Abstract
Fe4S4 clusters are very common versatile prosthetic groups in proteins. Their redox property of being sensitive to O2-induced oxidative damage is, for instance, used by the cell to sense oxygen levels and switch between aerobic and anaerobic metabolisms, as exemplified by the fumarate, nitrate reduction regulator (FNR). Using the hydrogenase maturase HydE from Thermotoga maritima as a template, we obtained several unusual forms of FeS clusters, some of which are associated with important structural changes. These structures represent intermediate states relevant to both FeS cluster assembly and degradation. We observe one Fe2S2 cluster bound by two cysteine persulfide residues. This observation lends structural support to a very recent Raman study, which reported that Fe4S4-to-Fe2S2 cluster conversion upon oxygen exposure in FNR resulted in concomitant production of cysteine persulfide as cluster ligands. Similar persulfide ligands have been observed in vitro for several other Fe4S4 cluster-containing proteins. We have also monitored FeS cluster conversion directly in our protein crystals. Our structures indicate that the Fe4S4-to-Fe2S2 change requires large structural modifications, which are most likely responsible for the dimer–monomer transition in FNR.
Keywords: FeFe-hydrogenase, X-ray crystallography
Iron–sulfur (FeS) clusters are among the most ubiquitous and functionally versatile prosthetic groups in nature. They can be involved in electron transfer, substrate binding and activation, be used as stress sensors to regulate gene expression or simply play a structural role (1). FeS clusters have variable compositions such as the common Fe2S2, Fe3S4, and Fe4S4 centers or the more unusual FeMo-cofactor Fe7S9Mo cluster of nitrogenase (2). When Fe4S4 clusters are redox active, they are also usually sensitive to oxidation and can be damaged (or disassembled) by oxygen. Escherichia coli uses this property to switch between aerobic and anaerobic metabolisms (3, 4). Indeed, at under about 0.5% oxygen concentration, the fumarate nitrate reduction regulator (FNR) can accommodate an Fe4S4 cluster and is a dimer that can bind specific palindromic DNA sequences to either repress or activate the expression of hundreds of genes (5). When exposed to higher oxygen levels, the Fe4S4 cluster is rapidly converted into an Fe2S2 cluster, causing FNR to dissociate into monomers that do not bind DNA. Finally, the Fe2S2 cluster slowly decomposes into two Fe3+ and two S2− ions, leading to the apo protein form of FNR (6). Thus, the cell response to oxygen and its ensuing commitment to aerobic metabolism are modulated by structural changes associated with the rapid conversion of the Fe4S4 cluster into an Fe2S2 cluster in FNR. The tridimensional structure of an FNR has not yet been determined. A very recent study combining Raman and UV-visible absorption/circular dichroism (CD) spectroscopies and mass spectrometry (7) indicates that Fe4S4-to-Fe2S2 cluster degradation does not proceed with concomitant release of one Fe2+, one Fe3+, and two S2− ions as was previously reported (8). Instead, that study argues that cluster conversion leads to an Fe2S2 cluster with retention of two sulfides as cysteine persulfide ligands according to the following reactions (7):
