Abstract
Angiogenesis is a complex cellular process involving multiple regulatory growth factors and growth factor receptors. Among them, the ligands for the endothelial-specific tunica intima endothelial receptor tyrosine kinase 2 (Tie2) receptor kinase, angiopoietin-1 (Ang1) and Ang2, play essential roles in balancing vessel stability and regression during both developmental and tumor-induced angiogenesis. Despite possessing a high degree of sequence identity, Ang1 and Ang2 have distinct functional roles and cell-signaling characteristics. Here, we present the crystal structures of Ang1 both unbound and in complex with the Tie2 ectodomain. Comparison of the Ang1-containing structures with their Ang2-containing counterparts provide insight into the mechanism of receptor activation and reveal molecular surfaces important for interactions with Tie2 coreceptors and associated signaling proteins. Using structure-based mutagenesis, we identify a loop within the angiopoietin P domain, adjacent to the receptor-binding interface, which confers the specific agonist/antagonist properties of the molecule. We demonstrate using cell-based assays that an Ang2 chimera containing the Ang1 loop sequence behaves functionally similarly to Ang1 as a constitutive Tie2 agonist, able to efficiently dissociate the inhibitory Tie1/Tie2 complex and elicit Tie2 clustering and downstream signaling.
Keywords: cellular signaling, Tie receptor tyrosine kinase, X-ray crystallography
Proper development of the cardiovascular system involves two highly integrated and dynamic processes: vasculogenesis and angiogenesis (1). Vasculogenesis involves the in situ differentiation of endothelial cells from precursor angioblasts and results in their subsequent migration and proliferation, leading to the formation of the endocardium of the heart, as well as the major primitive blood vessels (2–6). Angiogenesis occurs following vasculogenesis and results in the remodeling and extension of the endothelial tubes into the adult microvasculature, as well as in the growth and expansion of vessels into organs including the kidney and brain. Endothelial cells lining these newly formed vessels are surrounded by support cells, such as smooth muscle cells and pericytes, which serve to regulate blood flow and provide survival signals (7). Angiogenesis predominantly takes place during embryonic development, although it also recurs during wound repair in adulthood. Its role is essential for tumor development, as well as metastasis and, therefore, represents a viable target for therapeutic intervention (8).
The angiopoietins are a small set of growth factor ligands for the tunica intima endothelial receptor tyrosine kinase 2 (Tie2) endothelial-specific receptor tyrosine kinase and are critical for both development and pathological angiogenesis (9–11). The prototypic family member, angiopoietin-1 (Ang1), is a Tie2 agonist, whereas the highly homologous Ang2 is a context-dependent agonist/antagonist. All angiopoietin family members, Ang1 to -4, contain an amino-terminal “superclustering” and coiled-coil domain, followed by a carboxyl-terminal fibrinogen domain. Several studies demonstrate that the fibrinogen domain mediates the interaction with Tie2, whereas the coiled-coil and the superclustering motifs, on the other hand, are required for ligand oligomerization, a prerequisite for receptor clustering and activation (9–14). Indeed, all angiopoietins exist primarily as tetramers, hexamers, and higher-order oligomers in solution. Nonetheless, chimeric Ang ligands illustrate that only the highly conserved receptor-binding fibrinogen domain is necessary and sufficient for their unique functional signaling characteristics (13–15).
Recently, a unique molecular mechanism for the distinct signaling properties of the different Ang ligands was proposed when it was shown that the related orphan receptor tyrosine kinase Tie1 is a coreceptor for Tie2. Indeed, Tie1 and Tie2 form a complex on the endothelial cell surface that inhibits Tie2 signaling (15). The Tie1/Tie2 interactions are predominantly electrostatic in nature and are selectively modulated by the Ang ligands. Tie1/Tie2 dissociation occurs rapidly in the presence of the Tie2 agonist Ang1 concurrent with Tie2 clustering, phosphorylation, and activation. Alternatively, the receptor antagonist Ang2 has no effect on the inhibitory Tie1/Tie2 complex and is, therefore, unable to promote Tie2 activation. It remains unclear, however, exactly how the Ang ligands differentially effect the Tie1/Tie2 interactions and Tie2 activation despite the available structural models of Ang2 and the Ang2/Tie2 complex (12, 13).
To more precisely define the functional differences between Ang1 and Ang2, we determined and report the crystal structures of Ang1 alone, as well as in complex with the Tie2 ectodomain. Comparison of the Ang1-containing structures with their Ang2-containing counterparts provide mechanistic insight into receptor activation and reveal molecular surfaces important for interactions with Tie2 and associated signaling proteins. Using structure-guided mutagenesis, we further identify a short surface-exposed loop within the angiopoietin P domain, adjacent to the Tie1/Tie2 receptor-binding interface, that confers the specific agonist/antagonist properties of these biologically important molecules.
Results
Crystal Structure of Ang1.
