Abstract
Protein synthesis is targeted by numerous, chemically distinct antibiotics that bind and inhibit key functional centers of the ribosome. Using single-molecule imaging and X-ray crystallography, we show that the aminoglycoside neomycin blocks aminoacyl–transfer RNA (aa-tRNA) selection and translocation as well as ribosome recycling by binding to helix 69 (H69) of 23S ribosomal RNA within the large subunit of the Escherichia coli ribosome. There, neomycin prevents the remodeling of intersubunit bridges that normally accompanies the process of subunit rotation to stabilize a partially rotated ribosome configuration in which peptidyl (P)-site tRNA is constrained in a previously unidentified hybrid position. Direct measurements show that this neomycin-stabilized intermediate is incompatible with the translation factor binding that is required for distinct protein synthesis reactions. These findings reveal the functional importance of reversible intersubunit rotation to the translation mechanism and shed new light on the allosteric control of ribosome functions by small-molecule antibiotics.
A broad array of chemically diverse antibiotic compounds inhibit bacterial protein synthesis by targeting functional centers within the 2.4-MDa ribosome particle, an RNA–protein complex that is composed of a small (30S) and a large (50S) subunit1–3. A majority of these compounds operate by altering messenger RNA (mRNA) and tRNA interactions within the decoding, peptidyl transferase and GTPase centers of the small and large ribosomal subunits4, 5. Despite the effectiveness of such compounds in the clinical treatment of infectious disease, the growing prevalence of pathogens resistant to these agents6–8 has created a pressing need to identify new target sites in the ribosome with alternative modes of action that can be leveraged for the regulation of the translation machinery9, 10. In pursuit of this goal, we used single-molecule fluorescence and X-ray crystallographic methods to gain a deeper understanding of the mechanisms by which aminoglycoside-class antibiotics inhibit bacterial translation.
Aminoglycoside antibiotics in the 2-deoxystreptamine (2-DOS) family are broad-spectrum bactericidal agents that are used to treat Gram-negative bacterial infections. In vivo, these compounds have been shown to allosterically alter the mechanism of aa-tRNA selection during mRNA decoding on the ribosome11–13 by inducing local rearrangements in ribosomal RNA (rRNA) within the highly conserved helix 44 (h44) decoding site of the small (30S) subunit. Thus, 2-DOS aminoglycosides promote the inappropriate incorporation of near-cognate aa-tRNA (one mismatch with the mRNA codon) and noncognate aa-tRNAs (two or three mismatches with the mRNA codon) into the aminoacyl (A) tRNA binding site. Increased aa-tRNA misincorporation is thought to eventually exceed the capacity of the cell to cope with decreased translational fidelity14, ultimately resulting in cell death15. However, as reductions in translation fidelity alone typically have little effect on cell growth1, 16, the bactericidal nature of the action of 2-DOS aminoglycosides remains poorly understood.
In vitro, 2-DOS aminoglycosides also inhibit a range of distinct steps in the translation mechanism. These include mRNA and tRNA translocation, the directional movement of substrates with respect to both subunits of the intact (70S) ribosome1, 17 and ribosome recycling, the process of subunit separation after the termination phase of protein synthesis18. Although the physical origins of these aminoglycoside-induced effects are unclear, early biochemical studies revealed that aminoglycosides can bind to regions of the ribosome that are outside the canonical mRNA decoding site19, 20. Recently, the aminoglycoside neomycin (Fig. 1a) was shown crystallographically to bind to the bacterial ribosome within H69 of 23S rRNA in the large (50S) subunit21. Binding at this site was hypothesized to provide a potential explanation for the inhibition of both ribosome recycling and substrate (mRNA and tRNA) translocation17, 21.
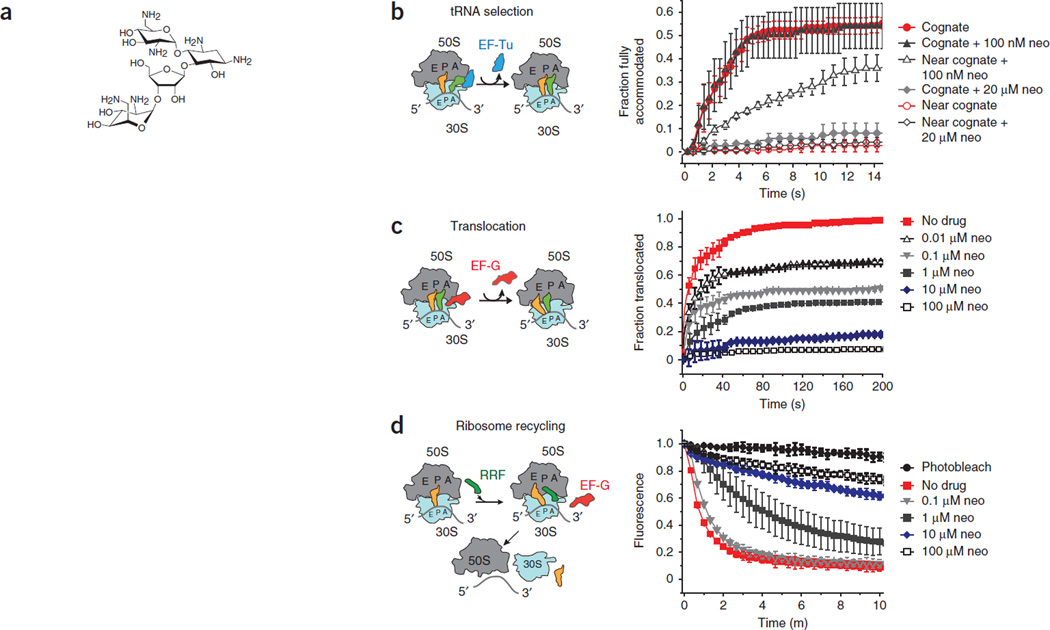
Figure 1.
Neomycin inhibits ribosome functions in vitro. (a) The chemical structure of neomycin. (b) EF-Tu (GTP)-catalyzed accommodation of Phe-tRNAPhe at the A site of wild-type ribosomes programmed with cognate (UUC) and near-cognate (UCU) codons monitored by FRET. The incorporation of cognate tRNA and near-cognate tRNA in the absence of antibiotics is shown. The incorporation of cognate tRNA and near-cognate tRNA in the presence of two concentrations of neomycin (neo) are shown. Experiments were conducted in triplicates, and the mean ± s.d. are plotted. (c) EF-G–catalyzed translocation, monitored by smFRET, normalized to a control group receiving no neomycin (no drug). The fractions of translocated molecules at various concentrations of neomycin are shown. Experiments were conducted in triplicates, and the mean ± s.d. are plotted. (d) Recycling of wild-type 70S ribosome complexes monitored by the disappearance of Cy5-labeled L1 fluorescence in 50S subunits from surface-immobilized wild-type ribosome complexes. Ribosome recycling at various concentrations of neomycin is shown. Under identical conditions, photobleaching was negligible. Experiments were conducted in triplicates, and the mean ± s.d. are plotted.
