL::ura4+ strain and in a strain harboring a mat3M::ura4+ reporter gene that measures silencing at the mat2/3 silent cassette (Figures S1A and S1F). RT-qPCR analysis showed that many of these mutants accumulate silenced transcripts depending on the heterochromatic region assessed (Figures S1B–S1G). The silencing defect observed in ddb1Δ cell mutants suggested a critical function of this E3 ligase subunit in heterochromatin formation, and its role in silencing was investigated further.
The Cul4-Ddb1Cdt2 Ubiquitin Ligase Promotes Silencing at Multiple Heterochromatic Domains
Cells lacking Ddb1 show a silencing defect at the inner most repeat (imr) and outer repeat (otr) elements of the pericentromeric region but are also impaired in silencing at the mat3M locus of the silent mating type cassette and a subtelomeric region (Figure 1). This result distinguishes Ddb1 from factors directly involved in the RNAi pathway as they only impact silencing at centromeres. Analysis of steady-state levels of mRNAs originating from the imr1L::ura4+ and mat3M::ura4+ loci, as well as endogenous heterochromatic sequences, showed modest (particularly at pericentric regions) but reproducible increases upon deletion of ddb1+ (Figure 1E) compared to control strains lacking Clr4 or Rik1. The largest fold-change was observed at the mat3M locus (Figure 1E). These changes in transcript levels were nonetheless sufficient to interfere with reporter gene silencing (Figures 1C and 1E). Thus, Ddb1 is required for efficient silencing but is presumably not a core component of the heterochromatin formation machinery.
Figure 1. Cul4-Ddb1Cdt2 Promotes Silencing at Major Heterochromatic Loci Independently of the S Phase Inhibitor Spd1.
(A) S. pombe heterochromatic domains with positions of the ura4+ reporter genes.
(B) Cul4-Ddb1Cdt2 cullin-RING ubiquitin ligase architecture.
(C and D) Reporter assays. N/S, nonselective; 5-FOA, 5′-fluoroorotic acid; –URA, without uracil.
(E) RT-qPCR analysis. Shown are transcript levels relative to wild-type (WT) ± standard error of the mean (SEM) of three independent experiments.
(F) ChIP analysis of H3K9me2 levels. Shown are mean values relative to WT ± SEM of three independent ChIP samples.
(G) ChIP analysis of histone H3 as in (F). Error bars represent variation from the mean of two independent experiments.
See also Figure S1.
To identify the relevant DCAF, we focused on the 105 WD40 repeat proteins present in S. pombe. We successfully knocked out 60 of the corresponding genes in the imrL::ura4+ reporter strain and screened this collection for mutants that phenocopy ddb1Δ. One mutant, cdt2Δ, displayed an identical phenotype to that of ddb1Δ cells in all assays (Figures 1C and 1E). Importantly, ddb1Δ cdt2Δ double mutants showed no additive silencing defect, indicating that ddb1+ and cdt2+ are epistatic and function in the same pathway (Figure 1C). Consistent with our findings, Cdt2 has previously been described as a substrate recognition factor of Cul4-Ddb1 involved in the degradation of chromatin-associated factors (Jin et al., 2006; Liu et al., 2005; Ralph et al., 2006).
Methylation of lysine 9 of histone H3 is a hallmark of heterochromatin. To study whether methylation of histone H3K9 is affected by the absence of Ddb1, we determined the profile of dimethylated H3K9 (H3K9me2) by performing chromatin immunoprecipitation (ChIP) experiments at various heterochromatic regions. To control for nonspecific effects of Ddb1 on growth, we used a ddb1Δ spd1Δ double mutant in which a known cell-cycle substrate of Ddb1, Spd1, is also absent—deletion of spd1+ suppresses the growth defect of the ddb1Δ mutant (see below). Consistent with the silencing defect seen at the pericentromeric and subtelomeric regions, we found a significant reduction in H3K9me2 levels at the cen-dg and the tlh1+/thl2+ loci (Figure 1F) but no changes in histone H3 levels (Figure 1G). In contrast, H3K9me2 levels were unaffected at the mating type locus in ddb1Δ spd1Δ cells (Figure 1F), despite the strong silencing defect seen at this heterochromatic locus (compare Figures 1C and 1E). Thus, the decrease in H3K9 methylation cannot generally explain the silencing defect of ddb1Δ cells, and the different heterochromatic domains seem to have distinct requirements for silencing.
Silencing Is Not Inhibited by the Cul4-Ddb1Cdt2 Substrate Spd1
To date, only two substrates of Cul4-Ddb1Cdt2 have been identified in S. pombe: Cdt1, which is required for licensing of replication origins (Ralph et al., 2006), and Spd1, which is an inhibitor of ribonucleotide reductase (Bondar et al., 2004); both substrates are degraded during S phase. Accumulation of Spd1 causes cell-cycle delay, abnormal cellular size, and a substantial growth defect in ddb1Δ cells (Bondar et al., 2004; Holmberg et al., 2005). As two recent studies linked the onset of S phase to RNAi-mediated assembly of heterochromatin (Chen et al., 2008; Kloc et al., 2008), we sought to test the hypothesis of whether the accumulation of Spd1 may be the reason for the silencing defect. To this end, we knocked out spd1+ in ddb1Δ or cdt2Δ cells and examined the phenotypes of the corresponding double mutants. Consistent with previous reports (Bondar et al., 2004; Holmberg et al., 2005), we observed a suppression of the slow growth phenotype in ddb1Δ spd1Δ and cdt2Δ spd1Δ cells on nonselective media (Figure 1D). In contrast, the silencing defects of ddb1Δ and cdt2Δ were unaffected in the double mutants (Figures 1D and 1E), with the exception of a partial alleviation of the silencing defect at a subtelomeric locus (Figures 1D and 1E). Thus, Spd1 is not the major target of Cul4-Ddb1Cdt2 in heterochromatin formation. Because cdt1+ is an essential gene, we could not test genetically a requirement for Cdt1 degradation in silencing. To avoid potential secondary effects that may arise from abnormal cellular morphology and slow growth associated with increased levels of Spd1, we used ddb1Δ spd1Δ cells instead of the single ddb1Δ mutant for the experiments described below.