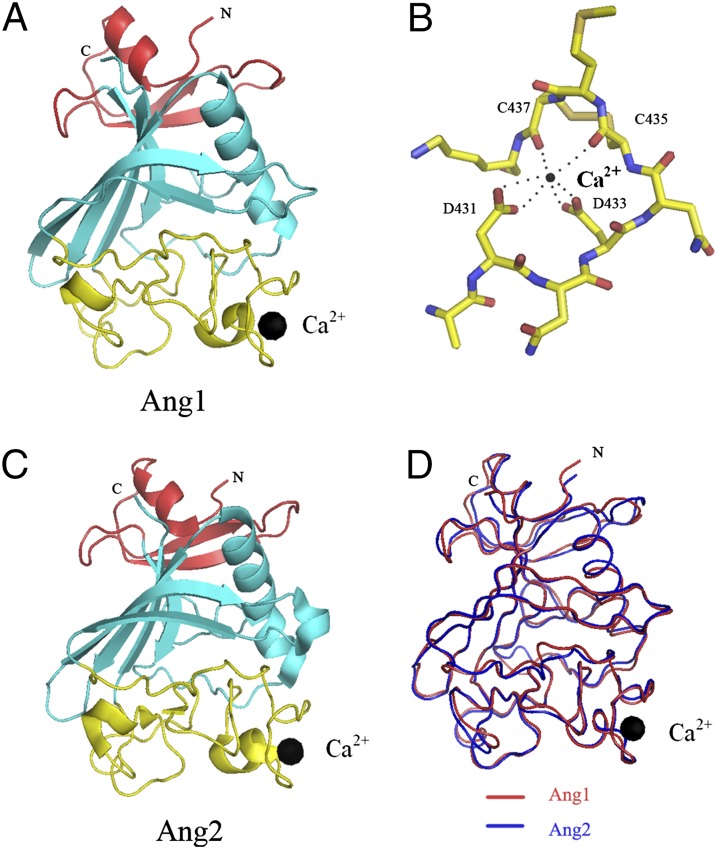
The structure of the receptor-binding fibrinogen-like domain of Ang1 [Ang1 receptor-binding domain (Ang1-RBD)] was determined by molecular replacement (Table S1 and Materials and Methods). The final model is refined to an R factor of 19.5% (Rfree of 25.1%) at 2.5-Å resolution. The structure is illustrated in Fig. 1 A and B. It is a compact protein with overall dimensions of ∼50 × 40 × 35 Å and has three domains A, B, and P (according to fibrinogen nomenclature) colored in red, cyan, and yellow, respectively. The P domain was identified previously as the site of receptor binding (12, 13). This region displays little secondary structure and contains mostly long, extended coil regions (Fig. S1). There are three conserved disulfide bonds in the Ang-RBDs, one of which plays a role in structurally coordinating and stabilizing a surface loop containing residues involved in calcium binding. However, the role of calcium in angiopoietin function is currently unknown.
Fig. 1.
Structure of the Ang1-RBD. (A) The refined model of Ang1-RBD with the individual subdomains shown in different colors (A domain, red; B domain, cyan; P domain, yellow). The black sphere represents the bound calcium atom. (B) Close-up view of the Ca2+-binding site. (C) Structure of the previously determined Ang2-RBD colored as in A. (D) Structural alignment in coil representation of the Ang1-RBD (shown in red) and the Ang2-RBD (shown in blue).
As expected, based on the high degree of homology (64% identity), Ang1 and Ang2 are structurally similar and can be superimposed with overall rmsds for equivalent Cα residues of 0.8 Å (Fig. 1 C and D). Indeed, the only significant structural differences between Ang1 and Ang2 are confined to the variable P domain, which forms the receptor-binding site. The residues forming the calcium binding site, as well as the disulfide bonds, are highly conserved among all angiopoietin family members.
Overall Structure of the Ang1/Tie2 Complex.
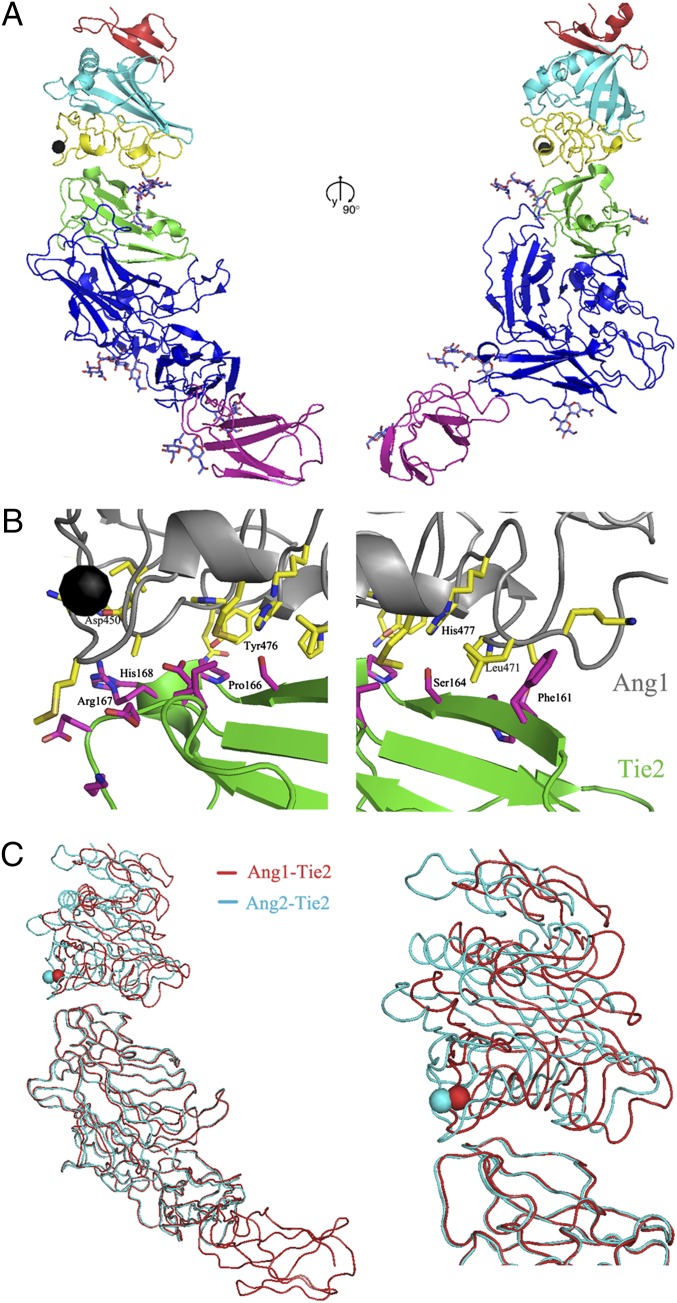
The structure of Ang1/Tie2 complex was determined using molecular replacement (Table S1 and Materials and Methods). The final model is refined to an R factor of 30.9% (Rfree of 33.4%) at 4.5-Å resolution. The Tie2/Ang1 complex is a 1:1 heterodimer in solution and the structure (Fig. 2). The complex contains the Ang1-RBD and the Tie2 ectodomain consisting of three Ig domains (Ig1 to Ig3), three EGF domains (EGF1 to EGF3), and one fibronectin type III (FNIII) domain. The Ang1/Tie2 complex has an elongated shape, with overall dimensions of 135 × 65 × 50 Å (Fig. 2A). Ang1 interacts exclusively with the Ig2 domain (colored in green) of Tie2 via its C-terminal (P) domain. The C terminus of Tie2 points toward the cellular membrane, on the opposite molecular end of the ligand-binding site. The ligand/receptor interface (Fig. 2B) buries 620 Å2 of molecular surface roughly 50% of which is hydrophobic.