Large-scale conformational changes within the ribosome contribute to every aspect of the translation cycle: initiation, elongation, termination and recycling22. Rearrangements at the subunit interface have been implicated in the joining of large and small subunits during initiation and the positioning of tRNA substrates during processive elongation reactions, as well as the binding of translation factors during termination and the release of the protein products23, 24. Conformational plasticity of the centrally located intersubunit bridge B2, formed by the interaction of H69 of the large subunit with h44 of the small subunit, has been posited to function as a potential regulator of translation processes25, 26. Recently reported ribosome structures have revealed that remodeling in this region—immediately proximal to the tRNA and mRNA binding sites—facilitates rotation of the small subunit with respect to the large subunit. This axis of movement affects almost every aspect of the translation mechanism27. Here, using a combination of single-molecule fluorescence and X-ray crystallographic methods27, we show that global inhibition of translation can be induced by neomycin through its specific interaction with H69 of the large subunit. Binding at this site allows the drug to allosterically control ribosome dynamics in a manner that affects multiple aspects of the translation mechanism.
RESULTS
Neomycin activities beyond the mRNA decoding site
Selection of cognate compared to near-cognate tRNA in mRNA decoding involves multiple kinetic steps that result in the accommodation of aa-tRNA into the ribosomal A site14, 28. To probe the effects of neomycin on mRNA decoding, we used pre–steady state single-molecule fluorescence resonance energy transfer (smFRET) to measure the rate and extent of aa-tRNA incorporation into the A site29 as a function of neomycin concentration. In line with the propensity of neomycin to induce translational miscoding10, 30, these smFRET measurements showed that low concentrations of neomycin (100 nM) substantially increased the rates of near-cognate aa-tRNA accommodation into the ribosomal A site (Fig. 1b). As expected from the results of the earlier studies, neomycin had negligible effects on the selection of cognate tRNA (Fig. 1b).
We did not find any evidence of neomycin-induced miscoding when we examined ribosomes containing the well-established A1408G resistance mutation in 16S rRNA (Supplementary Fig. 1a), which abrogates neomycin binding to the h44 decoding site31. However, when we performed identical experiments at a higher concentration of neomycin (20 µM), the selection of both cognate and near-cognate tRNA was strongly attenuated (Fig. 1b). Notably, the inhibition of both cognate and near-cognate tRNA selection at this elevated concentration of neomycin persisted in the context of the A1408G mutant ribosomes (Supplementary Fig. 1a). These findings suggest that the inhibition of aa-tRNA accommodation may involve a mechanism that is distinct from the neomycin-induced miscoding that results from binding within h44, where neomycin binds outside of the canonical mRNA decoding site.
In line with previous investigations17, single-molecule measurements of substrate translocation32 showed that neomycin also strongly inhibited the rate and extent of directional substrate movement with respect to the small subunit (Fig. 1c). Moreover, the neomycin-induced translocation inhibition profiles were similar for wild-type and A1408G ribosomes (compare Fig. 1c and Supplementary Fig. 1b). Neomycin also inhibited ribosome recycling, a process that is mediated by the ribosome recycling factor (RRF) and elongation factor G (EF-G)33. Again, the dependence of translocation inhibition on concentration was similar in wild-type and A1408G ribosomes (compare Fig. 1d and Supplementary Fig. 1c). Together, these findings implicate the binding of neomycin to one or more binding sites outside of the h44 decoding site as being responsible for the inhibition of substrate translocation and ribosome recycling.
Neomycin binding alters small-subunit rotation dynamics
To test whether neomycin inhibits translation by altering global conformational processes in the ribosome, we developed a new smFRET approach for detecting intersubunit rotation and the accompanying process of the formation of an ‘unlocked state’, both of which precede, and are crucial to, mRNA and tRNA translocation and ribosome recycling23, 32, 34. Formation of the unlocked state is achieved through an approximately 9° rotation of the small subunit with respect to the large subunit, which entails numerous rearrangements in key inter-subunit bridges, including B2 and B7a27. Related to this process, deacylated P-site tRNA also moves from its classical configuration to a hybrid configuration, and the L1 stalk—so named because it contains the L1 protein—compacts toward the subunit interface23, 27. Contacts between the 3′ CCA end of tRNA and the large subunit exit (E) site, as well as between the elbow domain of hybrid (P/E) tRNA and the L1 stalk, partially stabilize the unlocked state27.
Prior investigations revealed that each of these structural processes occur spontaneously in physiological solutions22. Consequently, the ribosome transitions into and out of the unlocked state35, 36 under equilibrium conditions. To probe the occurrence of these events using smFRET techniques, we introduced donor and acceptor fluorophores into peripheral regions of the large and small subunits of the ribosome that come into close proximity in the unlocked state. We did this by site-specifically attaching a Cy5 fluorophore to ribosomal protein L1 (ref. 36) in the large subunit and a Cy3 fluorophore to ribosomal protein S13 located within the small-subunit head domain (Fig. 2a and Online Methods). Using this labeling strategy, we probed ribosome complexes lacking A-site tRNA and bearing either deacylated P-site tRNAfMet or tRNAPhe, the substrates used in the tRNA selection and recycling studies, respectively.
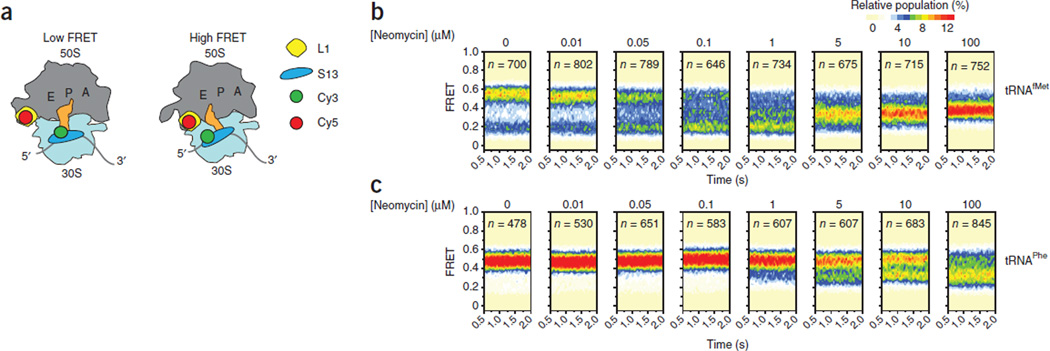
Figure 2.
Neomycin stabilizes an intermediate conformation of the ribosome. (a) Illustration showing the ribosome labeling strategy used for the single-molecule FRET experiments. (b,c) Single-molecule FRET trajectories, as shown in Supplementary Figure 2, summed into population FRET histograms to reveal the population behaviors of Cy3-labeled S13 (N terminus) and Cy5-labeled L1 (T202C) ribosome complexes bearing deacylated tRNAfMet (b) or deacylated tRNAPhe (c) in the P site.