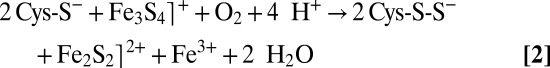 |
O2-induced Fe4S4 cluster degradation leading to Fe2S2 cluster with cysteine persulfide ligands does not seem to be restricted to FNR and is likely to be a general process, as it is also reported for other Fe4S4 cluster-containing proteins such as biotin synthase and aconitase (7, 9). Spectroscopic inference of species containing Fe2S2 cluster forms with only one or two cysteine persulfide ligands suggests successive oxidations but the mechanism involved is not fully understood. Here, we have used X-ray structures of HydE from Thermotoga maritima (Tm) as templates to shed light on the process of O2-induced Fe4S4 cluster disassembly in proteins. Our structural results provide direct evidence that the O2-induced oxidation of an Fe4S4 cluster can proceed via cysteine persulfide production and thus establishes a sulfur-centered mechanism for cluster degradation as a plausible biological process. TmHydE is one of the three essential FeFe-hydrogenase active site maturases (10, 11) which belongs to the radical S-adenosyl-L-methionine (SAM) protein superfamily (12, 13). Besides the conserved Fe4S4 cluster of radical SAM proteins, TmHydE has an additional, more exposed cluster-binding site defined by semiconserved Cys311, Cys319, and Cys322, in a Cx7Cx2C motif. Cys311 is located in a loop connecting β-strand 8 to α-helix 8 of the (βα)8 triose phosphate isomerase barrel of the enzyme, and Cys319 and Cys322 are located on the same side of α-helix 8 (11) (Fig. 1). HydEs are divided into two groups depending on whether the Cys ligands of the additional FeS cluster are present, but it is known that this cluster is not important for function (11). In the first TmHydE X-ray structure we solved, the additional cluster, located at the protein surface, was an Fe2S2 rhomb (11). Conversely, solution studies showed that after in vitro reconstitution TmHydE binds an Fe4S4 cluster at the additional site (14). Here, we report eight different structures of TmHydE that contain dissimilar species at their additional cluster site. These species are in agreement with several intermediate states, either postulated or previously described, in the Fe4S4 cluster degradation pathway occurring, for instance, in FNR. These observed intermediate states in our structures suggest a mechanism for Fe4S4 cluster degradation upon low-level oxygen exposure not restricted to FNR, but more general than previously thought.
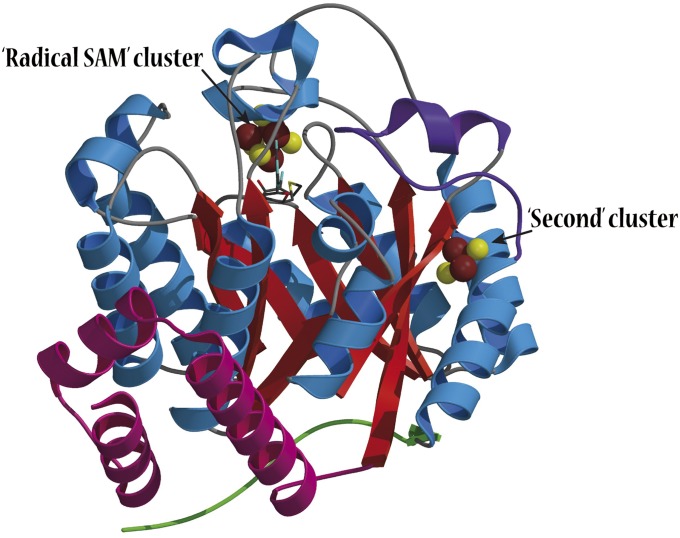
Fig. 1.
Structure of the HydE from T. maritima (PDB code 3CIW). The SAM-dependent Fe4S4 cluster is located at the top of the (βα)8 barrel (color codes are S in yellow, iron in brown, β-strands in red, and α-helices in blue). The SAM-independent additional Fe2S2 cluster is located on a side of the barrel at the surface of the protein. The loop containing the cysteine ligand Cys311 is depicted in purple. The N-terminal three helices and the C-terminal stretch are depicted in pink and green, respectively.