Cul4-Ddb1Cdt2 Controls the Levels of the JmjC Protein Epe1 by Regulating Its Protein Turnover
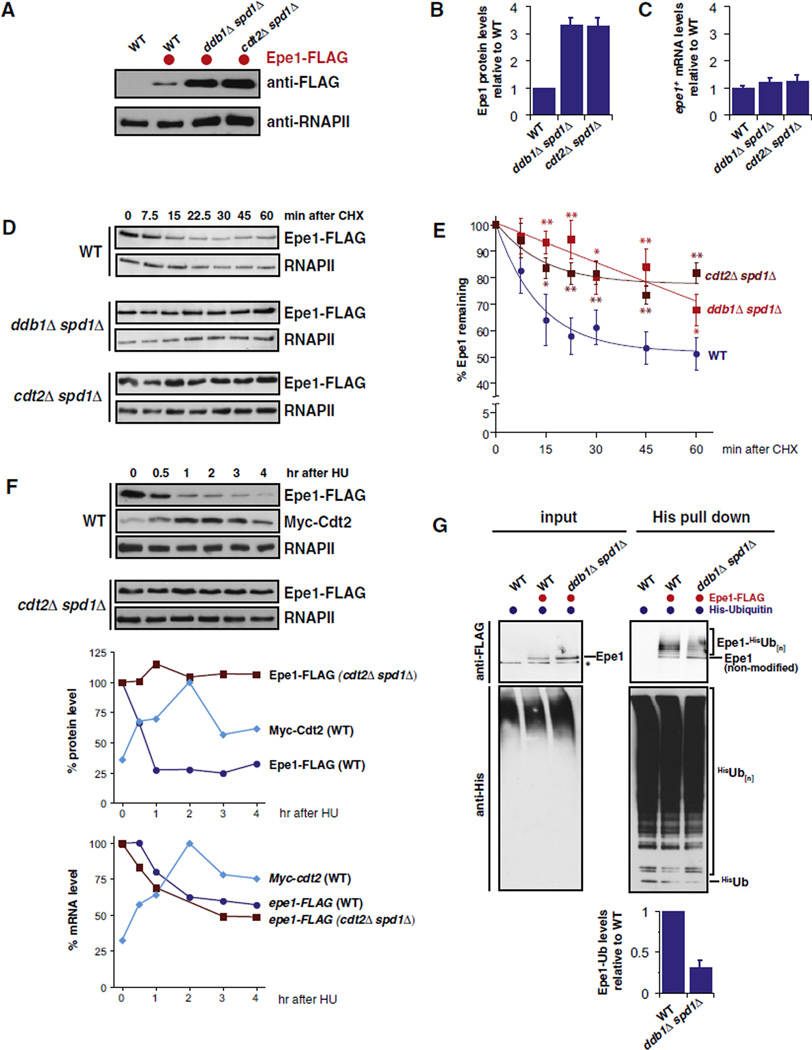
Considering the proteolytic role of Cul4-Ddb1Cdt2 in various systems (Jackson and Xiong, 2009), its requirement for proper heterochromatin formation may reflect the need for degrading an inhibitor of silencing that acts at pericentromeric regions, subtelomeric regions, and the silent mating type locus. Only one such antisilencing factor has been described in the S. pombe literature: Epe1 (Ayoub et al., 2003; Trewick et al., 2007; Zofall and Grewal, 2006). Having only a single obvious candidate to test, we therefore chose to focus on this factor as a possible target of Cul4-Ddb1Cdt2. Using homologous recombination, we epitope-tagged the endogenous epe1+ coding sequence with a CBP-23FLAG tag (Epe1-FLAG) and found that the steady-state level of the Epe1 protein was 3-fold higher in ddb1Δ spd1Δ and cdt2Δ spd1Δ mutants than in wild-type (WT) cells (Figures 2A and 2B). This increase in Epe1 protein levels was not due to changes in transcription or mRNA stability, as the mutants did not display a difference in epe1+ mRNA levels relative to WT cells (Figure 2C). These findings suggested that the degradation of Epe1 might be affected in the ddb1Δ and cdt2Δ mutants.
Figure 2. Cul4-Ddb1Cdt2 Promotes the Ubiquitylation and Degradation of the JmjC Protein Epe1.
(A) Western blot of C-terminally tagged Epe1 (Epe1-FLAG) expressed from its endogenous locus. Loading control: RNA polymerase II CTD repeat (RNAPII).
(B) Quantification of protein levels. Epe1-FLAG protein levels were normalized to RNAPII. Shown are mean values relative to WT with SEM of five independent biological experiments.
(C) epe1+ mRNA levels. Shown are transcript levels relative to WT with SEM from independent experiments (n = 4–5).
(D) Cycloheximide (CHX) chase experiments. For the ddb1Δ spd1Δ and cdt2Δ spd1Δ samples, half of the total protein amount was loaded to better visualize changes in the decay rates of Epe1. Loading control: RNAPII.
(E) Quantification of Epe1 decay. Epe1 protein levels were normalized to RNAPII and plotted versus time after CHX addition (time = 0 was set to 100%). Data-are represented as mean ± SEM of independent experiments (n = 7–14) and fitted for exponential decay. Single and double asterisks indicate p values of < 0.05 and < 0.01, respectively (Student’s t test).
(F) Protein levels after treatment with hydroxyurea (HU). Epe1-FLAG and Myc-Cdt2 were expressed from their endogenous loci and analyzed at the designated time points after HU treatment (20 mM) for protein (top panels) and mRNA (lower graph) levels. Upper graph: levels of Epe1-FLAG and Myc-Cdt2, normalized to RNAPII and plotted as percentage of the relative maximum protein level. Lower graph: mRNA levels of epe1-FLAG and Myc-cdt2 plotted as percentage of the maximum of mRNA level.
(G) In vivo ubiquitylation of Epe1-FLAG in WT and ddb1Δ spd1Δ cells expressing 6His-ubiquitin. Input fraction (0.005%) and precipitated 6His-ubiquitin conjugates were analyzed by anti-FLAG (upper panels) and anti-His (lower panels) immunoblotting. Negative control: WT cells expressing untagged Epe1.
To examine the half-life of Epe1 protein, we performed cycloheximide chase experiments. As shown in Figures 2D and 2E, we observed for WT cells that Epe1 is initially rapidly degraded, followed by a slower turnover after 20–30 min. This degradation kinetics suggests that distinct pools of Epe1 exist in the cell, which may be turned over by different pathways. In agreement with the increased steady-state protein levels, we found that Epe1 is stabilized in ddb1Δ spd1Δ cells and in cdt2Δ spd1Δ cells (Figure 2E). This result implies that Cul4-Ddb1Cdt2 promotes degradation of Epe1 in vivo. Since Cdt2 is transcriptionally induced during S phase (Liu et al., 2005), we examined whether Epe1 levels decreased in a Cdt2-dependent fashion upon the induction of an S phase arrest using hydroxyurea (HU). Indeed, we observed that, upon addition of HU to asynchronous cultures, Epe1 levels decreased in WT cells but not in cdt2Δ cells (Figure 2F). epe1+ mRNA levels dropped modestly during the time course, but there was no difference in this phenotype between WT and cdt2Δ cells, indicating that the Cdt2-dependent drop in protein levels was due to turnover rather than an indirect effect of Cdt2 on epe1+ mRNA levels. Similarly, we found that the HU-induced turnover of Epe1 was blocked in cells lacking Ddb1 (Figure S2).