Consistent with the intrinsically dynamic nature of the ribosome, we found that these ribosome complexes exchanged between two predominant configurations (a low-FRET (0.19 ± 0.01) and a high-FRET (0.55 ± 0.01) (mean FRET values ± s.d.) state) on the subsecond time scale (Supplementary Fig. 2a,b). As we anticipated from previous investigations of native ribosome complexes36, 37, we also found evidence of at least one short-lived intermediate-FRET configuration (Supplementary Fig. 2c–f). We probed the structural origins of these FRET states using the addition of translation factors known to stabilize distinct ribosome conformations. For example, release factor 1 (RF-1) favors a locked ribosome configuration in which P-site tRNA occupies its classical state (P/P) and the L1 stalk adopts an open position38–40. Consistent with this notion, we found that RF-1 strongly stabilized the low-FRET state (Supplementary Fig. 3a). In line with the known propensity of both factors to stabilize a rotated configuration of the small subunit and a closed L1 stalk position23, 34, 41, 42, RRF and EF-G each stabilized the high-FRET state (Supplementary Fig. 3b,c). Together, these studies stipulated that transitions between low- and high-FRET states provide information on the process of small-subunit rotation.
Antibiotic control of subunit rotation dynamics
We next tested whether small-molecule compounds known to alter the translation mechanism also affect subunit rotation dynamics. We first examined the effects of the 4,6-linked, 2-DOS antibiotic kanamycin, which stabilizes deacylated P-site tRNA and peptidyl A-site tRNA in classical configurations within the pre-translocation complex17. The effect of kanamycin on the pre-translocation complex has been attributed to its stabilization of interactions between the mRNA codon and the tRNA anticodon in the A site resulting from the drug binding to the h44 decoding site17, 43. Notably, although the complexes examined here lack peptidyl A-site tRNA, we did find that kanamycin stabilized the low-FRET ribosome configuration (Supplementary Fig. 3d). This finding argues that the interactions of kanamycin with h44 restrict the conformational changes that are required for small-subunit rotation. By contrast, we found that the peptide antibiotic viomycin, whose crystallographic binding site also includes h44 (ref. 44), stabilized the high-FRET state (Supplementary Fig. 3e). This result is consistent with previous investigations showing that ribosomes favor hybrid (P/E) tRNA and rotated ribosome configurations in the presence of viomycin45, 46. The smFRET results obtained here suggest that kanamycin and viomycin stabilize the structurally defined locked and unlocked ribosome configurations, respectively. Together, these observations support the notion that antibiotic-induced remodeling of the small-subunit decoding region can affect the properties of bridge B2 in a manner that has an impact on subunit rotation17, 27.
Notably, in identical complexes, neomycin exerted a bimodal effect on subunit rotation dynamics (Fig. 2b). Consistent with the proposed effects of neomycin on binding in the h44 decoding site, the predominant effect of low concentrations of neomycin (<1 µM) was to stabilize the low-FRET, locked ribosome configuration17. However, at higher concentrations of neomycin (>1 µM), an intermediate-FRET configuration of the ribosome clearly emerged, whose FRET value (0.37 ± 0.01) fell between those assigned to the locked and unlocked states. Close inspection of the smFRET data revealed that stabilization of the intermediate-FRET state began to emerge even at low concentrations of neomycin (<1 µM). We also observed a similar intermediate-FRET configuration at comparable neomycin concentrations in complexes containing tRNAPhe in the P site (Fig. 2c). Here, however, the bimodal nature of the effect of neomycin was substantially masked by the inherent propensity of this complex to favor the unlocked state36. Collectively, these data argue that neomycin can stabilize an intermediate ribosome configuration that is in transition between the locked and unlocked states36. The functional data presented here (Fig. 1) suggest that such a configuration may be incompatible with tRNA selection, translocation and ribosome recycling.
Crystal structure of neomycin-bound recycling complex
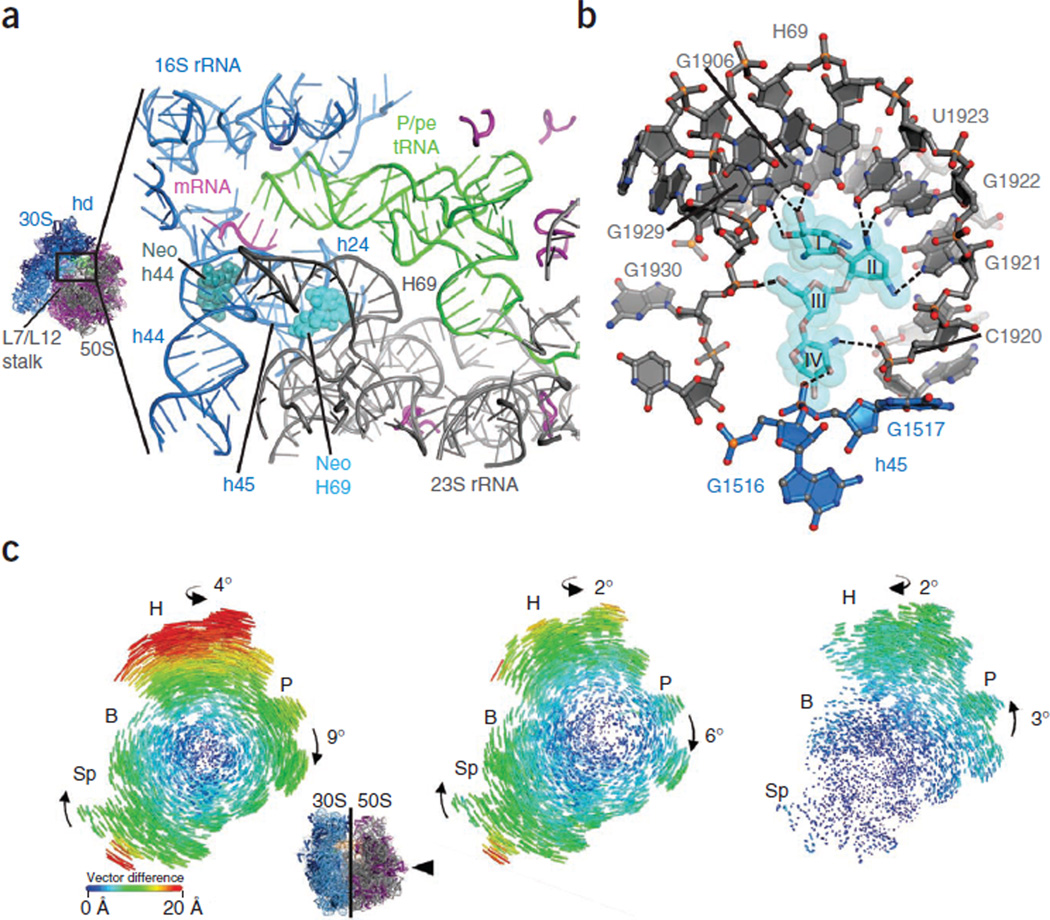
To gain structural insights into this intermediate ribosome configuration, we used neomycin to soak crystals containing 70S ribosomes in either the unrotated or the fully rotated state and containing bound P/P or P/E deacylated tRNAPhe, respectively27. We used these crystals to measure diffraction data to a resolution of 3.5 Å (Table 1). After refinement of the 70S ribosome structures with tRNA removed from the models, we found that electron density maps of the unbiased Fobs–Fcalc difference contained a clear, positive electron density for neomycin. In the unrotated ribosome, neomycin was bound in its canonical h44 decoding site (Supplementary Fig. 4a). In the rotated ribosome, neomycin was also present in the h44 site. However, we also found unambiguous positive density for neomycin at a site near the base of H69 in the large subunit (Supplementary Fig. 4b), which is a region of the intersubunit bridge B2 where H69 interacts with helices h24 and h45 of the small-subunit rRNA (Fig. 3a). In this site, interactions between neomycin and the ribosome bridge the subunit interface such that rings I–III of neomycin contact residues within the H69 major groove and ring IV mediates contacts with h45 of the small subunit (Fig. 3b).