Fig. 2.
Structure of the Ang1/Tie2 complex. (A) Two 90° orientations of the refined model of the Ang1/Tie2 complex. The Tie2 Ig2 domain is colored green, whereas the remaining domains are colored in blue, with the exception of the FNIII-1 domain, which is colored in magenta; Ang1 is colored as in Fig. 1. The N-acetyl-β-D-glucosamine moieties are shown in ball-and-stick format. (B) Close-up view of the Ang1/Tie2 interface. Ang1 is shown in gray, whereas Tie2 is in green. Residues at the interface are colored yellow (for Ang1) or magenta (for Tie2). (C) Structural alignment in coil representation of the Ang1/Tie2 (red) and Ang2/Tie2 (blue) complexes. The FNIII repeat seen in red at the bottom in the Ang1/Tie2 complex was not present in the Ang2/Tie2 complex structure previously reported by our groups (13).
The overall structure of Tie2 in the Ang1/Tie2 complex is similar to that of the unbound receptor with an rmsd between equivalent Cα positions of 1.1 Å. The Tie2 construct used in this study contains the N-terminal Tie2 FNIII domain (FNIII-1), in addition to the Ig and EGF domains visualized in the previously reported Tie2-containing structures. The FNIII-1 domain (colored in magenta on Fig. 2A) is well structured and, as expected, is not involved in ligand binding, positioned well away from Ig2. It adopts a typical Ig-like fold, structurally most closely homologous to the FNIII domains of receptor tyrosine phosphatase μ (RPTPμ) and neural cell adhesion molecule 2 (NCAM2) (16, 17). The association between Ig3 and FNIII-1 is inflexible, which facilitates the overall rigid elongated molecular architecture of the Tie2 ectodomain.
Comparison of the structures of bound and free Ang1 reveals that the Ang1-RBD undergoes no significant changes upon Tie2 binding and the two structures can be superimposed with an rmsd between equivalent Cα positions of 0.3 Å.
Ang1/Tie2 Interface.
The ligand–receptor interface is confined to the top of the Tie2 Ig2 domain, which interacts with the P domain of Ang2 near the Cα-binding site (Fig. 2). The Ang1-Tie2–binding interface is continuous, burying ∼1,300 Å2 of molecular surface. Specifically, loops β5-α5, α5-β7, β7-β8, and α6-β9, strand β8, and helix α6 in P domain of Ang1-RBD interact with the loops B-C, C-C′, and F-G, strand C, and strand C′ in Ig2 domain of Tie2. The core of the interface is dominated by van der Waals interactions between nonpolar side chains, although several peripheral hydrogen bonds are also involved in stabilizing the Ang1/Tie2 complex. The structure of the Ang1/Tie2 complex reveals that the calcium ion is located close to the Ang1/Tie2-binding interface but is not directly involved in receptor binding.
Overall, the Ang1/Tie2 interface is very similar to the Ang2/Tie2 interface. Indeed from the 13 Tie2-contacting residues in Ang1, 6 residues are conserved between Ang1 and Ang2. The conserved Ang2 residues Lys469, Lys473, and Tyr476 have been identified by mutagenesis as important for Tie2 binding (12), and, indeed, our structures document that they form the core of the Ang/Tie2 interface. Of the seven differences, two are conservative substitutions of hydrophobic residues that result in subtle rearrangements of the Tie2/Ang van der Waals contacts but do not significantly affect the ligand-receptor interface (Ang1 Met436–Ang2 Leu434, Ang1 Leu471–Ang2 Phe469). A close comparison of the Ang1/Tie2 and Ang2/Tie2 structures further reveals that the remaining five interface residues that differ between Ang1 and Ang2 also do not significantly affect the Ang/Tie2 binding. Thus, these substitutions have little effect on the ligand–receptor–binding interface.
Tie2/Ang1 Complex Is Structurally Similar to the Tie2/Ang2 Complex.
The individual angiopoietins exert different biological effects on Tie2-expressing cells as a function of their unique abilities to influence the heterodimeric association of Tie1 and Tie2 on the cell surface (15). It is not unexpected, therefore, that the overall architecture and receptor–ligand interactions are largely conserved between the two structures. Comparison of the Ang1/Tie2 and Ang2/Tie2 structures indeed reveal that Ang1 and Ang2 bind the Tie2 receptor in a very similar manner. The Tie2 receptor ectodomains in the two complexes can be superimposed with an rmsd between equivalent Cα positions of 0.6 Å (Fig. 2C). The only difference in the overall structure of the two Ang/Tie complexes comes from the slight translation of the bound angiopoietin on top of the Tie2 Ig2, as illustrated in Fig. 2C. Upon superimposition of the Tie2 Ig2 domain, the positions of the individual Cα atoms of Ang1 are shifted from 1.5 to 6 Å compared with their Ang2 counterparts. However, this subtle shift does not seem to affect the binding affinities or the on and off rates for the individual ligands and is unlikely to account for the distinct signaling properties of the two angiopoietins.
Identification of Molecular Surface Regions Essential for Ang1 and Ang2 Functional Differences.