To further analyze the role of Cul4-Ddb1Cdt2 in the regulation of Epe1, we examined whether Epe1 is ubiquitylated in vivo and whether ubiquitylation is diminished in the ddb1Δ spd1Δ mutant. In order to enrich for ubiquitylated Epe1 conjugates, we coexpressed N-terminally His-tagged ubiquitin (His-Ub) in WT and ddb1Δ spd1Δ cells, both expressing Epe1-FLAG, and performed pull-down experiments against the His-tag under denaturing conditions. When the precipitated His-Ub conjugates were analyzed by anti-FLAG immunoblots, we detected distinct, Epe1-FLAG-specific bands that show a slower migration pattern, indicating that a fraction of Epe1 is modified by ubiquitin (Figure 2G). Notably, whereas the levels of nonmodified Epe1 are increased in the ddb1Δ spd1Δ cells compared to WT cells, the corresponding ubiquitin conjugates are significantly decreased in the mutant (Figure 2G). To quantify the decrease in Cul4-Ddb1Cdt2-dependent Epe1 ubiquitylation, we determined the ratio of Epe1-ubiquitin conjugates (pull-down samples) to nonmodified Epe1 (input) and found that the relative level of ubiquitylated Epe1 was about 3-fold reduced in the ddb1Δspd1Δ mutant compared to WT cells (Figure 2G). Collectively, these results demonstrate that Epe1 is ubiquitylated and degraded in a Cul4-Ddb1Cdt2-dependent manner. The remaining amount of Epe1-ubiquitin conjugates observed in ddb1Δ spd1Δ cells suggests that other ubiquitylation routes exist and is consistent with our findings that degradation of Epe1 is not entirely abrogated in cells lacking Cul4-Ddb1Cdt2.
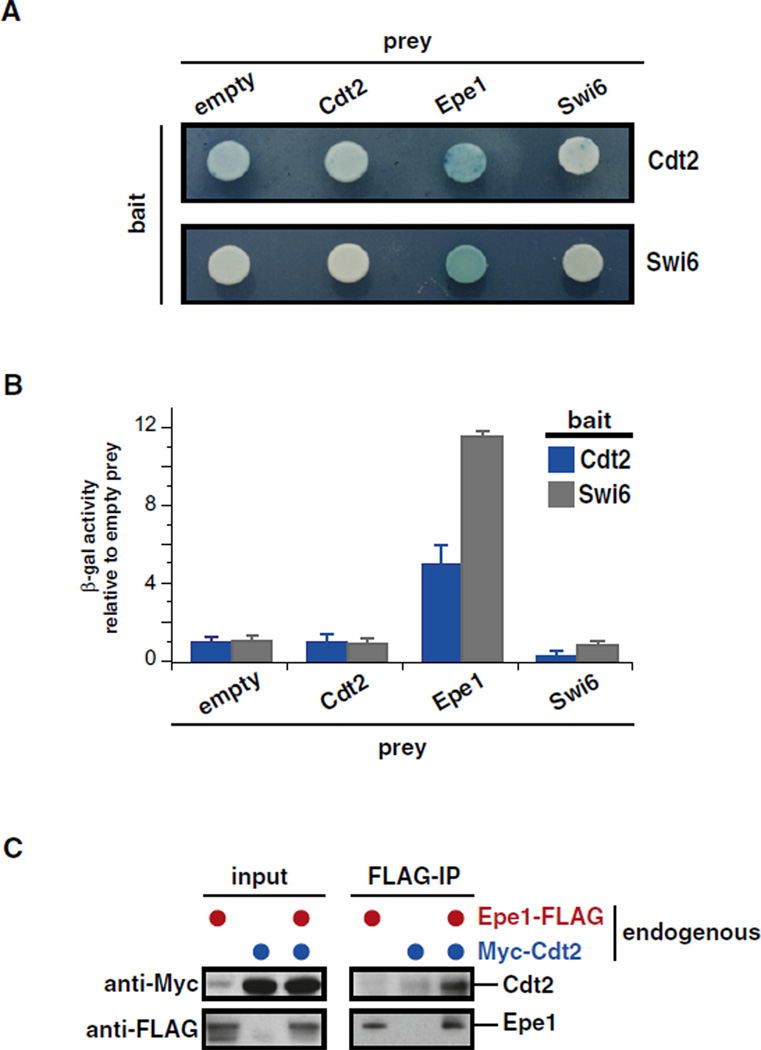
As Cdt2 is a substrate recognition component of Cul4-Ddb1 ubiquitin ligases, we tested whether it binds to Epe1. We first examined by two-hybrid analysis whether Cdt2 and Epe1 interacted. Indeed, we found that a Cdt2-lexA DNA-binding domain bait fusion interacted with an Epe1-B42 activation domain prey fusion but not a control prey fusion (Figures 3A and 3B). As might be expected, this interaction appeared to be weaker than the interaction between Epe1 and Swi6 (Figures 3A and 3B). Because the Epe1 DNA-binding fusion protein activated transcription strongly in the absence of a prey, it could not be used to examine interactions. We next generated S. pombe strains harboring epitope-tagged versions of Epe1 and Cdt2 expressed from their endogenous loci. Coimmunoprecipitation experiments on whole-cell extracts derived from these strains confirmed a biochemical interaction between Epe1 and Cdt2 (Figure 3C). These data support the view that Cul4-Ddb1Cdt2 directly recognizes Epe1 to promote its ubiquitylation and degradation in vivo.
Figure 3. Cdt2 Physically Interacts with Epe1.
(A) Plate yeast two-hybrid analysis. Photographs of plates were taken 1 (bottom panel) or 2 days (top panel) after exposure to X-gal.
(B) Quantitative yeast two-hybrid analysis. β-gal activity was normalized to the empty prey for each bait and plotted for Cdt2 (blue) and Swi6 (gray). Error bars: standard deviation (SD) of three replicates.
(C) Coimmunoprepcipation of Cdt2 with Epe1. Strains expressing endogenous levels of Epe1-CBP-2×FLAG, Myc13-Cdt2, or both were subjected to anti-FLAG immunoprecipitation. Input and immunoprecipitated material were analyzed by anti-Myc (top panel) and anti-FLAG (bottom panel) immunoblots. Note that the anti-Myc antibody slightly crossreacts with an unspecific band that comigrates with Myc13-Cdt2 seen in the untagged anti-Myc control lane of the input fraction.