Results
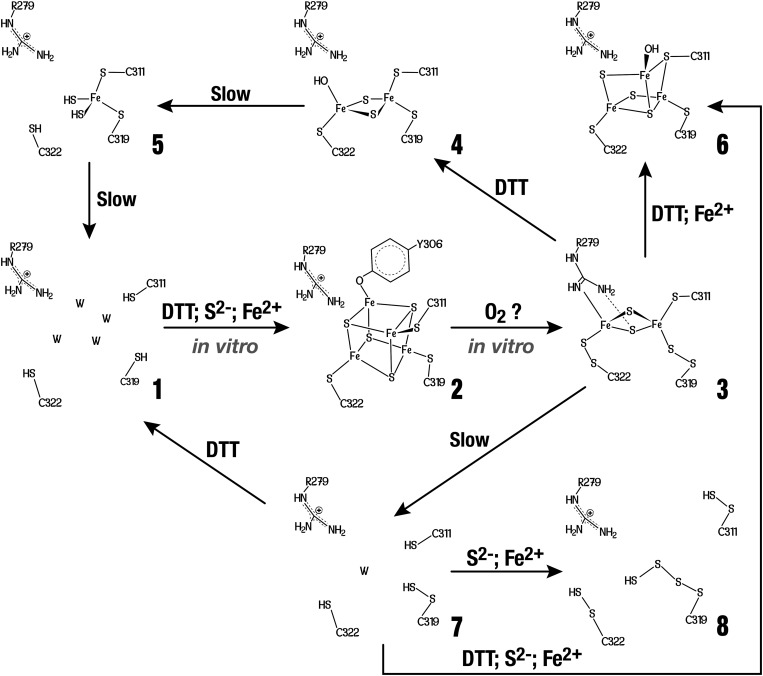
As mentioned above, in vitro anaerobically reconstituted TmHydE has been reported to contain an Fe4S4 center at its additional cluster-binding site in solution (14). However, when crystallized without DTT, the additional site coordinates an Fe2S2 cluster bound by the semiconserved Arg279, Cys311, and persulfided forms of Cys319 and Cys322 (species 3 in Fig. 2 and Fig. S1). The Nη2 atom of Arg279 interacts directly with one of the iron ions (d = 2.3 Å), whereas Nη1 makes a hydrogen bond with one of the sulfide ions (d = 3.2 Å). Freshly reconstituted samples normally produce crystals of this form after 1 wk in the glove box. Persulfide formation must result from either traces of O2 present in the glove box or transient exposure to air during the transfer of the frozen protein sample to it, because when intentionally oxygenated buffers are used these crystals appear more rapidly. Although in these experiments it is not possible to quantify the amount of O2 that reached our crystals, it must be extremely low as indicated by the high stability of the SAM-dependent Fe4S4 cluster under the same conditions. Indeed, when SAM is substituted by 5′-deoxyadenosine, this cluster is slowly converted to an Fe3S4 center in weeks without any further decay (Fig. S2). In the meantime, the additional cluster is rapidly converted into species 3. The latter is very stable and only decays after several months, leading to 7 (Fig. 2).
Fig. 2.
Postulated degradation and assembly of an FeS cluster based on our crystal structures. Unless specified, all reactions were performed in our crystals. W in species 1 and 7 indicates solvent molecules. X-ray structures of TmHydE containing species 1 to 8 were determined at 1.35, 2.9, 1.45, 1.6, 1.71, 1.7, 1.45, and 1.25 Å resolution, respectively (Table S2).
When crystals containing 3 are treated with DTT before flash cooling, a different Fe2S2 cluster, coordinated by Cys311 Cys319, and Cys322, is observed (4 in Fig. 2). There are no major structural changes at the protein level between 3 and 4 and the DTT treatment only causes the cluster to slightly move as a rigid body in its binding site. Conversely, the electron density indicates that the Fe fourth ligand may be OH−, as Arg279 is clearly too far to be a ligand. We cannot exclude a mixture of OH− and SH− at that position because our structures unambiguously show that sulfide ions are released from the cysteine persulfide ligands upon DTT treatment. X-ray structures indicate that this cluster form decays within weeks in the glove box. This process, which starts with the loss of one iron atom, is followed by the dissociation of the sulfide ions and finally results in the vacant-site species 1 (Fig. 2). Species 5 represents an intermediate form in the degradation of 4 (Fig. 2 and Fig. S1). X-ray structures from crystals flash-cooled at different times after DTT addition show that the slow disassembly of the additional cluster starts with the most solvent-exposed iron ion followed by the sulfide ions and, finally, by the second iron ion (5 to 1 in Fig. 2). The slow decomposition of the Fe2S2 form is likely to be a nonredox process and may result from slow ligand exchange with solvent.