Our complex crystal structures document that Ang1 and Ang2 bind in a very similar manner to Tie2, revealing that altered ligand presentation is not responsible for the angiopoietins distinct biological activities. Alternatively, it appears more likely that angiopoietin surface residues outside the receptor-binding region are involved in defining their precise biological function, presumably by mediating direct or indirect interactions with coreceptors, such as Tie1. In this regard, the angiopoietin fibrinogen domain presents an extensive molecular surface (∼9,600 Å2) capable of mediating interactions with potential binding partners. Because candidate residues involved in mediating these interactions are likely not conserved between agonist and antagonist, we postulated a close examination of the RBD molecular surface in context of sequence conservation could potentially identify regions of interest.
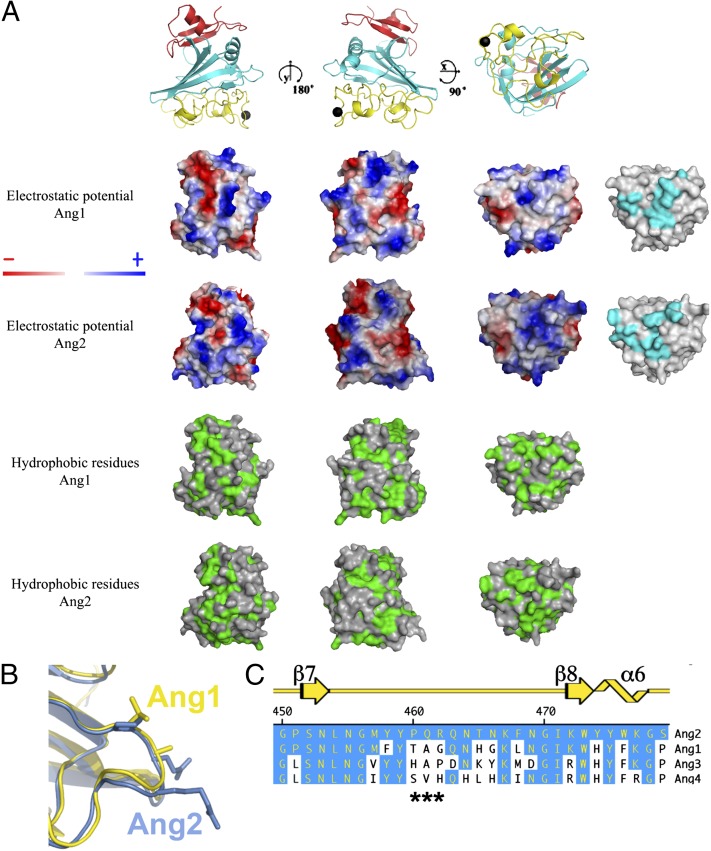
As highlighted in Fig. S2, the angiopoietins display considerable sequence homology, which is particularly evident within the receptor-binding P domain (12). Furthermore, the overall surface charge and hydrophobicity are relatively similar, although several regions with distinct chemical properties are unique to each ligand (Fig. 3). However, a small stretch of residues within the β6-β7 loop attracted our attention based upon the apparent lack of conservation, proximity to the receptor-binding interface, and location relative to the acidic Tie2 interface responsible for Tie1 binding (15). The β6-β7 loop, composed of residues 463–465 in Ang1 and 461–463 in Ang2, forms a small surface-exposed loop on the opposite face of the calcium-binding motif as highlighted in Fig. 3 B and C.
Fig. 3.
Conservation between Ang1-RBD and Ang2-RBD. (A) Electrostatic surface potential of Ang1-RBD (top row) and Ang2-RBD (second row from the top) and surface hydrophobicity of Ang1-RBD (third row from the top) and Ang2-RBD (last row). Secondary structures of Ang1-RBD are illustrated above the image to aid in model orientation. Surfaces residues involved in receptor recognition are shown in cyan for Ang1 and Ang2 on the right. (B) Close-up view of the β7-β8 loop in Ang1 (yellow) and Ang2 (blue). (C) Primary sequence alignment of the β7-β8 loop within Ang1 to -4. Regions that are conserved relative to Ang2 are shown in blue. Secondary structure is depicted in yellow at the top.
We decided to test whether an Ang2 chimera containing the Ang1 β6-β7 loop residues would behave functionally as a Tie2 agonist, more similar to Ang1 than to Ang2. Therefore, we mutated the P-Q-R sequence within Ang2 to that found in Ang1, T-A-G. Because these mutations are in a surface-exposed loop, they are unlikely to affect the overall folding of the protein. The Ang2-TAG chimera was expressed and found to behave biochemically well, as judged by gel-filtration chromatography. Its functional potential was assessed in three distinct assays: ability to modulate Tie1/Tie2 interactions, as observed by FRET microscopy; ability to cluster Tie2 in vivo; and ability to elicit functional activation of the Tie2-signaling pathway.
Chimeric Ang2-TAG Is Biologically Indistinguishable from Ang1.
To evaluate our Ang-fragment crystallizable (Fc) variants for functional activity, we analyzed their ability to cluster a Tie2-mCherry receptor stably expressed in EA.hy 926 endothelial cells. It is known from prior experiments that only Ang1 can significantly cluster and activate Tie2 in endothelial cells (15). Moreover, we have demonstrated previously (15) that Ang1 can dramatically change the localization of Tie2-mCherry after 30 min, with the receptor transitioning from diffuse membrane staining to forming discrete puncti on the cell surface. Tie2-mCherry localization was, therefore, monitored by fluorescence confocal microscopy following angiopoietin addition. Fig. 4 shows images of cells at 30 min after ligand addition. As observed previously, Tie2-mCherry exhibits punctate staining within 30 min in the presence of native Ang1 but remains unaltered in the presence of native Ang2 (Fig. 4 B and C). Full-length native angiopoietins form heterogeneous clusters and are difficult to express and purify. Therefore, for our studies, we took advantage of an Ang-RBD fusion to an Fc fragment to manipulate ligand stoichiometry. Under these conditions, Ang multimerization can be manipulated from their dimeric state induced by the IgG Fc domain, which is incapable of eliciting Tie2 activation under normal conditions, to tetramers and higher-order aggregates, which can induce Tie2 signaling in the presence of an anti-Fc antibody. In accordance, Davis and coworkers reported that addition of anti-Fc IgG to Ang1-Fc, and not Ang2-Fc, is essential to induce Tie2 phosphorylation (9, 14).