Cul4-Ddb1Cdt2 Confines Epe1 to Heterochromatin Boundaries
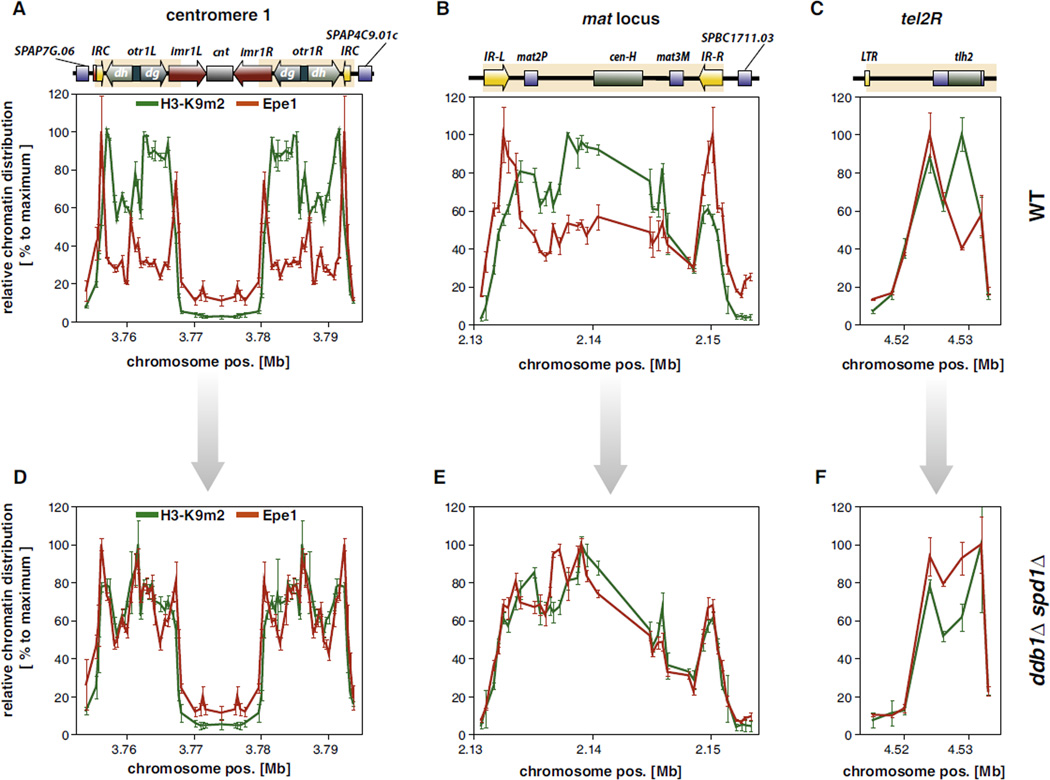
To determine whether Cul4-Ddb1Cdt2 affects the levels of Epe1 on chromatin, we performed extensive ChIP experiments in cells expressing Epe1-FLAG. In agreement with a previous study (Zofall and Grewal, 2006), we found that Epe1 can be detected in WT cells at sites within the pericentromeric region (Figure 4A), the silent mating type locus (Figure 4B), and the right telomeric end of chromosome 2 (tel2R, Figure 4C). In addition, Epe1 is present at a meiotic gene, mei4, but not at other nearby euchromatic genes (Figure 4D). It is important to note that, however, the pattern of Epe1 within heterochromatin is not uniform. In agreement with its function in boundary formation, Epe1 is enriched at the margins of heterochromatin with distinct peaks coinciding with the heterochromatic boundaries flanking the outer repeats (at the IRC elements) and inner most repeats of centromere 1, the left and right boundaries of the silent mating type locus (IR-R/L), and the telomere-distal side of the telomeric tlh2+ locus. When we explored the chromatin profile of Epe1 in the ddb1Δ spd1Δ mutant, we observed a strong accumulation of Epe1 at all heterochromatic domains as well as the meiotic mei4 gene (Figures 4A–4D). These increases in chromatin-associated Epe1 were also observed in cdt2Δ spd1Δ cells (Figure 4; lower panels) but absent in spd1Δ single mutants (Figure S3). Importantly, the accumulation of Epe1 in mutant cells was not confined to the boundaries but was seen in heterochromatic regions that are relatively depleted of Epe1 in WT cells. In particular, we observed for chromatin-bound Epe1 an increase up to 7-fold in the body of the mat2/3 silent locus but only 2-fold at the IR-R/L boundary elements (Figure 4B). These results demonstrate that the altered Epe1 levels on chromatin do not merely reflect the increase in cellular Epe1 levels but indicate a significant change in the chromosomal distribution of Epe1 in absence of Cul4-Ddb1Cdt2.
Figure 4. Deletion of ddb1+ Causes Accumulation of Epe1 within Heterochromatin Domains.
ChIP analysis of Epe1 at centromere 1 (A), the silent mating type region (B), the subtelomeric region of telomere 2 (C), and a meiotic gene locus (D) in WT (blue) and ddb1Δ spd1Δ cells (red). Upper panels: ChIP signals normalized to act1+. Lower panels: fold enrichment of Epe1 in ddb1Δ spd1Δ (red) and cdt2Δ spd1Δ (dark red) relative to WT. Data are represented as mean ± SEM of three independent experiments.
See also Figure S3.
To understand the mechanisms that determine the heterochromatic distribution of Epe1, we compared its chromatin profile with the pattern of H3K9me2. Previous work demonstrated that Epe1 is recruited to H3K9 methyl marks by the HP1 proteins Swi6 and Chp2 (Sadaie et al., 2008; Zofall and Grewal, 2006), which bind preferentially to di- and trimethylated H3K9 and show a virtually identical chromatin distribution to H3K9me2 (Noma et al., 2001; Sadaie et al., 2008). Surprisingly, we found that in WT cells the chromatin distributions of Epe1 and H3K9me2 are quite disparate at every heterochromatic domain tested (Figures 5A–5C), implying that the recruitment to heterochromatin is not sufficient to explain the specific chromatin profile of Epe1. In striking contrast, the profiles of Epe1 and H3K9me2 are nearly indistinguishable in the ddb1Δ spd1Δ mutant for the centromere and the silent mating type locus (Figures 5D and 5E); both profiles become similar for the subtelomeric tel2R region as well (Figure 5F). These findings strongly suggest that although H3K9me2 mediates the initial recruitment of Epe1 to heterochromatin via HP1 proteins, the distribution of Epe1 within heterochromatic domains, and in particular its restriction to boundaries, is shaped by its removal from specific heterochromatic regions by the action of the Cul4-Ddb1Cdt2 complex.
Figure 5. Epe1 Is Confined to Heterochromatic Boundaries in Wild-Type but Spreads through Entire Heterochromatin Domains in Cells Lacking Ddb1.
Relative chromatin distribution of Epe1 (red) and H3K9me2 (green) within heterochromatic regions in WT (A–C) and ddb1Δ spd1Δ cells (D–F). ChIP for Epe1-FLAG and H3K9me2 were performed as described in Figure 1F and Figure 4. ChIP data were act1+ normalized and median centered. Data are represented as mean ± SEM of three independent experiments relative to the maximum (100%) of each heterochromatic region.