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c(Å) | 212.18,433.90, 608.83 |
| Resolution (Å)a | 70–3.3 (3.58–3.48) |
| Rmergea | 23.1 (77.8) |
| I/σIa | 5.76 (1.64) |
| Completeness (%)a | 95.2 (89.2) |
| Redundancya, b | 5.6 (3.4) |
| Refinement | |
| Resolution (Å) | 70–3.3 |
| Number of reflections | 792,717 |
| Rwork/Rfree | 0.206/0.255 |
| Number of atoms | |
| Protein/RNA | 291,292 |
| Ligand/ion | 1,248 |
| Water | 1,727 |
| B-factors | |
| Protein/RNA | 130.50 |
| Ligand/ion | 91.81 |
| Water | 95.67 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.141 |
Values in parentheses are for the highest resolution shell.
Data were measured from 16 crystals.
Figure 3.
Neomycin contacts within H69 of 23S rRNA and bridging interactions with h45 of 16S rRNA induce global rearrangements in the 70S ribosome. (a) Overview of neomycin binding sites in h44 (dark green) of the small ribosomal subunit (blue) and in H69 (light blue) of the large ribosomal subunit (gray). tRNA is shown in green, mRNA is shown in red, and L proteins are shown in purple. The head domain (hd) of the 30S subunit and proteins L7/L12 of the 50S subunit are also indicated. (b) Neomycin interactions with the H69 binding site. Neomycin and rRNA contacts > 3.5 Å are shown as dashed lines. Ring II of neomycin packs against the major groove face of residues G1921, G1922 and U1923, whereas rings I, III and IV participate mainly in backbone contacts. (c) Effects of neomycin binding to H69 on intersubunit rotation in the ribosome. Inset, view of the 30S subunit from the perspective of the 50S subunit. Difference in the vector shifts between equivalent RNA phosphorus atoms and protein Cα atoms in the unrotated compared to the fully rotated state (left); the unrotated compared to the intermediate-rotated state with neomycin bound to H69 (middle); and the fully rotated compared to the intermediate-rotated state with neomycin bound to H69 (right). The vectors are color coded as indicated in the scale. Ribosomes were superimposed using the 50S subunit as the frame of reference27. 30S head domain, H; 30S body, B; 30S platform, P; 30S spur, Sp.
Neomycin binding to H69 was accommodated by substantial alterations in the extent of the small-subunit rotation. In contrast to the fully rotated state, the overall extent of small-subunit rotation was markedly attenuated (Fig. 3c). In addition, motions of the small-subunit body along the subunit rotation trajectory were partially uncoupled from those of the platform and head domains. Whereas the small-subunit spur region moved to the same extent as in the fully rotated structure (approximately 20 Å), the small-subunit platform domain in the neomycin-bound state rotated to a lesser extent (approximately 6° compared to 9°). Lateral motions of the small-subunit head domain were also smaller in the neomycin-bound state than in the fully rotated state (approximately 15 Å compared to 20 Å, respectively). The swivel-like rotation of the small-subunit head domain in the direction of translocation was likewise reduced (approximately 2° compared to 4°).
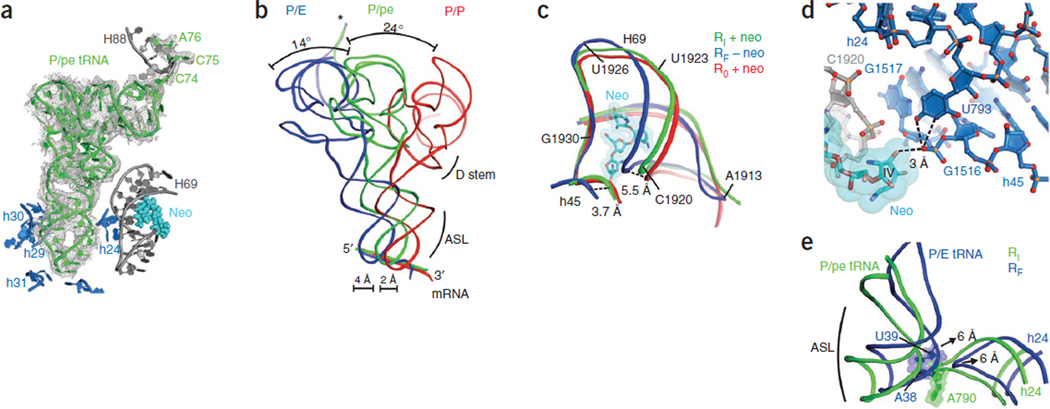
Notably, electron density maps of the Fobs–Fcalc difference also placed P-site tRNA in a position that was intermediate between the classical (P/P) and hybrid (P/E) configurations27, 47 (Fig. 4a,b). In this new configuration, the tRNA anticodon stem loop maintained the same key interactions within the small-subunit P site in both the unrotated and fully rotated ribosome structures (Supplementary Fig. 4c). The 3′ CCA terminus of the tRNA also remained within the large-subunit E site27, 47 (Supplementary Fig. 4d). However, because of the intermediate extent of small-subunit rotation (Fig. 3c), the tRNA anticodon was only partially displaced in the direction of translocation (approximately 2 Å compared to 6 Å in the fully rotated state) (Fig. 4b). The P-site tRNA elbow domain also clearly occupied a position that was intermediate between the classical (P/P) and hybrid (P/E) configurations27 and that prevented its physical interaction with the L1 stalk in the crystal. In keeping with nomenclature previously established for other intersubunit hybrid tRNA positions48, we define this state as a P/pe hybrid configuration. These structural rearrangements are in good agreement with the FRET value observed for the neomycin-induced intermediate ribosome configuration (Fig. 2 and Supplementary Fig. 5) and provide key structural insights into events underpinning the process of subunit rotation.