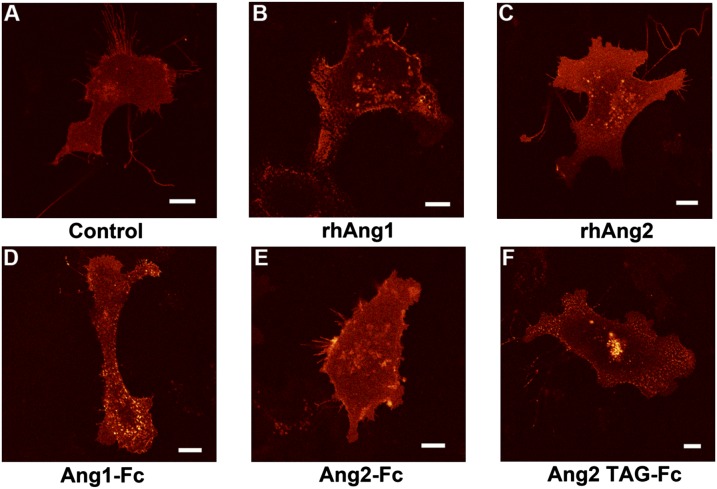
Fig. 4.
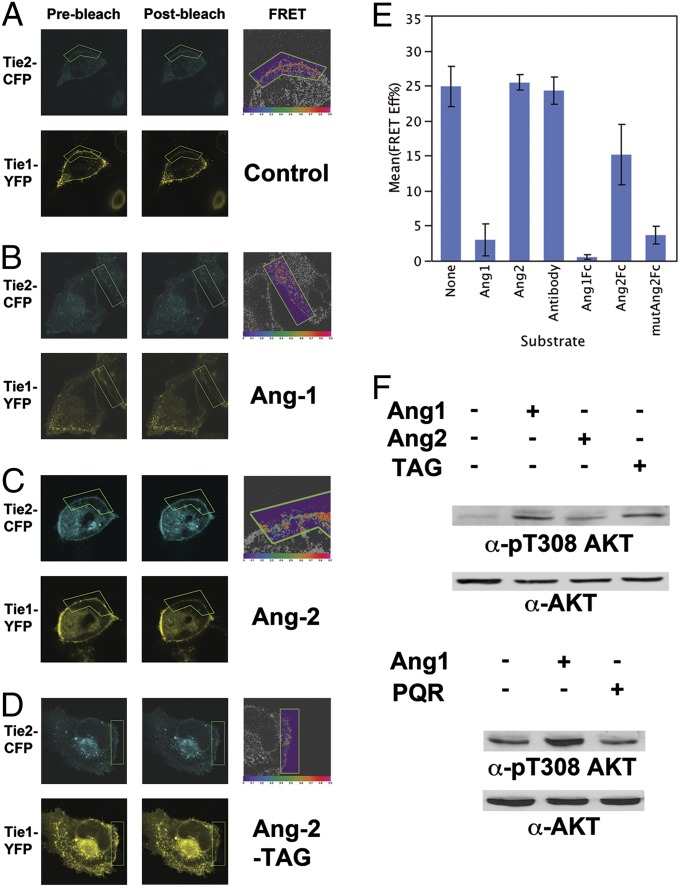
Ang1 and chimeric Ang2-TAG promote Tie2 clustering in endothelial cells. Stable EA.hy 926 endothelial cells expressing Tie2-mCherry (full-length) were stimulated with anti-Fc IgG (control) (A), recombinant human Ang1 (B), or Ang2 (C) or Fc fusions of Ang1 (D), Ang2 (E), or Ang2-TAG (F) (premixed with equimolar amounts of anti-Fc IgG cross-linking antibody) and followed for 30–60 min postaddition by confocal fluorescence microscopy. (Scale bars: 10 μm).
When added to cells, Ang1-Fc, indeed, functioned similar to native recombinant Ang1 and was able to cluster Tie2-mCherry within 30 min (Fig. 4D). Alternatively, addition of equivalent amounts of Ang2-Fc did not significantly affect Tie2 localization within 60 min (Fig. 4E). Interestingly, however, Ang2-TAG led to discrete Tie2 clusters within the membrane in a similar timeframe to that seen for both Ang1-Fc and native Ang1 (Fig. 4F). These findings suggest that, unlike Ang2, the Ang2-TAG chimera can activate Tie2 signaling and, analogous to Ang1, manipulate and disrupt the preexistent Tie1/Tie2 complex on the cell surface.
To test this hypothesis, we used an in vivo FRET-based proximity assay. The tyrosine kinase domains of Tie1 and Tie2 were replaced by the monomeric (m) green fluorescent protein variants, mCFP and mYFP, respectively. We have previously shown that Tie1 and Tie2 directly interact through opposing electrostatic interfaces within their ectodomains and that FRET between Tie1-mCFP and Tie2-mYFP can serve as a convenient and facile tool to monitor this interaction in vivo (15, 18). For our experiments, epithelial cells lacking endogenous receptors were transiently transfected with expression vectors for Tie1-CFP and Tie2-YFP. These modified Tie receptors localized to the cell surface and readily associated through their ectodomains in the absence of ligand. Building on our earlier results, we postulated that only the Ang1-Fc, but not Ang2-Fc, would be able to cause a disruption in FRET efficiency between Tie1-mCFP and Tie2-mYFP within 30 min of ligand addition. As a control, cells were treated with the clustering anti-Fc IgG antibody in the absence of ligand. Under these conditions, and as illustrated in Fig. 5A and graphically in Fig. 5E, no change was observed in the initial FRET efficiency between mCFP and mYFP after 30 min. Similarly, upon addition of Ang2-Fc, the average FRET efficiency remained high and was determined to be 15.2% using the acceptor photobleaching methodology (Fig. 5 C and E). Importantly, Tie2 localization remained unchanged under both conditions. In contrast, a dramatic loss in FRET was observed after treatment of cells with Ang1-Fc (Fig. 5 B and E). As illustrated in Fig. 5B, after 30 min, a large change in Tie2 localization occurred, with mYFP emission concentrated in discrete signaling clusters or foci. However, in agreement with our previous study, Tie1 localization remained relatively unaffected on the cell surface despite the decrease in the average FRET efficiency to 0.49%. These observations confirm that the Ang1-Fc fusion protein can disrupt Tie1/Tie2 complexes in a similar manner to the native Ang1 ligand. Both Ang1 ligands appear to induce Tie2 activation by clustering the Tie2 receptor and disturbing the inhibitory Tie1/Tie2 complex. Finally, we evaluated the effect our mutant Ang2-TAG protein has on modulating the Tie interactions. As illustrated in Fig. 5D and graphically in Fig. 5E, following treatment with Ang2-TAG, Tie2-mCFP localization is altered on the cell surface, resulting in a corresponding loss in FRET efficiency between Tie1 and Tie2. The average FRET efficiency after 30 min declined to 3.6%. Collectively, these observations demonstrate that Ang2-TAG functions more akin to a Tie2 receptor agonist, than antagonist.