Regulation of Epe1 by Cul4-Ddb1Cdt2 Acts in Parallel with the CK2-Swi6 Pathway
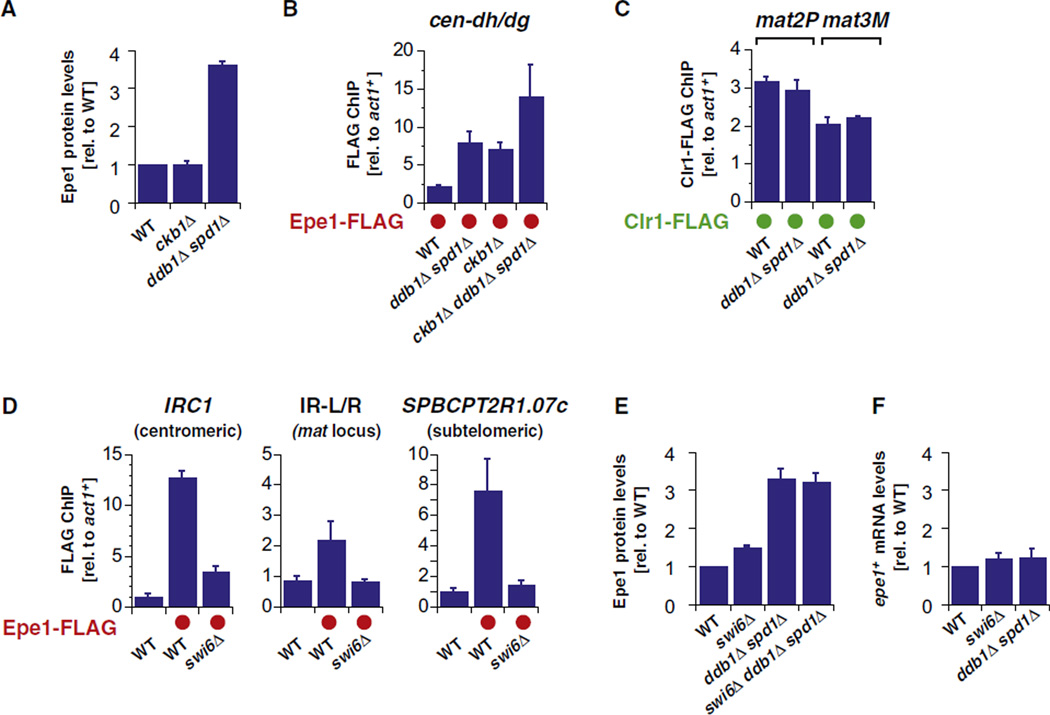
Because Epe1 is tethered to heterochromatin by silencing factors, we tested whether the modification state of heterochromatin influenced its turnover. Phosphorylation of Swi6 by CK2 has been shown recently to inhibit the association of Epe1 with heterochromatin and to promote the binding of the SHREC effector complex (Shimada et al., 2009). Because CK2 mutants and cells lacking Cul4-Ddb1Cdt2 both display increased association of Epe1 with heterochromatin, we considered the hypothesis that they function in a single pathway in which phosphorylation of Swi6 by CK2 triggers the turnover of Epe1. This hypothesis makes three predictions: (1) Epe1 protein should accumulate in mutants of CK2, e.g., cells lacking its regulatory subunit Ckb1, (2) double mutants lacking ckb1Δ and the ubiquitin ligase should show the same increase in Epe1 association with heterochromatin as the single mutants, and (3) mutants lacking Cul4-Ddb1Cdt2 should display a decrease in the binding of SHREC to heterochromatin seen in ckb1Δ mutants. As shown in Figures 6A–6C and Figure S4A, we obtained data that contradicted each of these predictions. Epe1 does not accumulate in ckb1Δ cells (Figure 6A), the double mutants show more Epe1 association with heterochromatin than the single mutants (Figure 6B and Figure S4A), and SHREC occupancy is unaffected in ligase-deficient cells (Figure 6C and Figure S4B). These data indicate that the two mechanisms operate in parallel (rather than in a single pathway) to regulate Epe1.
Figure 6. Cul4-Ddb1Cdt2 -Dependent Degradation of Epe1 Acts Independently of HP1 Phoshorylation by Casein Kinase II.
(A) Epe1-FLAG protein levels from quantified western blots (normalized to RNAPII). Shown are mean values relative to WT with SEM from five independent experiments (except ddb1Δ spd1Δ with n = 2; error shows the variation from the mean).
(B) ChIP analysis of Epe1-FLAG levels at centromere 1 (region between cen-dh and -dg). ChIP signals were normalized to act1+. Shown are mean values with SD of three parallel IP samples of one representative experiment.
(C) ChIP analysis of Clr1-FLAG levels at the silent mating type region. Shown are mean values of two independent experiments with error bars representing the variation from the mean.
(D) ChIP analysis of Epe1-FLAG levels at the outer boundary of centromere 1 (left panel), at inverted repeats of silent mating type region (middle panel), and at subtelomeric locus telomere-distal of tlh2 (right panel) in WT and swi6Δ cells. Shown are mean values with SD of three parallel IP samples of one representative experiment.
(E) Epe1-FLAG protein levels. For comparison, the level of Epe1-FLAG in ddb1Δ spd1Δ (from Figure 2B) is also shown. Shown are mean values relative to WT with error bars (SEM) from independent experiments (n = 4–5).
(F) epe1-FLAG mRNA levels. For comparison, the level of epe1-FLAG in ddb1Δ spd1Δ (from Figure 2C) is displayed. Shown are mean values relative to WT with error bars (SEM) from independent experiments (n = 4–8).
See also Figure S4.
Given that Swi6 phosphorylation by CK2 is not required for Epe1 turnover, we examined whether Swi6 was required for Epe1 regulation. We first confirmed and extended previous data demonstrating that Swi6 is required for the association of Epe1 with heterochromatin, finding that at boundaries, Epe1 association was either completely (IR-L/R) or nearly completely (IRC1) eliminated in swi6Δ cells (Figure 6D and Figures S4C and S4D). Next we tested whether Epe1 levels accumulate to those seen in ddb1Δ and cdt2Δ mutants when swi6+ is deleted. We found only a subtle increase in Epe1 levels in swi6Δ cells, indicating that Swi6 is not critical for Epe1 turnover (Figures 6E and 6F). These results demonstrate that heterochromatin association is not required for Epe1 turnover. Nonetheless, given that Epe1 is a chromatin-bound protein (Sadaie et al., 2008; Shimada et al., 2009; Zofall and Grewal, 2006), it seems likely that its ubiquitylation and its regulation occur in the context of chromatin (see Discussion).