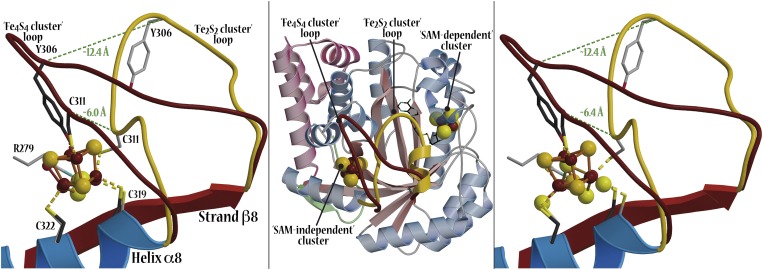
When crystals containing 7 are treated with ferrous ions and sulfide, no iron bound to the protein is observed but Cys311 and Cys22 of the additional cluster site are modified as persulfides and Cys319 as polysulfide (8 in Fig. 2 and Fig. S1). Treating similar crystals with ferrous ions, sulfide, and DTT leads to another unusual species (6 in Fig. 2 and Fig. S1) that can be described as an Fe2S2 cluster onto which an Fe–S unit is attached. This cluster resembles a standard Fe3S4 center except that Cys311 bridges two iron ions. Modeling indicates that the structural changes required to allow Cys311 to become a classic terminal Fe4S4 cluster ligand are prevented by crystal packing contacts (not shown). This problem was circumvented by the growth of a different crystal form of anaerobically purified TmHydE, treated with DTT, sulfide, and ferrous or ferric ions (Materials and Methods, Table S1). This form that was obtained by increasing the NaCl concentration in the crystallization solution contains a regular additional Fe4S4 cluster, as previously described in solution (14), with Tyr306 as its fourth ligand (2 in Fig. 2 and Fig. S1). Fig. 3 shows that shifts of over 6.0 Å for the Cα of Cys311 and 12.4 Å for the Cα of Tyr-306 are involved in the transition between the Fe2S2-containing and the Fe4S4-containing forms. This change requires a complete rearrangement of the loop that connects strand β8 to helix α8 and contains Cys311 (Fig. 3). Thus, the Fe4S4-to-Fe2S2 cluster conversion (2 to 3) is a conformationally drastic one. In the reverse direction, addition of DTT and Fe2+ (but not of S2-) converts 3 into 6 (Fig. 2 and Fig. S1). The peculiar Fe3S3 cluster in 6 probably results from the aborted assembly of an Fe4S4 cluster hindered, as mentioned above, by crystal packing interactions. A major conclusion from this observation is that there is no need to add sulfide ions to obtain the observed Fe3S3 cluster because they are already present as part of persulfide ligands in 3.
Fig. 3.
Structural differences between SAM-independent Fe4S4 and Fe2S2 cluster-containing TmHydE. (Left) Superposition of the additional cluster sites containing species 2 and 4 (Fig. 2). Loop containing Cys311 is depicted in brown for the Fe4S4-cluster structure and in gold for the Fe2S2-cluster structure. Dashed line indicates the shift of the Cα atom of Cys311 and Tyr-306 between the two structures. (Center) View of the whole structure. Loop containing Cys311 covers the barrel cavity when the protein contains an Fe2S2 cluster (gold) and moves away from it when the protein contains an Fe4S4 cluster (brown). (Right) Superposition between species 2 and 3. Only minor differences are observed in the conformation of the Cys311-containing loop between species 3 and 4, indicating that conversion of species 2 into 3 or 4 should induce similar conformational changes.