Fig. 5.
Ang1 and chimeric Ang2-TAG dissociate Tie1/Tie2 complexes on the cell surface and stimulate Tie2 signaling. (A) U2OS cells were transfected with both Tie1-CFP and Tie2-YFP and analyzed by confocal microscopy following stimulation with vehicle (A), Ang1-Fc (B), Ang2-Fc (C), or the Ang2-TAG-Fc chimera (D). Thirty minutes postaddition, regions of interest (ROIs) on the membrane were subjected to acceptor photobleaching (green box) and images were taken before (prephotobleach) and after (postphotobleach) bleaching to calculate FRET efficiencies. Images on the right are false-colored according to FRET efficiency (red, high; purple, low). Average FRET efficiencies are graphically illustrated in E. Values for control, Ang-1, and Ang-2 experimental stimulations are in agreement with previous studies (15). (F) Endothelial cells were stimulated at ∼80% confluence with vehicle, Ang-1, Ang-2, or Ang-2-TAG (Upper) or with vehicle, Ang-1, or Ang1-PQR (Lower) for 30 min. Whole-cell lysates were probed for both activated and total AKT protein levels.
To more thoroughly assess the functional signaling properties of the individual ligands, we next assayed their ability to induce endogenous Tie2 downstream signaling cascades in endothelial cells by following the activation, or phosphorylation, of v-akt murine thymoma viral oncogene homolog (AKT). AKT is an immediate downstream effector of Tie2 signaling (19). As opposed to Tie2 phosphorylation, which is difficult to track and yields only modest (two- to threefold) changes in response to ligand, even when using the commercially available anti-pY992 Tie2 antibody, AKT phosphorylation is more pronounced and can be easily and conveniently followed using excellent phospho-specific antibodies. For our experiments, EA.hy 926 cells were grown to ∼80% confluence, serum-starved for 6 h, and incubated with equivalent amounts of full-length ligand for 15 min before cellular harvest. Whole-cell lysates were subsequently probed by Western blot with anti-pT308 AKT antibodies and normalized for total protein content using anti-AKT antibody. As illustrated in Fig. 5F, AKT phosphorylation increases substantially over background levels in the presence of Ang1 or the Ang2-TAG chimera but not in the Ang2 or control stimulated cells, demonstrating that in addition to its ability to cluster and disrupt the Tie1/Tie2 complexes, Ang2-TAG can stimulate functional Tie2 downstream signaling.
Finally, to validate and confirm our conclusions, we constructed the complementary variant to Ang2-TAG, which we term Ang1-PQR. Ang1-PQR contains the corresponding three residues (P-Q-R) found within Ang2 and would be predicted to function analogously to Ang2, as a Tie2 antagonist. Indeed, in the presence of Ang1-PQR, endogenous Tie2 is not activated and in contrast to wild-type Ang1, basal AKT phosphorylation remains low (Fig. 5F).
Chimeric Ang2-TAG Is Structurally Sound.
To further reveal any molecular alterations that occur in Ang2-TAG, we determined the Ang2-TAG crystal structure by molecular replacement using the Ang2 structure as a search model. The final model was refined to an R factor of 20.2% (Rfree of 22.6%) at 1.9 Å. As illustrated in Fig. S2, the overall architecture is unchanged and Ang-2 can be superimposed on Ang2-TAG with an rmsd of 0.5 Å for all Cα atoms. Perhaps not unexpectedly, the most prominent difference between corresponding Cα positions was observed for the R462G substitution in the β7-β8 loop, which was shifted by 1.8 Å outward from the Tie2 receptor-binding interface (Fig. S2B). The similarity between these two structures reveals the PQR/TAG substitution does not induce any large conformation changes to account for the difference in Tie2 activation and, instead, suggests that the observed difference in ligand activity is a result of altered ability to modulate interactions of the Tie/Ang complex with other proteins.
Discussion
Although the individual angiopoietins share significant sequence homology, they have very distinct signaling properties. To understand angiopoietin differences at the atomic level, we determined the crystal structures of Ang1 and the Ang1/Tie2 complex and compared them with the previously determined Ang2 and Ang2/Tie2 structures, respectively. Ang1 and Ang2 share significant structural similarity and form remarkably similar complexes with Tie2. Indeed, residues involved in receptor–ligand interactions are mostly conserved and, therefore, ligand binding to Tie2 is not substantially different between Ang1 or Ang2.