Regulation of Epe1 Is Sufficient to Explain the Role of Cul4-Ddb1Cdt2 in Heterochromatin Formation
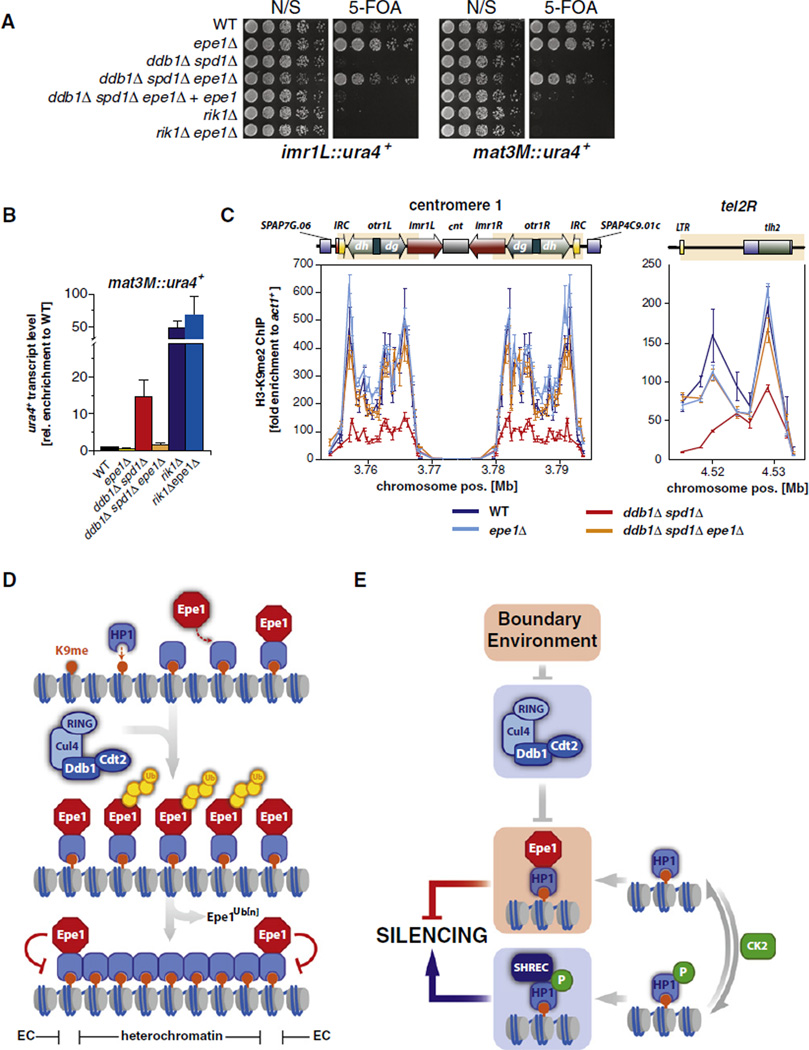
Next, by deleting epe1+ in a ddb1Δ spd1Δ strain, we examined whether the misregulation of Epe1 accounts for the defects in heterochromatin formation observed in cells lacking Ddb1. Indeed, by using silencing reporter assays, we found that the silencing defect of ddb1Δ spd1Δ cells was suppressed in the ddb1Δ spd1Δ epe1Δ triple mutant at the pericentromeric region and the mating type locus (Figure 7A). This suppression was specific for the Cul4-Ddb1Cdt2 pathway, as deletion of epe1+ did not suppress the silencing defect of cells lacking Rik1, the Ddb1 paralog in the Clr4-associated ubiquitin ligase Cul4-Rik1Dos1. Furthermore, the suppression of the silencing defect of ddb1Δ spd1Δ was due to the loss of Epe1, as complementation of the epe1Δ mutation by reintroducing epe1+ completely reverted the suppression phenotype (Figure 7A). Consistent with these silencing reporter assay results, RT-qPCR measurements revealed that the levels of ura4+ transcripts originating from the mat3M::ura4+ locus were reduced in ddb1Δ spd1Δ epe1Δ cells to WT levels (Figure 7B).
Figure 7. Deletion of epe1+ Suppresses the Silencing Defect of Cells Lacking Ddb1.
(A) Reporter gene assays. N/S, nonselective; 5-FOA, 5′-fluoroorotic acid.
(B) RT-qPCR of ura4+ transcript levels derived from mat3M::ura4+. Shown are mean values relative to WT ± SEM of three independent experiments.
(C) ChIP analysis of H3K9me2 at centromere 1 and the right arm subtelomeric region of chromosome 2. Shown are mean values ± SD of three parallel IP samples of one representative experiment.
(D) Model for boundary formation through recruitment of Epe1 to heterochromatin by HP1 proteins and its subsequent removal from central heterochromatic domains by Cul4-Ddb1Cdt2. See text for details.
(E) Independent pathways regulate Epe1 at chromatin. See text for details.
See also Figure S5.
In agreement with a previous study (Trewick et al., 2007), we found that epe1Δ single mutants display a quantitative increase in centromeric transcripts (Figure S5), precluding a similar analysis at these regions. We instead probed the suppression of the ddb1Δ-associated silencing defects by investigating the level of H3K9me2 at pericentromeric and telomeric regions, which are decreased in cells lacking Ddb1 (Figure 1F). Remarkably, we observed that H3K9me2 levels were restored to WT levels in a ddb1Δ spd1Δ epe1Δ mutant at the pericentromeric region (Figure 7C). The H3K9me2 defect was also suppressed in this triple mutant at tel2R to levels seen in an epe1Δ single mutant. These results indicate that the reduced levels of H3K9me2 at these heterochromatic loci are caused by misregulation of Epe1 in cells lacking Cul4-Ddb1Cdt2. Collectively these findings demonstrate that degradation of Epe1 is sufficient to explain the requirement of Cul4-Ddb1Cdt2 for silencing.
DISCUSSION
Our study identified a regulatory mechanism required for proper boundary architecture and heterochromatic silencing in S. pombe. This mechanism involves the conserved ubiquitin ligase Cul4-Ddb1Cdt2, which targets the JmjC protein Epe1 for ubiquitin-dependent degradation. Epe1 antagonizes the spread of heterochromatin and has a potential role in boundary formation (Ayoub et al., 2003), yet it is found within heterochromatic domains and associates directly with the H3K9me-binding protein Swi6 (Zofall and Grewal, 2006). This paradoxical finding raises the fundamental question of how Epe1 is precluded from interfering with heterochromatin formation. Our findings demonstrate that Cul4-Ddb1Cdt2 controls the chromosomal landscape of Epe1 in a manner that substantially restricts its accumulation to heterochromatic boundaries by limiting its spreading into the bodies of heterochromatic domains. This heterochromatin-shaping function of Cul4-Ddb1Cdt2 is required for silencing.
A Conserved Ubiquitin Ligase Promotes Silencing by Targeting a Silencing Inhibitor
We identified Ddb1 and Cdt2 as silencing factors in targeted knockout screens for pericentromeric silencing and demonstrated their requirement for the integrity of other heterochromatic domains. Mutants of ddb1+ and cdt2+ are indistinguishable in their silencing defects and are epistatic to each other (Figure 1). Ddb1 and Cdt2 are highly conserved proteins (25% and 26% identity, 47% and 44% similarity, respectively, between the fission yeast and human homologs). Both proteins were originally identified as a heterodimeric factor recruited to DNA upon damage by ultraviolet irradiation (UV) (Dualan et al., 1995; Keeney et al., 1993), and mutations in the DCAFs DDB2 and CSA are associated with the human diseases Xeroderma pigmentosum complementation group E (XP-E) and the Cockayne Syndrome (CS), respectively (O’Connell and Harper, 2007). Although more than 50 different DCAFs have been identified (Lee and Zhou, 2007), the number of known substrates is significantly smaller, reflecting the difficulty of identifying substrates of ubiquitin ligases. Notably, the known substrates are predominantly chromatin-associated proteins, suggesting a specialized role for Cul4-Ddb1 ligases in nuclear processes (O’Connell and Harper, 2007). Here we show that Cul4-Ddb1Cdt2 targets Epe1 in vivo (Figure 2) and that the putative substrate recognition subunit Cdt2 interacts with Epe1 (Figure 3). In WT cells, Epe1 is polyubiquitylated and degraded by an initial rapid and a late slow decay. Conversely, in cells lacking Ddb1 or Cdt2, Epe1 is significantly stabilized. Ubiquitylation of Epe1 is not completely abolished in ddb1Δ mutant cells, and only the rapid decay component is abrogated in the mutants, suggesting that other ligases likely also target Epe1. Nonetheless, the regulation of Epe1 by Cul4-Ddb1Cdt2 appears to be sufficient to explain the role of the ligase in silencing: The silencing defect at the mat3M locus and the decrease of H3K9 methylation at pericentromeric regions in ddb1Δ mutants are completely suppressed by removal of Epe1.