Discussion
Many of our X-ray crystal structures are in full agreement with the very recent Raman resonance (RR) spectroscopic study of Zhang et al. and consequently may be relevant to FeS cluster interconversion in FNR (7). As opposed to the additional cluster, the SAM-related Fe4S4 cluster of TmHydE was stable for a long time inside the glove box. This is most likely explained by the protection afforded to the latter by the Cx3Cx2C motif-containing loop. Conversely, the additional cluster is fully exposed to solvent and hence to traces of O2 either in the glove box or during sample transfer to it. This unanticipated situation is actually advantageous because it mimics what should happen for example to E. coli in the rectal segment of the intestinal tract when oxygen levels begin to rise and FNR is inactivated, switching off anaerobic metabolism. The very slow reaction of the additional FeS cluster with traces of O2 allowed us to trap intermediate species in our protein template that would have been difficult to obtain otherwise. Thus, the Fe2S2 cluster with two cysteine persulfide ligands proposed from RR spectroscopic data in FNR is equivalent to 3 (Fig. 2 and Fig. S1). In addition, when treated with ferrous ions and DTT, this FNR Fe2S2 species is readily converted to a regular Fe4S4 cluster (7), in a process similar to the 3 to 6 transition in Fig. 2. According to reaction 1, the first step of O2-induced Fe4S4 cluster degradation corresponds to iron oxidation, leading to an intermediate Fe3S4⌉+ cluster with concomitant release of one ferrous ion and a superoxide radical species (15). Conversely, species 3 with cysteine persulfide ligands demonstrates that subsequent oxidative damage proceeds preferentially via sulfur rather than iron oxidation. Furthermore, the intermediate species with only one persulfide ligand observed in FNR indicates that successive oxidation steps are possible (7). At variance with what is proposed by Zhang et al. for FNR (reaction 2), instead of the addition of a new oxygen molecule, we advance that the next step corresponds to the reaction of the superoxide anion resulting from reaction 1 with one of the cysteine ligands forming a cysteine persulfenate. Such species that would be transient in our case has been observed in the crystal structure of cysteine dioxygenase (16). The expected conversion of the cysteine persulfenate to sulfinate does not occur, maybe due to the sulfide-rich environment of the adjacent iron ion. Instead, this species is converted to cysteine persulfide with production of one hydrogen peroxide molecule and release of a second ferrous ion, as previously observed in FNR (17, 18). Reaction of a superoxide ion, instead of molecular oxygen, with the Fe3S4⌉+ intermediate would be consistent with the observation that in FNR only 0.37 superoxide ions per damaged Fe4S4 cluster were detected from reaction 1 (15). Substantial amounts of hydrogen peroxide were also produced (15), which could subsequently oxidize a second cysteine ligand to cysteine sulfenate. Reaction of this species with the last available hydrogen sulfide ion would generate a second persulfide, thus leading to a form equivalent to 3. Such mechanism would explain why only substoichiometric amounts of H2O2 are produced by FNR and is consistent with the recent proposition that some FNR molecules have only one cysteine persulfide ligand (7, 15, 17). Furthermore, our mechanism is supported by the observation that both O2 and H2O2 can provoke an Fe4S4-to-Fe2S2 cluster transition in FNR. However, the resulting Fe2S2 cluster has a different circular dichroism spectrum depending on the oxidant used (17). This difference is probably due to the expected number of cysteine persulfide ligands produced: two when using O2 and one when using H2O2. The observation that in FNR one ferric iron is released during the 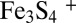 to
to 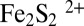 cluster conversion may be explained by the oxygen-independent reaction 3 (8):
cluster conversion may be explained by the oxygen-independent reaction 3 (8):
This nonredox reaction may compete with sulfide oxidation and could be favored in vitro by the presence of strong ferrous or ferric ion chelators (8), suggesting an alternative nonbiological pathway for oxygen-induced FeS cluster degradation.
Addition of a reductant and extra ferrous ions converts species 3 back to 2 (or 6 in our crystals). Both our structural results and those reported by Zhang et al. (7) using RR spectroscopy raise the possibility of a persulfide-mediated Fe4S4 cluster reparation mechanism that, avoiding de novo sulfide synthesis by the iron–sulfur cluster (ISC) system, would be less costly to the cell. Cysteine persulfide ligands as an O2-induced degradation product of Fe4S4 clusters appear to be generally distributed, being also proposed in biotin synthase and other radical SAM enzymes (7).