In additional support of this observation, we previously identified Tie1 as a critical regulator of the functional differences between the angiopoietin ligands (15). Here, we further demonstrate that angiopoietin residues outside of the receptor-binding site influence the Tie1/Tie2 interactions and mediate the angiopoietin functional differences. Indeed, analysis of the surface properties, including amino acid conservation, suggested several potentially important surfaces within Ang2. Investigating these candidate regions using structure-based mutagenesis in combination with cell-based signaling assays, we identified one chimeric ligand, Ang2-TAG, which displayed the predicted phenotypic switch from antagonist to agonist. In contrast to Ang2, Ang2-TAG was able to cluster Tie2, disrupt Tie1/Tie2 interactions, and stimulate Tie2 signaling in the presence of Tie1 within endothelial cells. To validate our findings with Ang2-TAG, we demonstrate that the corresponding Ang1 chimera, Ang1-PQR, functions as a Tie2 receptor antagonist.
Based on our current knowledge of Tie1/Tie2 interactions, which are likely dominated by electrostatic contacts, it is not unexpected that a charged surface residue is among those involved in ligand differentiation. Indeed, the TAG mutant replaces an arginine with glycine and suggests that Ang2 binding may electrostatically stabilize the Tie1/Tie2 complex.
Conclusion
In conclusion, we demonstrate that three critical residues within the angiopoietin fibrinogen domain are necessary to confer the biological activity of an angiopoietin ligand from Tie2 antagonist (Ang2) to Tie2 agonist (Ang1), thus illustrating that limited alterations in protein–protein interactions and interfaces may have broad implications for protein function. In light of these findings, we predict that therapeutics targeting this region within Ang2 would be both highly specific and beneficial in blocking Tie2-induced angiogenesis.
Materials and Methods
Cloning and Mutagenesis.
Human Ang1-RBD (residues 279–498) was cloned into a modified pAcGP67 baculovirus expression vector (BD Bioscience), with GP67 secretion signal and human Fc fragment as the C-terminal tag. Recombinant baculovirus was generated by cotransfecting the expression plasmid along with linearized BaculoGold DNA (Pharmingen) into SF9 cells. The human Ang2-RBD (residues 276–496) or Tie2 ectodomain (residues 1–541) was cloned as an IgG fusion protein into a modified pcDNA3.1 vector (Invitrogen) for constitutive overexpression in a human embryonic kidney (HEK)293 cell line as described previously (12). Expression constructs for full-length angiopoietins with a C-terminal myc and FLAG epitope tags were obtained from Origene. Human recombinant Ang1 and Ang2 were purchased from R&D Systems.
Mutations within Ang2 coding regions were introduced by site-directed mutagenesis (QuikChange Multi; Stratagene). To confirm the presence of the desired mutations, both DNA strands were sequenced.
Protein Expression and Crystallization.
Large-scale protein expression was performed from stably expressing HEK293 cells in a BioFlo310 bioreactor (New Brunswick Scientific) or roller-bottle culture with typical yields averaging 10 mg/L for Ang2-TAG and Tie2. Fusion proteins were purified as described (12, 13). Full-length angiopoietins were purified via anti-myc (9E10) affinity chromatography and eluted with low-pH buffer [100 mM glycine (pH 3.0), 150 mM NaCl] before buffer exchange with HBS [20 mM Hepes (pH 7.0), 150 mM NaCl].
For Ang1-RBD expression, Hi5 cells were infected with baculoviruses at a multiplicity of infection of ∼10. The clarified harvested media, containing the secreted fusion protein, was collected 72 h postinfection and loaded onto a protein A-Sepharose affinity column. Recombinant protein was eluted with low-pH buffer containing 150 mM NaCl and 100 mM glycine (pH 3.0). The Fc tag was cleaved by thrombin proteolysis and removed by Protein A-Sepharose. Ang1-RBD was further purified by gel-filtration chromatography. Purified Ang1-RBD was concentrated to 12 mg/mL in HBS and crystallized by hanging-drop vapor diffusion at room temperature against a well solution of 3.9 M sodium formate and 100 mM Tris (pH 8.5). Native crystals were frozen in a cryo-buffer consisting of the mother liquor with an additional 25% (vol/vol) glycerol.
Following purification, Ang2-TAG was concentrated to 15 mg/mL in HBS and crystallized by hanging-drop vapor diffusion at room temperature against a well solution of 1.0 M Na/K-tartrate, 0.1 M Tris (pH 7.0), 0.2 M LiSO4. Crystals were frozen in a cryo-buffer consisting the mother liquor with an additional 20% (vol/vol) ethylene glycol.
For production of the Ang1/Tie2 complex, purified Tie2 and Ang1-RBD were mixed in a 1:2 molar ratio and incubated on ice for 1 h. The complex was purified on a SD200 gel-filtration column (GE Healthcare) and concentrated to 10 mg/mL in HBS. The complex was crystallized by sitting-drop vapor diffusion at room temperature against a well solution of 1.6 M NaH2PO4, 0.4 M K2HPO4, and 0.1 M phosphate-citrate (pH 4.2). Thin, long needle-like crystals grew over the course of 2–3 d, with a maximum size of 300 × 30 × 30 μm. For data collection, the crystals were frozen in a cryo-buffer consisting of the mother liquor with an additional 25% (vol/vol) glycerol. X-ray diffraction data were collected at beamline 24ID-C (Northeastern Collaborative Team, Advance Photon Source) and processed with DENZO and SCALEPACK (20). Crystallographic details are listed in Table S1.
The structures of Ang1-RBD and the Ang1/Tie2 complex and Ang2-TAG were determined using molecular replacement and the Collaborative Computational Project 4 (CCP4) program AMoRe (21, 22). The structures of Ang2-RBD [Protein Data Bank (PDB) ID code 1Z3S] and the Ang2/Tie2 complex (PDB ID code 2GY7) determined by our group were used as a search model. Subsequent refinement proceeded with iterative rounds of model adjustments (using the molecular graphics program O), molecular dynamics, and energy minimization in the Crystallography and NMR System (CNS) or the CCP4 program REFMAC5 (23–25). For Ang2-TAG, residues 467–476 were omitted in the initial search model to keep the model as unbiased as possible.
Cell Manipulations and Transfections.