Sculpting Heterochromatin by Preventing the Internal Spread of a Silencing Inhibitor
Cul4-Ddb1Cdt2 affects the Epe1 levels on chromatin consistent with its known role in regulating other chromatin-associated substrates. We observed that Epe1 is located predominantly at the heterochromatic boundaries in WT cells, in agreement with the notion that Epe1 plays a role in boundary formation (Ayoub et al., 2003; Zofall and Grewal, 2006). In striking contrast, Epe1 accumulates to high levels in the bodies of heterochromatic domains in cells lacking Cul4-Ddb1Cdt2. Although our results show that turnover of Epe1 does not require its association with heterochromatin, several pieces of evidence suggest that its regulation likely takes place on chromatin (Figures 7D and 7E). First, we confirmed previous findings that show that Epe1 does not appear to have affinity for boundaries in the absence of Swi6; thus, increasing Epe1 levels per se would not be expected to result in its enrichment at boundaries. Second, changes in Epe1 levels at chromatin are not uniform in cells lacking the ubiquitin ligase but instead show a distinct pattern: a strong accumulation of Epe1 within the bodies of the heterochromatic domains but only a modest increase of Epe1 at the boundaries (Figure 4). This is not because the association of Epe1 with boundary chromatin is saturated under these conditions, as we have found that cells also lacking ckb1Δ display even higher levels of Epe1 on chromatin (Figure S4A). Third, there is only a 3-fold increase of the total pool of Epe1 in ddb1Δ and cdt2Δ mutant cells, whereas the chromatin-bound Epe1 accumulates up to 7-fold (Figure 4). Fourth, whereas the distribution of Epe1 differs substantially from the chromatin profile of H3K9me2 in WT cells, its chromatin localization is nearly identical to the H3K9me2 pattern in absence of Ddb1 and no longer shows a preference to the boundaries (Figure 5). Taken together, these findings strongly suggest that Epe1 by itself does not have any particular affinity to boundary elements, but rather that its removal from the body of heterochromatin explains its relative enrichment at boundaries.
A corollary to this model is that Epe1 must be protected from removal by Cul4-Ddb1Cdt2 at boundaries. Much evidence points to a role for nuclear envelope tethering as a requirement for boundary function (Ishii and Laemmli, 2003; Noma et al., 2006; Yusufzai et al., 2004). It is thus possible that subnuclear localization of boundary regions limits their accessibility to Cul4-Ddb1Cdt2 or the proteasome. Such a mechanism together with the ability of Swi6 to recruit Epe1 to heterochromatin could explain the enrichment of Epe1 observed at boundaries.
Posttranslational modification or the presence of auxiliary factors could also play a role in directing Cul4-Ddb1Cdt2 to Epe1. Mutants defective in phosphorylation of Swi6 by CK2 display increased accumulation of Epe1 and decreased accumulation of the SHREC ATPase/HDAC complex on chromatin (Shimada et al., 2009). Together with our observations, these published data would be compatible with a model in which phosphorylation of Swi6 triggers Epe1 turnover. However, our analysis demonstrates decisively that Swi6 phosphorylation and ubiquitylation of Epe1 by Cul4-Ddb1Cdt2 act in different pathways to regulate the heterochromatin association of Epe1 (Figure 7E). That this protein is subjected to multiple layers of regulation is striking and emphasizes the concept that tightly regulating this antisilencing factor is critical for maintaining heterochromatic domains.
Regulation of the Activity of Epe1 by Defining Its Distribution within Heterochromatin
The barrier function of Epe1 correlates with its spatial restriction to the boundaries. Conversely, when Epe1 accumulates within heterochromatic domains due to the absence of Cul4-Ddb1Cdt2, lack of phosphorylation of Swi6, or overexpression of Epe1, it acts as an antagonist of silencing (Shimada et al., 2009; Zofall and Grewal, 2006). Interestingly, mutants of epe1+ affect Pol II-dependent transcription through heterochromatin and are perturbed in their levels of heterochromatic siRNAs (Trewick et al., 2007; Zofall and Grewal, 2006). These observations may point to an additional role of Epe1 besides its barrier function that is associated with the RNAi-dependent pathway of heterochromatin formation. Indeed, we observed within the body of heterochromatic regions detectable amounts of Epe1 above background levels (Figure 4). These low levels of chromatin-bound Epe1 may represent the pool that is deposited prior to its removal by Cul4-Ddb1Cdt2. Considering that the processes of Pol II-dependent transcription through heterochromatin and siRNA formation are restricted to S phase (Chen et al., 2008; Kloc et al., 2008) and are also affected by Epe1 (Zofall and Grewal, 2006), it is possible that targeting of Epe1 by Cul4-Ddb1Cdt2 is temporally controlled. This notion is supported by the finding that Cdt2, which itself is an unstable protein, is expressed only within a short time window during S phase (Liu et al., 2005; Oliva et al., 2005). In such a scenario, initial tethering of Epe1 to Swi6 would stimulate the binding of Pol II to heterochromatin and thus the formation of siRNAs during S phase; subsequent removal of Epe1 by Cul4-Ddb1Cdt2 would then allow assembly of heterochromatin.
General Role of CRLs in Silencing
The general significance of ubiquitylation in regulating heterochromatin formation is highlighted by the specialized CRL Cul4-Rik1Dos1/2, which is associated with the histone methyltransferase Clr4 in the CLRC complex and is required for silencing. The biologically relevant substrate of this E3 and its specific role in heterochromatin formation have not been elucidated. Orthologs of Rik1 have not been identified in other eukaryotes so far; however, the requirement of coupling E3 activity with H3K9 methylation seems to be conserved. A recent study demonstrated that mutants of Cul4 and Ddb1 homologs in N. crassa are completely deficient in H3K9 methylation analogous to rik1 mutants in S. pombe (Zhao et al., 2010). Moreover Cul4 is associated with the corresponding H3K9 histone methyltransferase, suggesting that a homologous Cul4-Ddb1DCAF complex replaces the role of Cul4-Rik1Dos1 in this fungal species (Zhao et al., 2010). Intriguingly, Ddb1 and Cullin-4A were also found to be components of the CEN-complex, which associates with the centromere-specific histone H3 CENP-A in human cells (Obuse et al., 2004), suggesting a conserved role in chromatin regulation. Whether CRLs of N. crassa and mammals target inhibitory substrates analogous to Epe1 remains to be investigated.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Techniques
Standard media and genome engineering methods were used. 5-FOA media contained 1 g/l 5′-fluoroorotic acid. Synthetic complete (SC) media minus the corresponding amino acid were used for drop-out media. EMM-leu media were used for growing strains harboring pREP1 plasmids. Strains are listed in Table S2.
Library Construction and Screen
Gene disruptions were performed in an imr1L(NcoI)::ura4 otr1R(SphI)::ade6K P(h+) reporter strain (Ekwall et al., 1999).