Figure 4.
Position of tRNAPhe in the intermediate-rotated and neomycin-bound ribosome. (a) Electron density map of the Fobs–Fcalc difference for P/pe tRNA in the neomycin-bound, intermediate-rotated ribosome configuration, with its position relative to rRNA elements of the small (blue) and large (gray) subunits and neomycin (light blue spheres) bound to H69 shown. The electron density map is contoured at 2.5 s.d. from the mean. (b) The intermediate position of P-site tRNA (P/pe, green) is between the classical (red) and hybrid (blue) configurations in the extent of both T-stem and anticodon displacement toward the E site. The 3′ CCA end of P/pe tRNA (asterisk) occupies the large-subunit E site. The dihydrouridine (D) stem and the anticodon stem loop (ASL) are indicated. (c) Superposition of the positions of H69 and h45 in unrotated (R0; P/P tRNA), fully rotated (RF; P/E tRNA) and intermediate-rotated (RI; P/pe tRNA) ribosome configurations showing the position of neomycin (neo) in the RI structure. (d) Neomycin (light blue) binding to H69 (gray) stabilizes the small-subunit platform conformation through interactions between neomycin ring IV, the G1517 backbone in h45 (blue) and the U793 residue in h24 (dark blue). Dashed lines indicate possible hydrogen-bond contacts between ring IV, G1517 and U793. (e) In the intermediate RI configuration (green), h24 of the small ribosomal subunit sterically blocks movement of P-site tRNA into the P/E configuration of the fully rotated RF ribosome. Arrows indicate the direction and extent of tRNA and h24 movements during the transition from the intermediate RI configuration to the fully rotated RF configuration.
Formation of the unlocked state, which precedes both substrate translocation and ribosome recycling, requires a 9° rotation of the small subunit that is accompanied by a lateral compression of the major groove of H69 (ref. 27). Notably, neomycin binding to the rotated ribosome in the crystals reversed the compression of H69 (Fig. 4c). The position of neomycin within the major groove of H69, buttressed by the interactions of ring IV with the phosphate of G1517 within h45 and the Watson-Crick face of U793 in h24 of the small subunit (Fig. 4d), sterically blocks H69 compression. Thus, neomycin alters bridge B2 rearrangements that normally accompany the transition from the unrotated to the rotated state (Fig. 4c), preventing movement of the ribosome into the fully rotated state. Concomitantly, h24 of 16S rRNA impedes P-site tRNA movement into its P/E hybrid position (Fig. 4e), resulting in its intersubunit hybrid (P/pe) position.
These crystallographic data suggest that neomycin can allosterically inhibit global aspects of the translation mechanism through its interactions with the H69 binding site. Neomycin does this by preventing normal bridge B2 remodeling events that accompany the small-subunit rotation and the repositioning of tRNA into the P/E hybrid site that precede both substrate translocation and ribosome recycling27, 36. Neomycin-induced stabilization of the intermediate configuration also provides a structural rationale for its inhibition of tRNA selection, as this process probably requires an unrotated configuration of the ribosome. Collectively, these observations provide a compelling argument for a central role of dynamics in the translation mechanism and for the notion that productive-factor–binding interactions with the ribosome may operate through a conformational selection-type mechanism17, 32, 49–51.
Conformational selection and the translation mechanism
The synthesis phase of translation hinges on the cyclical binding of EF-G and elongation factor thermal unstable (EF-Tu) to overlapping sites at the leading edge of the ribosome48, 52, 53. Yet, despite the fact that the cellular concentrations of both elongation factors exceed their Km values for ribosome binding, EF-Tu and EF-G do not competitively inhibit translation. A simplified conformational selection model of the translation mechanism posits that the unrotated, locked configuration of the ribosome is competent to productively engage EF-Tu in a ternary complex with aa-tRNA and GTP, whereas the fully rotated, P/E hybrid ribosome configuration is competent to productively engage EF-G. Such a model predicts that intermediate configurations of subunit rotation should not productively bind either factor. Consistent with this notion, both EF-G and EF-Tu sterically clash with elements of the large and small subunits when docked to the intermediate ribosome configuration (Fig. 5a,b). Direct smFRET measurements also showed that neomycin prevents productive elongation factor interactions at the A site (Fig. 5c,d). These data suggest that small-molecule antibiotics, such as neomycin, can effectively inhibit or alter the translation mechanism by allosterically controlling ribosome conformation and, thus, ribosome interactions with essential translation components.
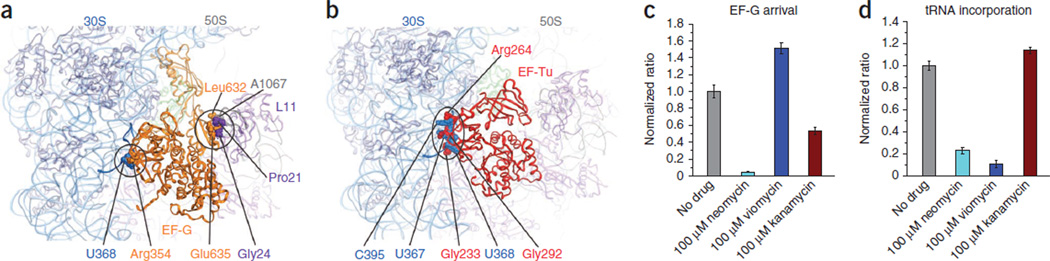
Figure 5.
The intermediate ribosome configuration is incompatible with productive EF-G and EF-Tu binding. (a) The intermediate ribosome structure superimposed on the Thermus thermophilus ribosome structure containing EF-G (gold)48. (b) The intermediate ribosome structure superimposed on the T. thermophilus ribosome structure containing EF-Tu (red)53. In a and b, specific points of steric clash between the elongation factors and the intermediate ribosome configuration are indicated. EF-G domain II clashes with the 30S body near h15 (blue spheres), and EF-G domain V clashes with the L7/L12 stalk protein L11 (purple spheres) and residue A1067 within H43 of 23S rRNA (gray). Domain II of EF-Tu clashes with the 30S body. Superpositions were carried out using the PyMOL ‘pair_fit’ function61. (c) The normalized ratio of individual pre-translocation complexes showing EF-G binding events in the absence of drugs (gray) or in the presence of neomycin (cyan), viomycin (blue) or kanamycin (maroon). All data are shown as the mean ± s.d. from three independent experiments. (d) The normalized ratio of individual empty A-site (after initiation) complexes showing ternary complex binding events in the absence of drugs (gray) or in the presence of neomycin (cyan), viomycin (blue) or kanamycin (maroon). All data are shown as the mean ± s.d. from at least three independent experiments.