Our observation that species 4 slowly decomposes into 5 and then 1, most probably via a nonredox process (Fig. 2 and Fig. S1), agrees well with the previous observation that iron depletion itself can mediate the holo to apo transition in cytoplasmic aconitase (19). Our study also shows that the transition from the Fe4S4 cube to the Fe2S2 rhomb would require a major reorganization of the coordination site in TmHydE. The loop rearrangement resulting from this reorganization implies that an Fe4S4-to-Fe2S2 cluster conversion will necessarily trigger significant protein structural changes. Equivalent changes may be responsible for O2-induced dimer dissociation in FNR (20). Similarly, the transition between holo and apo cytoplasmic aconitase has been structurally characterized and shown to involve large structural rearrangements (21, 22). Finally, an early study (9) showed that controlled oxidation of apo-aconitase resulted in the formation of both persulfides and polysulfides, a situation illustrated here by 7 and 8 (Fig. 2 and Fig. S1).
Our results may also shed light on the sequence of events during in vivo ISC-dependent FeS cluster assembly in the complex formed by the L-cysteine desulfurase IscS and the scaffold IscU. The order of arrival of iron and sulfide to the IscU assembly site has not been clearly established. This scaffold protein can bind iron, but only at lower-than-physiological temperatures (23, 24). Conversely, several groups have reported that S0, transferred from the cysteine desulfurase, can form a Cys-S–S species in the scaffold protein, in the absence of iron (25, 26). However, it has been reported that the persulfided scaffold does not bind iron (24). The latter observation applies also to the oxidized TmHydE 7 species where Cys311 and persulfided Cys319 form a long S–S bond (Fig. S1). Addition of sulfide and Fe2+ to 7 produces 8, a species that contains persulfided and polysulfided cysteines but does not bind iron (Fig. 2 and Fig. S1). This observation implies that free Fe2+ is not able to reduce the S–S bond of the modified Cys319 in 7. This result favors in turn a concerted mechanism for Fe–S cluster assembly by ISC, where two ferrous ions bind simultaneously the first persulfide S0 from the active Cys residue of IscS at the assembly site of IscU and reduce it to S2−. We have already suggested such mechanism based on the Archeoglobus fulgidus (IscS–IscU)2 complex structure (27).
Conclusion
The various FeS cluster structures found in our TmHydE crystals can be ordered in a coherent sequence that provides a solid base for understanding cluster degradation and assembly (1 through 5 in Fig. 2 and Fig. S1). The plasticity of the additional FeS cluster coordination site has proved to be well suited for a systematic characterization of different protein-bound FeS species using X-ray crystallography. Taken together, our results and the RR spectroscopic studies of Zhang et al. (7) provide a convincing structural and functional interpretation of the Fe4S4-to-Fe2S2 cluster conversion upon O2 exposure in FeS cluster-containing proteins. The main proposition derived from our work is the existence of FeS cluster direct repair mechanisms involving the reduction of persulfide ligands in an Fe2S2 cluster similar to the 3 to 6 transition we have observed in our crystals. Without the packing constraints in the high-resolution crystal structure, 3 should have readily converted to the Fe4S4 cluster-containing species 2 observed in the high-salt crystal form (Figs. 2 and 3) following reaction 4:
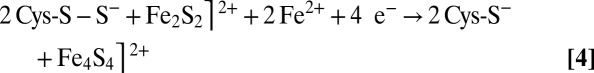 |
The requirement of very significant conformational changes in going from 3 to 2 clearly illustrates the extensive structural rearrangement expected in the dimer–monomer transition of FNR. Finally, the resistance of the 7 species to reduction by free Fe2+ favors the concerted arrival of iron and sulfur to the scaffold protein during ISC-dependent Fe2S2 cluster biogenesis.