HEK293, U2OS, and EA.hy 926 cells (a gift from Cora-Jean Edgell, retired) were grown in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. TIME (hTERT immortalized microdermal endothelial) cells (ATCC) were grown in endothelial basal media-2 (EBM-2) media supplemented with the EGM-2 MV SingleQuot (Lonza). Cells were consistently transfected at 80–90% confluence in 35-mm glass bottom culture dishes (MatTek) (for imaging) or six-well plates (for biochemical analysis) using Lipofectamine 2000 (HEK293), FuGENE HD (U2OS), or FuGENE 6 (EA.hy 926) (Invitrogen and Roche). For coexpression, equimolar concentrations of Tie1 and Tie2 vector DNA were used.
Cellular Imaging.
Cultures were grown to ∼80% confluence, serum-starved for 2–6 h before the addition of 400 ng/mL Ang-1, -2, -2-TAG, or vehicle (PBS). The FRET-based spatial proximity assay and all fluorescence imaging experiments were based upon previous studies and performed essentially as described (15, 18).
Tie2 Activation Assays Cellular Imaging.
For analysis of Tie2 activation, full-length ligand constructs (pcDNA3.1 vector only, Ang-1, -2, -1-PQR, and -2-TAG) were transfected into HEK293 cells and incubated for 48 h. Media containing secreted ligand were analyzed for ligand concentration via Western blot. Ea.hy 926 or TIME cells were grown to ∼80% confluence and serum-starved for 4 h before the addition of equal concentrations of ligand media for 30 min. Cells were lysed in HBST [20 mM Hepes (pH 7.4), 150 mM NaCl, 0.1% Triton X-100] in the presence of PhosSTOP and Complete protease and phosphatase inhibitors (Roche). Equal amounts of protein were resolved by SDS/PAGE and transferred to nitrocellulose for Western blotting. Endogenous AKT was analyzed with anti-AKT (Cell Signaling) or anti–phospho-specific AKT T308 (Cell Signaling).
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (NIH) Grants 1R01CA127501 (to W.A.B.) and 1R01HL077249 (to D.B.N.), as well as pilot project funding from the Massey Cancer Center and School of Medicine [Virginia Commonwealth University (VCU)] (to W.A.B.). Microscopy was performed at the VCU Department of Neurobiology and Anatomy Microscopy Facility, supported, in part, by NIH National Institute of Neurological Disorders and Stroke Center Core Grant 5P30NS047463. X-ray diffraction studies were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by National Center for Research Resources Grant 5P41RR015301-10, National Institute of General Medical Sciences Grant 8 P41 GM103403-10, and Department of Energy Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the RCSB Protein Data Bank (accession nos. 4JZC, 4JYO, and 4KOV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216890110/-/DCSupplemental.
References
- 1.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146(6):873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J, D’Amore PA. Blood vessel formation: What is its molecular basis? Cell. 1996;87(7):1153–1155. doi: 10.1016/s0092-8674(00)81810-3. [DOI] [PubMed] [Google Scholar]
- 3.Jones N, Iljin K, Dumont DJ, Alitalo K. Tie receptors: New modulators of angiogenic and lymphangiogenic responses. Nat Rev Mol Cell Biol. 2001;2(4):257–267. doi: 10.1038/35067005. [DOI] [PubMed] [Google Scholar]
- 4.Sato TN. Vascular development: Molecular logic for defining arteries and veins. Curr Opin Hematol. 2003;10(2):131–135. doi: 10.1097/00062752-200303000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL. Angiogenesis in ischemic and neoplastic disorders. Annu Rev Med. 2003;54:17–28. doi: 10.1146/annurev.med.54.101601.152418. [DOI] [PubMed] [Google Scholar]
- 6.Yancopoulos GD, et al. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407(6801):242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 7.Zeeb M, Strilic B, Lammert E. Resolving cell-cell junctions: Lumen formation in blood vessels. Curr Opin Cell Biol. 2010;22(5):626–632. doi: 10.1016/j.ceb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Bhat A, Woodnutt G, Lappe R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer. 2010;10(8):575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]
- 9.Davis S, et al. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87(7):1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 10.Maisonpierre PC, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277(5322):55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela DM, et al. Angiopoietins 3 and 4: Diverging gene counterparts in mice and humans. Proc Natl Acad Sci USA. 1999;96(5):1904–1909. doi: 10.1073/pnas.96.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barton WA, Tzvetkova D, Nikolov DB. Structure of the angiopoietin-2 receptor binding domain and identification of surfaces involved in Tie2 recognition. Structure. 2005;13(5):825–832. doi: 10.1016/j.str.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Barton WA, et al. Crystal structures of the Tie2 receptor ectodomain and the angiopoietin-2-Tie2 complex. Nat Struct Mol Biol. 2006;13(6):524–532. doi: 10.1038/nsmb1101. [DOI] [PubMed] [Google Scholar]
- 14.Davis S, et al. Angiopoietins have distinct modular domains essential for receptor binding, dimerization and superclustering. Nat Struct Biol. 2003;10(1):38–44. doi: 10.1038/nsb880. [DOI] [PubMed] [Google Scholar]
- 15.Seegar TC, et al. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell. 2010;37(5):643–655. doi: 10.1016/j.molcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aricescu AR, et al. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317(5842):1217–1220. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 17.Kulahin N, et al. Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule. Structure. 2011;19(2):203–211. doi: 10.1016/j.str.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Seegar T, Barton W. Imaging protein-protein interactions in vivo. J Vis Exp. 2010;(44):e2149. doi: 10.3791/2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeBusk LM, Hallahan DE, Lin PC. Akt is a major angiogenic mediator downstream of the Ang1/Tie2 signaling pathway. Exp Cell Res. 2004;298(1):167–177. doi: 10.1016/j.yexcr.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 22.Trapani S, Navaza J. AMoRe: Classical and modern. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):11–16. doi: 10.1107/S0907444907044460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skubak P, Murshudov GN, Pannu NS (2004) Direct incorporation of experimental phase information in model refinement. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2196–2201. [DOI] [PubMed]
- 24.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.