DISCUSSION
The intrinsically dynamic nature of the ribosome enables the translating particle to spontaneously alternate between globally distinct functional configurations. The nature of these dynamics directly affects translation factor binding and translational activity. Correspondingly, the underlying energy landscape22 of these dynamics must be considered as a central feature of translational control. Previous data supporting this conclusion include the observation that P-site tRNA deacylation after peptide-bond formation controls subunit rotation23, 54 and that translation elongation processes may be regulated by the structural elasticity of the tRNA ligands of the ribosome32, 41, 55. We now show that neomycin binding at the base of H69 allosterically regulates ribosome dynamics and the mechanism of translation by controlling the process of subunit rotation.
The observation of a second aminoglycoside binding site within intersubunit bridge B2 helps explain the pleiotropic effects of neomycin— and, perhaps, related 2-DOS aminoglycosides—on the translation mechanism1. However, as the three-ring 4,6-linked 2-DOS aminoglycoside kanamycin does not lead to intermediate ribosome configurations, inhibitory effects associated with the H69 binding site may be restricted to aminoglycosides containing a ring IV substituent. This also highlights the potential for disease-associated H69 perturbations to allosterically control translation processes56. Future efforts should be aimed at exploring whether other small-molecule compounds target this site to inhibit the translation mechanism. Efforts must also be directed to understanding whether the bactericidal nature of aminoglycosides is tied to the binding of the drug to H69.
Miscoding effects alone are insufficient to explain the mechanism of aminoglycoside-induced cell killing57, 58; rather, cell death is associated with elevated drug concentrations in the cell during late stages of drug influx and translational catastrophe59. In line with our smFRET subunit rotation data (Fig. 2b), the functional studies presented here (Fig. 1 and Supplementary Fig. 1) suggest that neomycin binding to H69 begins to block the essential translation reactions even at low concentrations (>10 nM). This observation implies that the effects of neomycin on subunit rotation through H69 may feature prominently in the mechanism of translation inhibition at the minimum inhibitory concentrations of wild-type cells (approximately 1.5–5 µM)31, 60. In this view, the partial cellular resistance to neomycin conferred by the A1408G mutation in the h44 decoding site may arise from a loss of miscoding-induced, late-stage drug influx into the cell, a synergistic loss of the drug binding to H69 as a result of h44 alterations or both. These models may be delineated by future investigations aimed at determining the intracellular concentrations of neomycin in wild-type and A1408G cells immediately before cell death. The physical proximity of h44 and H69 binding suggest that the functional coupling of these sites may be nontrivial. Investigations of this kind will not only enable a deeper understanding of the full potential of H69 as a drug target but also reveal whether the allosteric regulation of ribosome dynamics and conformation can be mediated in a species-specific manner as a means for therapeutic translation control.
ONLINE METHODS
Generation of site-specifically labeled 30S subunits and 50S subunits
Ribosomal protein S13 was PCR cloned from E. coli strain K12 genomic DNA into the pPROEX HTb vector with a TEV-protease–cleavable histidine (His)6 tag and a 12-residue peptide encoding the S6 epitope for the Sfp phosphopantetheinyl transferase reaction62 (amino acid sequence, GDSLSWLLRLLN) fused at the N terminus (N-Sfp). After transformation of this plasmid into an E. coli ΔS13 knockout strain63, cells were cultured and ribosomes were harvested as previously described49. Pure 30S subunits were isolated by sucrose gradient centrifugation in a low-magnesium buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 10 mM NH4Cl, 0.5 mM EDTA, 6 mM β-mercaptoethanol (BME) and 1 mM MgCl2). 30S subunits containing Sfp-tagged S13 were isolated from this population by cobalt affinity chromatography (Clontech). Then, the Sfp tag was enzymatically labeled, and the His6 tag was enzymatically removed in a buffer containing 20 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2 and 6 mM BME. Twenty micromolar N-Sfp-S13 30S subunits, 5 µM TEV protease, 250 µM Cy3–coenzyme A (CoA) and 25 µM Sfp enzyme were incubated for 24 h at 18 °C. Sfp enzyme, TEV protease and unbound Cy3-CoA were then removed by filtration over a 100K membrane (Millipore). Before 70S complex formation, ribosomes were buffer exchanged into Tris-polymix buffer32. 50S subunits labeled with Cy5-L1 (T202C) were prepared and purified as previously described36.
Preparation of L1 S13 FRET ribosome complexes with tRNAfMet or tRNAPhe in the P site
Cy3-S13 30S and Cy5-L1 50S subunits were heat activated at 42 °C for 10 min in Tris-polymix Mg2+ buffer32. Ribosomes were then initiated with fMet-tRNAfMet or NAc-Phe-tRNAPhe, as previously described32.
Single-molecule L1 S13 FRET assay
All single-molecule FRET experiments were performed at room temperature in Tris-polymix with 5 mM Mg2+ buffer, as previously described49, and in which oxygen scavenging and triplet-state quenching systems were used64. After surface immobilization65, the ribosome-bound, P-site tRNA was deacylated by incubation with 2 mM puromycin for 10 min at room temperature. The smFRET data were acquired by directly exciting the Cy3 fluorophore at 532 nm (LaserQuantum) while the Cy3 and Cy5 intensities were simultaneously recorded in Metamorph (Molecular Devices) with a 40-ms integration time. The data were analyzed in MATLAB (MathWorks) and plotted in Origin (OriginLab), as previously described65.
Ribosome purification and crystallization
Ribosomes lacking protein S1 were purified from E. coli strain MRE600 using sucrose gradient centrifugation, as previously described66. Ribosomes were crystallized at 18 °C using microbatch 96-well plates and buffers containing 4.0–5.0% 2-methyl-2,4-pentanediol (MPD), 4.1–4.5% PEG 8000, 4.0 mM MgCl2, 380 mM NH4Cl, 5.7 mM putrescine, 5.0 mM spermidine, 10 mM Tris, 40 mM MES, pH 6.5–7.0, and 0.25 mM EDTA. Ribosome complexes were formed by incubating 4 µM deacylated tRNAPhe and 8 µM mRNA of sequence 5′- GGCAAGGAGGUAAAAUUCUACAAA-3′ (Thermo Scientific) with 2 µM ribosomes at 37 °C for 15 min. Eight micromolar RRF was then added, and the samples were incubated for an additional 15 min at 37 °C. Before crystallization, samples were subjected to ultrafiltration to remove excess ligands.
Data collection and processing
Ribosome crystals were stabilized with crystallization buffer containing 7.0% MPD, 7.0% PEG 8000 and 24% PEG 400, pH 4.8, to allow cryo-cooling of the crystals to liquid nitrogen temperatures. During the last cryo-cooling step (PEG 400 24%), neomycin (Sigma-Aldrich) was added at a 100 µM concentration to the cryo-protection buffer, and crystals were incubated at 4 °C with neomycin containing cryo-protection buffer. After 2 h of incubation, the concentration of neomycin was reduced to 2 µM by cryo-protection buffer exchange, and crystals were frozen with liquid nitrogen after 1–4 d of incubation at 4 °C. Diffraction data were measured from crystals cooled to 100 K using 0.1–0.3° oscillations at the Advanced Light Source (beamlines 8.3.1 and 12.3.1), each of which is equipped with an ADSC Q315 area detector. Data were reduced using XDS67, yielding the statistics shown in Table 1.