Materials and Methods
Protein Purification and Crystallization.
HydE was purified either aerobically, as previously described (14), or the purification was carried out anaerobically following the same protocol. Crystals were obtained as described previously (11). All of the crystallization experiments were carried out in glove boxes with oxygen concentrations of about 2–5 ppm. Great care was taken to remove oxygen from the various solutions by sparging them with the glove box atmosphere, except when aerated solutions were intentionally used. The sample leading to the Fe4S4 cluster-containing structure at the additional site was obtained using a different protocol. In this case, a streptavidin tag was added at the N terminus of the amino acid sequence between Met1 and Thr2 (new N-terminus sequence: MWSHPQFEKAST-). The Strep-tag-containing HydE construct was obtained following the QuikChange site-directed mutagenesis kit strategy (Stratagene) using plasmid pTmHydE (Rubach et al., ref.14), Phusion polymerase, and the following primers: Forward: 5′-CACCCGCAGTTCGAAAAGGCAAGCACCGGTAGAGAAATTCTGGAAAAA-3′; Reverse: 5′-CTTTTCGAACTGCGGGTGGCTCCACATGGTATATCTCCTTCTTAAAGTTAAACAAAA-3′. This produced the pStrepTmHydE construct. The correctness of the cloned DNA sequence was confirmed by sequencing the entire gene. Protein expression was carried out following the protocol previously described for the untagged protein (14). After cell disruption in the glove box, the crude extract was treated directly with 1 mM FeCl3, 5 mM DTT, and 1 mM Na2S during 60 min, with mild stirring, to reconstitute the FeS clusters. The crude extract was subsequently cleared by ultracentrifugation at 165,000 × g during 30 min at 4 °C. The clear supernatant was loaded onto a streptavidin-agarose column buffered with 100 mM Tris pH 8.0; 150 mM NaCl; and 5 mM DTT. After extensive washing of the column, the protein was eluted by adding 2.5 mM desthiobiotin, which was subsequently removed using a GE Healthcare HiPrep 26/10 desalting column equilibrated with 100 mM Tris pH 8.0; 150 mM NaCl. HydE was then concentrated to 16 mg/mL, as determined by the Rose Bengal method (28), using an Amicon concentrator with a 10-kDa cutoff membrane. This HydE sample contained 8 iron atoms per protein molecule as determined by the method of Fish (29). Crystallization was performed as previously described (11), except that the protein sample contained 100 mM Tris pH 8.0 and 150 mM NaCl instead of 50 mM Tris pH 8.0 solution without NaCl. Large hexagonal brownish crystals were produced after 1 d. In the absence of NaCl, we obtained orthorhombic crystals like those previously reported (11). This shows that salt, and not the presence of the Strep-tag, is responsible for the change in space group. Table S1 summarizes the crystallization and cryoprotecting conditions used for each X-ray structure presented.
Data Collection and X-Ray Structure Refinement.
Data were collected at the European Synchrotron Radiation Facility and Swiss Light Source and processed with the XDS package (30). Model refinements were carried out using REFMAC5 (31) from the Collaborative Computational Project No. 4 package (32). The set of flagged reflections for Rfree calculations was the same used previously (11) although the initial set was extended to 1.25 Å resolution for the datasets diffracting that far. All of the data processing and refinement statistics are presented in Table S2. Manual model building was performed using COOT (33). Identification of Fe versus S atoms was based on the observed anomalous scattering electron density peaks for the corresponding atoms.
Supplementary Material
Acknowledgments
We thank Prof. Michael Johnson and Dr. Isabelle Artaud for fruitful discussions and the Commissariat à l’Energie Atomique (CEA), the Centre National de la Recherche Scientifique, and the Université Grenoble-Alpes for institutional support. We also thank the staff of the European Synchrotron Radiation Facility beamlines ID14, ID23, and ID29 (France) and Swiss Light Source PXI beamline (Switzerland) for help with data collection. R.R is the recipient of an “Irtelis” Fellowship from the CEA. This work was financed by Contract ANR-08-BLAN-0224-01 from the French Agence Nationale pour la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CIW, 3IIX, 4JXC, 4JY8, 4JY9, 4JYD, 4JYE and 4JYF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302388110/-/DCSupplemental.