Yeast Two-Hybrid Analysis
Plasmids containing fusion proteins of Swi6, Cdt2, and Epe1 (described in Table S3) were transformed into EGY48 (Golemis et al., 2009). Cultures were grown overnight in SC-his-trp-ura +2% raffinose, plated onto SC-his-trp-ura +1% raffinose +2% galactose, and grown for 2 days at 30°C. Cells were permeabilized by chloroform and overlayed with top agar containing X-gal as described (Richter et al.,2007). For liquid assays, overnight cultures were diluted 1:20 and grown in SC-his-trp-ura+1% raffinose +2% galactose for another 4 hr. β-galactosidase liquid assays were performed as described (Shock et al., 2009), except that 20 µl each cell culture and permeabilization buffer were used.
Chromatin Immunoprecipitation
ChIP experiments were performed essentially as described (Nobile et al., 2009). Unless otherwise noted, cells were crosslinked with 1% formaldehyde for 20 min at 30°C. To increase the ChIP sensitivity, in Figure 6D and Figures S4B–S4F, crosslinking was performed by subsequent treatment of 10 mM dimethyl adipimidate and 1.5% formaldehyde as described (Kurdistani and Grunstein, 2003), except that formaldehyde crosslinking was restricted to 30 min. Epe1-FLAG, Clr1-FLAG, and anti-H3K9me2 were immunoprecipitated with 2–5 µg antibody (anti-FLAG, Sigma F3165; anti-H3K9me2, Abcam ab 1220) from lysates corresponding to 50–75 optical density 600 (OD600) (Epe1-FLAG, Clr1-FLAG) and 15–25 OD600 (H3K9me2) of cells. Immunoprecipitated DNA was quantified by real-time PCR (qPCR) with primers listed in Table S4 and normalized against act1+.
RNA Extraction and RT-qPCR Analyses
RT-qPCR experiments were carried out as previously described (Rougemaille et al., 2008), except that RNA samples were DNaseI-treated with DNA-free kit (Ambion). Ten micrograms of RNA was used in standard RT reactions using oligo[(dT)20-N] primers. cDNAs were quantified by qPCR with the primers listed in Table S4 and normalized against act1+.
Immunotechniques
For examination of protein levels, extracts were prepared under denaturing conditions (Knop et al., 1999). Cycloheximide (CHX) chase experiments were performed as described (Braun et al., 2002) except that 0.15 µg/ml CHX was used as final concentration. Lysates corresponding to 1 OD600 of cells were analyzed by immunoblotting with anti-FLAG (Sigma, P3165) and anti-RNA polymerase II carboxy-terminal domain (CTD) repeat (Abcam ab817) antibodies diluted 1:1000 and 1:8000, respectively, in blocking solution (LI-COR). For detection and quantification, an infrared imaging system (Odyssey, Li-COR) and the corresponding software were used. Details of coimmunoprecipitation experiments can be found in the Extended Experimental Procedures.
Ubiquitin Pull-Down Experiments
Expression of nmt1 promoter-driven 6His-ubiquitin (pREP1-6His-Ubi) was performed as described. Thirty to Forty-five minutes prior to harvest, cells were treated with 5 mM NEM added directly to the growth medium. Protein extraction and binding of ubiquitin conjugates were done under denaturing conditions essentially as described (Sacher et al., 2005). Further details can be found in the Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Rape, Stefan Jentsch, Geeta Narlikar, David Morgan, members of their labs, and Ulrike Boettcher for critical reading of the manuscript. We are grateful to Danesh Moazed, Karl Ekwall, and Takashi Toda for strains, plasmids, and protocols. S.B., M. Rougemaille, and S.S. were supported by postdoctoral fellowships of the German Research Foundation (BR 3511/1-1), the Human Frontier Science Program, and the Leukemia and Lymphoma Society, respectively. J.F.G. was supported by an NIH/NIGMS IMSD predoctoral fellowship (R25-GM56847). This work was supported by a grant to H.D.M. from the National Institutes of Health (GM071801) and a scholar’s award to H.D.M. from the Leukemia and Lymphoma Society.
Footnotes
Note that a fraction of nonubiquitylated Epe1 can also be detected in the pull-down samples, probably due to the presence of several His-residue clusters within the Epe1 protein. Graph below shows the mean values of the ubiquitylation level of Epe1 relative to WT of three independent experiments (error bars = SEM). Total Epe1-ubiquitin conjugates (without the nonmodified Epe1 fraction) was quantified by densitometry of anti-FLAG western blots and normalized for the input level. See also Figure S2.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and four tables and can be found with this article online at doi:10.1016/j.cell.2010.11.051.
S.B. and H.D.M. designed the study. S.B., M. Rowley, M. Rougemaille, and S.S. constructed and characterized the deletion library and strains. S.B. and M. Rowley performed the screen. J.F.G. designed and performed the experiments shown in Figure 3. S.B. performed the experiments shown in Figure 1, Figure 2, Figure 4, Figure 5, and Figure 6. S.B. and H.D.M. wrote the manuscript. All authors contributed to editing the manuscript.
REFERENCES
- Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- Ayoub N, Noma K-I, Isaac S, Kahan T, Grewal SIS, Cohen A. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T, Ponomarev A, Raychaudhuri P. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J. Biol. Chem. 2004;279:9937–9943. doi: 10.1074/jbc.M312570200. [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, Fitzgerald PC, Grewal SIS. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. Current Protocols in Protein Science. 2009;Chapter 19(Unit19.12) doi: 10.1002/0471140864.ps1902s14. [DOI] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E-JE, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Ishii K, Laemmli UK. Structural and dynamic functions establish chromatin domains. Mol. Cell. 2003;11:237–248. doi: 10.1016/s1097-2765(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Keeney S, Chang GJ, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum. E. J. Biol. Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida S-h, Shimoda C, Holmberg C, Nielsen O, Carr AM. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi Y-h, Chandrasekharan MB, Sun Z-W, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault J-S, Nantel A, Andes DR, Johnson AD, Mitchell AP. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000133. e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Noma K-I, Cam HP, Maraia RJ, Grewal SIS. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006;7:1134–1139. doi: 10.1038/sj.embor.7400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 2007;26:2454–2464. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Shankar S, Braun S, Rowley M, Madhani HD. Ers1, a rapidly diverging protein essential for RNA interference-dependent heterochromatic silencing in Schizosaccharomyces pombe. J. Biol. Chem. 2008;283:25770–25773. doi: 10.1074/jbc.C800140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Jentsch S. Identification of SUMO-protein conjugates. Methods Enzymol. 2005;399:392–404. doi: 10.1016/S0076-6879(05)99027-7. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J-I. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Shimada A, Dohke K, Sadaie M, Shinmyozu K, Nakayama J-I, Urano T, Murakami Y. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009;23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock TR, Thompson J, Yates JR, Madhani HD. Hog1 mitogen-activated protein kinase (MAPK) interrupts signal transduction between the Kss1 MAPK and the Tec1 transcription factor to maintain pathway specificity. Eukaryot. Cell. 2009;8:606–616. doi: 10.1128/EC.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z-W, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y-i, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Shen Y, Yang S, Wang J, Hu Q, Wang Y, He Q. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J. Biol. Chem. 2010;285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SIS. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.