Molecular replacement and structure refinement
The two copies of the 70S ribosome in the crystallographic asymmetric unit were located using rigid-body refinement in PHENIX68 of the well-ordered E. coli ribosome from the recent atomic-resolution structure determination27. The tRNAs were removed from the molecular replacement model to reduce possible model bias during refinement. Corrections to the resulting model were carried out in Coot69 and side chain rotator tools in KiNG70. Refinement was carried out in PHENIX, including the use of pucker-specific target parameters. The resulting structural models were then refined using rounds of manual rebuilding in Coot or KiNG, as well as positional refinement in PHENIX, including the use of a new functionality for automatic assignment of hydrogen-bond restraints using the Saenger basepair types71. Electron density maps were generated from the PHENIX output directly. RNA rebuilding concentrated on the tRNAs, H69 and h44 (neomycin binding sites), and neomycin structures were inserted and refined by using Coot and PHENIX. Neomycin occupancies were determined by matching neomycin atomic displacement parameters to the mean value of neighboring rRNA, and then the group occupancy of each neomycin was refined in PHENIX. After final refinement, the Coot program tool ‘Find ligand’ was used to search meaningful (>3 s.d. above the mean) electron density features from Fobs–Fcalc difference electron density maps with the structure of neomycin as the target. Using this approach, evidence for additional neomycin binding sites was found near A482 (H24), G551 (H25), G1157 (H41), U1240 (H46), A1858 (H68) and C2674 (H95) in 23S rRNA, and near C658 (h22), G902 (h27) and U1420 (h44) in 16S rRNA. However, the conformational change from the fully rotated to the intermediate ribosome configuration described here cannot be rationalized by neomycin binding at these sites.
Superpositions
Comparisons to atomic-resolution structures of the ribosome were carried out by the ‘pair_fit’ command in PyMOL61. From the chosen atom pairs, disordered or moving regions of 23S rRNA were not used (for example, the L1 stalk, the L7/L12 stalk, H38 and H69) in the superpositions. Superpositions were performed using ribose C1′ positions or phosphorus atoms in nucleotides. The angles of rotation of the 30S subunit domains were calculated essentially as described previously27, 37. Angles given for the rotation of the head domain were calculated from 30S subunit structures superimposed by means of their platform domains. A rotation of 0° is defined as centering the head domain over the 30S P site, as seen in the structure of the unrotated ribosome27. Superpositions of P/P, P/pe and P/E tRNAs used the C1′ atoms of nucleotides 31–39 in the anticodon stem loop27. The comparisons of tRNA bending angles used the glycosidic bond of position 31 near the end of the anticodon stem loop and the glycosidic bond of nucleotide 63 in the superimposed tRNAs27. The bending angles calculated in this way were 24° for P/P tRNA compared to P/pe tRNA and 14° for P/pe tRNA compared to P/E tRNA.
Figure preparation
All structure figures were made using the program PyMOL61.
Single-molecule tRNA incorporation assay
The process of tRNA selection on the ribosome was monitored using single-molecule FRET by stropped-flow injection of a 10 nM solution of ternary complex (EF-Tu·GTP·aa-tRNA) containing Phe-tRNAPhe (Cy5-acp3U47) to surface-immobilized ribosome complexes containing deacylated tRNAfMet (Cy3-s4U8) in the P site, as previously described29. Fluorescence and FRET traces were extracted from the video and selected for analysis using automated analysis software implemented in MATLAB (MathWorks) using the segmental k-means algorithm72. Complete accommodation of Phe-tRNAPhe into the A site was marked by the first observation of a 400-ms dwell in a high (0.55 ± 0.61) FRET state, structurally assigned to the classical pre-translocation complex configuration through previous investigations29. Individual experiments, in which approximately 100 accommodation events were obtained, were performed in triplicate. The mean extent of accommodation and s.d. are plotted as a function of time. Wild-type and mutant ribosomes for these experiments were purified as previously described17.
Single-molecule translocation assay
Fluorescence-based, single-molecule translocation assays were carried out as previously described32 on either wildtype or A1408G mutant pre-translocation ribosome complexes. Experiments were performed in Tris-polymix buffer (pH 7.5, 5 mM Mg(OAc)2) and analyzed as previously described32, 49, plotting the fraction of translocating molecules as a function of time.
Single-molecule ribosome recycling assay
Ribosome recycling assays were performed and analyzed as previously described27 in Tris-polymix buffer (pH 7.5, 5 mM Mg(OAc)2). Subunit release was monitored by directly exciting Cy5 fluorophore on L1 protein (S55C). Large-subunit dissociation from the surface-immobilized 70S subunit leads to an abrupt loss of fluorescence from the image plane. The fluorescence intensities of an array of molecules was recorded with time-lapse imaging by acquiring one image (40-ms integration time) every 20 s for 20 min.
Monitoring FRET between EF-G or EF-Tu and the ribosome
To monitor EF-G productively engaging the pre-translocation complex, EF-G was labeled with Cy5 fluorophore and stop-flow delivered to surface-immobilized ribosome complexes bearing deacylated tRNAfMet in the P site and Cy3-labeled dipeptidyl fMet-Phe-tRNAPhe (Cy3-acp3U47) in the A site, as previously described32, 49. Pre–steady state smFRET measurements of this kind were taken in the absence and presence of 20 µM neomycin at a 100-ms time resolution. Fluorescent traces were analyzed with MATLAB (MathWorks). All molecules with a signal-to-noise ratio >3 were inspected individually. Productive EF-G binding events were marked by the appearance of FRET (≥0.2), which is indicative of domain IV of EF-G entering the A site32. The interaction of ternary complex (EF-Tu·GTP·aa-tRNA) with the ribosome was similarly monitored in pre–steady state experiments collected at a 15-ms time resolution in the absence and presence of 20 µM neomycin. Here, productive binding was marked by the appearance of FRET (>0.15) between the ternary complex containing Phe-tRNAPhe (Cy5-acp3U47) and deacylated tRNAfMet (Cy3-s4U8) within the P site of surface-immobilized 70S ribosome complexes29. Complete accommodation was marked by the appearance of a relatively stable (>200 ms) high-FRET state (0.55 ± 0.61). Fluorescent traces were analyzed with MATLAB (MathWorks). All molecules with a signal-to-noise ratio >3 were inspected individually. Each tRNA selection and translocation experiment was performed in triplicate, and the average number and s.d. of each event type were calculated and plotted in Origin (OriginLab).