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Drennan CL, Peters JW. Surprising cofactors in metalloenzymes. Curr Opin Struct Biol. 2003;13(2):220–226. doi: 10.1016/s0959-440x(03)00038-1. [DOI] [PubMed] [Google Scholar]
- 3.Crack JC, Green J, Thomson AJ, Le Brun NE. Iron-sulfur cluster sensor-regulators. Curr Opin Chem Biol. 2012;16(1-2):35–44. doi: 10.1016/j.cbpa.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Fleischhacker AS, Kiley PJ. Iron-containing transcription factors and their roles as sensors. Curr Opin Chem Biol. 2011;15(2):335–341. doi: 10.1016/j.cbpa.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. Quantitative assessment of oxygen availability: Perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J Bacteriol. 2002;184(5):1402–1406. doi: 10.1128/JB.184.5.1402-1406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achebach S, Selmer T, Unden G. Properties and significance of apoFNR as a second form of air-inactivated [4Fe-4S].FNR of Escherichia coli. FEBS J. 2005;272(16):4260–4269. doi: 10.1111/j.1742-4658.2005.04840.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, et al. Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein. Proc Natl Acad Sci USA. 2012;109(39):15734–15739. doi: 10.1073/pnas.1208787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crack JC, et al. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J Am Chem Soc. 2008;130(5):1749–1758. doi: 10.1021/ja077455+. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy MC, Beinert H. The state of cluster SH and S2- of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J Biol Chem. 1988;263(17):8194–8198. [PubMed] [Google Scholar]
- 10.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem Rev. 2007;107(10):4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 11.Nicolet Y, et al. X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima. J Biol Chem. 2008;283(27):18861–18872. doi: 10.1074/jbc.M801161200. [DOI] [PubMed] [Google Scholar]
- 12.Nicolet Y, Drennan CL. AdoMet radical proteins—from structure to evolution—alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 2004;32(13):4015–4025. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29(5):1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubach JK, Brazzolotto X, Gaillard J, Fontecave M. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermatoga maritima. FEBS Lett. 2005;579(22):5055–5060. doi: 10.1016/j.febslet.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 15.Crack JC, Green J, Cheesman MR, Le Brun NE, Thomson AJ. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci USA. 2007;104(7):2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons CR, et al. A putative Fe2+-bound persulfenate intermediate in cysteine dioxygenase. Biochemistry. 2008;47(44):11390–11392. doi: 10.1021/bi801546n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crack J, Green J, Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279(10):9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 18.Sutton VR, Mettert EL, Beinert H, Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ Cluster. J Bacteriol. 2004;186(23):8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185(1):221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271(5):2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 21.Dupuy J, et al. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14(1):129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Walden WE, et al. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314(5807):1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 23.Agar JN, et al. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J Biol Inorg Chem. 2000;5(2):167–177. doi: 10.1007/s007750050361. [DOI] [PubMed] [Google Scholar]
- 24.Nuth M, Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis: Characterization of iron nucleation sites for assembly of the [2Fe-2S]2+ cluster core in IscU proteins. J Am Chem Soc. 2002;124(30):8774–8775. doi: 10.1021/ja0264596. [DOI] [PubMed] [Google Scholar]
- 25.Smith AD, et al. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123(44):11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 26.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem. 2001;276(48):44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 27.Marinoni EN, et al. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew Chem Int Ed Engl. 2012;51(22):5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- 28.Elliott JI, Brewer JM. The inactivation of yeast enolase by 2,3-butanedione. Arch Biochem Biophys. 1978;190(1):351–357. doi: 10.1016/0003-9861(78)90285-0. [DOI] [PubMed] [Google Scholar]
- 29.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.