Monitoring FRET between S13 and A-site tRNA
The efficiency of tRNA selection on the ribosome was monitored using single-molecule FRET by stropped-flow injection of a 10 nM solution of ternary complex (EF-Tu·GTP·aa-tRNA) containing Phe-tRNAPhe (Cy5-acp3U47) to surface-immobilized ribosome complexes containing Cy3-labeled S13 and deacylated tRNAfMet in the P site, as previously described29. After a 30-s incubation time, the ternary complex was washed out with Tris-polymix Mg2+ buffer. The smFRET data were acquired by directly exciting the Cy3 fluorophore at 532 nm (Laser Quantum) while simultaneously recording Cy3 and Cy5 intensities in Metamorph (Molecular Devices) with a 100-ms integration time. Fluorescence and FRET traces were extracted from the video, and traces from each movie were selected according to the following criteria: signal-to-noise ratio >5, background intensity <1,500 and Cy3 blinking <1. All selected traces were then manually inspected for the appearance of a stable 0.15–02 FRET state, which indicated Cy5-tRNAPhe incorporation into the A site. The ratio of the number of molecules showing A-site tRNA incorporation to the total number of molecules inspected for all experiments (no drug, 100 µM neomycin, 100 µM viomycin or 100 µM kanamycin) were normalized to that of the group receiving no drug. Three independent experiments were performed under each condition, and the mean with s.d. were plotted using Origin (OriginLab).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Green (Johns Hopkins University) for providing the S13 knockout mouse strain, T. Suzuki (University of Tokyo) for providing the pKK3535 ribosome plasmids, K. Hamadani (University of California, Berkeley) for the RRF expression vector and M. O’Connor (University of Missouri-Kansas City) for helpful discussions throughout the course of this work. We also acknowledge helpful discussions and insights provided by all members of the Blanchard and Cate laboratories and J. Headd (Lawrence Berkeley National Laboratory) for help with PHENIX refinement. This work was supported by the US National Institutes of Health (2R01GM079238 to S.C.B., 1R01GM65050 to J.H.D.C. and National Cancer Institute grant CA92584 for the SIBYLS and 8.3.1 beamlines at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory), the Human Frontiers in Science Program (RGY0088), the National Science Foundation (0644129) and the US Department of Energy (DE-AC0376SF00098 for the SIBYLS and 8.3.1 beamlines at the ALS, Lawrence Berkeley National Laboratory). M.B.F. is a trainee in the Weill Cornell/Rockefeller University/Sloan-Kettering Tri-Institutional MD/PhD Program supported by US National Institutes of Health Medical Scientist Training Program grant GM07739. M.R.W. is supported by US National Institutes of Health National Research Service Award fellowship 5F31DC012026-02.
Footnotes
Accession codes. The structures reported here have been deposited in the Protein Data Bank with codes 4GAQ, 4GAR, 4GAS and 4GAU.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
L.W. and M.R.W. prepared dye-labeled ribosomes, tRNAs and translation factors and performed the smFRET imaging and analyzed the results. A.P. crystallized, collected, processed and refined X-ray data. L.W. and M.B.F. performed and analyzed bulk functional assays. L.W. and A.P. made the figures. R.B.A. prepared reagents crucial for smFRET experiments. S.C.B. and J.H.D.C. designed the study. All authors discussed the results and contributed to the writing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ. The Molecular Basis of Antibiotic Action. 2nd edn. John Wiley & Sons Ltd.; 1981. [Google Scholar]
- 2.Wilson DN. The A–Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard SC, Cooperman BS, Wilson DN. Probing translation with small-molecule inhibitors. Chem. Biol. 2010;17:633–645. doi: 10.1016/j.chembiol.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 5.Tenson T, Mankin A. Antibiotics and the ribosome. Mol. Microbiol. 2006;59:1664–1677. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 6.Nathan C. Antibiotics at the crossroads. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 7.von Nussbaum F, Brands M, Hinzen B, Weigand S, Habich D. Antibacterial natural products in medicinal chemistry—exodus or revival? Angew. Chem. Int. Edn. Engl. 2006;45:5072–5129. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- 8.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llano-Sotelo B, Hickerson RP, Lancaster L, Noller HF, Mankin AS. Fluorescently labeled ribosomes as a tool for analyzing antibiotic binding. RNA. 2009;15:1597–1604. doi: 10.1261/rna.1681609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David-Eden H, Mankin AS, Mandel-Gutfreund Y. Structural signatures of antibiotic binding sites on the ribosome. Nucleic Acids Res. 2010;38:5982–5994. doi: 10.1093/nar/gkq411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodnina MV, et al. GTPases mechanisms and functions of translation factors on the ribosome. Biol. Chem. 2000;381:377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 12.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 13.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 14.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- 16.Kurland CG. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 17.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat. Chem. Biol. 2010;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirokawa G, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J. Biol. Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- 20.Dahlberg AE, Horodyski F, Keller P. Interaction of neomycin with ribosomes and ribosomal ribonucleic acid. Antimicrob. Agents Chemother. 1978;13:331–339. doi: 10.1128/aac.13.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borovinskaya MA, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 22.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem. Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, Gonzalez RL. Jr. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu. Rev. Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz-Meoz RF, Green R. Helix 69 is key for uniformity during substrate selection on the ribosome. J. Biol. Chem. 2011;286:25604–25610. doi: 10.1074/jbc.M111.256255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulkley D, Johnson F, Steitz TA. The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge B2a of the ribosome. J. Mol. Biol. 2012;416:571–578. doi: 10.1016/j.jmb.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 29.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 31.Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat. Struct. Mol. Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirokawa G, et al. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G–bound ribosome. EMBO J. 2010;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc. Natl. Acad. Sci. USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat. Struct. Mol. Biol. 2009;16:861–868. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 40.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr. Opin. Struct. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc. Natl. Acad. Sci. USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent. tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 44.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 47.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc. Natl. Acad. Sci. USA. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. RNA. 2011;17:2189–2200. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan YL, Wool IG. The integrity of the sarcin/ricin domain of 23 S ribosomal RNA is not required for elongation factor-independent peptide synthesis. J. Mol. Biol. 2008;378:12–19. doi: 10.1016/j.jmb.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 51.Whitford PC, et al. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc. Natl. Acad. Sci. USA. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat. Struct. Mol. Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fei J, Richard AC, Bronson JE, Gonzalez RL. Jr. Transfer RNA–mediated regulation of ribosome dynamics during protein synthesis. Nat. Struct. Mol. Biol. 2011;18:1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 57.Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- 58.O’Connor M, Goringer HU, Dahlberg AE. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 1992;20:4221–4227. doi: 10.1093/nar/20.16.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hobbie SN, et al. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 2006;50:1489–1496. doi: 10.1128/AAC.50.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The PyMOL molecular graphics system. Version 1.3. Schrödinger, LLC; 2002. [Google Scholar]
- 62.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 63.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA: tRNA complex. Mol. Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 64.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blaha G, et al. Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 2000;317:292–309. doi: 10.1016/s0076-6879(00)17021-1. [DOI] [PubMed] [Google Scholar]
- 67.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen VB, Davis IW, Richardson DC. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 2009;18:2403–2409. doi: 10.1002/pro.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 72.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.