Abstract
A 3D atomistic model of a plant cellulose synthase (CESA) has remained elusive despite over forty years of experimental effort. Here, we report a computationally predicted 3D structure of 506 amino acids of cotton CESA within the cytosolic region. Comparison of the predicted plant CESA structure with the solved structure of a bacterial cellulose-synthesizing protein validates the overall fold of the modeled glycosyltransferase (GT) domain. The coaligned plant and bacterial GT domains share a six-stranded β-sheet, five α-helices, and conserved motifs similar to those required for catalysis in other GT-2 glycosyltransferases. Extending beyond the cross-kingdom similarities related to cellulose polymerization, the predicted structure of cotton CESA reveals that plant-specific modules (plant-conserved region and class-specific region) fold into distinct subdomains on the periphery of the catalytic region. Computational results support the importance of the plant-conserved region and/or class-specific region in CESA oligomerization to form the multimeric cellulose–synthesis complexes that are characteristic of plants. Relatively high sequence conservation between plant CESAs allowed mapping of known mutations and two previously undescribed mutations that perturb cellulose synthesis in Arabidopsis thaliana to their analogous positions in the modeled structure. Most of these mutation sites are near the predicted catalytic region, and the confluence of other mutation sites supports the existence of previously undefined functional nodes within the catalytic core of CESA. Overall, the predicted tertiary structure provides a platform for the biochemical engineering of plant CESAs.
Keywords: rosette cellulose synthase complex; molecular modeling; protein structure prediction; GlycosylTransferase Family 2; ß-1,4-glucan polymerization
Cellulose fibrils within plant cell walls provide the foundation for plant structure and are renewable biomaterials that account for most of the world’s biomass. Despite the importance of plant cellulose to nature and industry, we have little insight into the 3D structure of proteins required for plant cellulose biosynthesis. This deficiency arose due to experimental barriers in purification of active enzyme, recombinant expression, and crystallization of any plant cellulose synthase (CESA). However, manipulating the physical properties of cellulose through biochemical engineering of CESA structure offers many prospects for improved biomaterials. For example, moderate reduction of cellulose crystallinity increases the efficiency of saccharification (1), a process important for biofuels production from lignocellulosic biomass. However, the capacity for directed enzyme design requires an understanding of CESA protein structure/function relationships.
CESA is a membrane-bound Glycosyltransferase Family 2 (GT-2) enzyme (2) that catalyzes β-1,4-glucan (cellulose) chain polymerization using UDP-glucose as substrate (3). Although CESA proteins typically arrange themselves into multimeric cellulose synthase complexes (CSC), which are required for the production of multichain cellulose microfibrils, the CSCs of land plants and related algae are uniquely organized as six-lobed circular “rosettes” containing a still-undefined number (e.g., 18–36 in number) of CESAs (3). In contrast, bacteria, other algae, and tunicates have linear CSCs that correlate with synthesis of cellulose fibrils with different physical structures (4). Accordingly, there are differences in CSC organization and the resulting properties of cellulose fibrils between, for example, bacteria and plants.
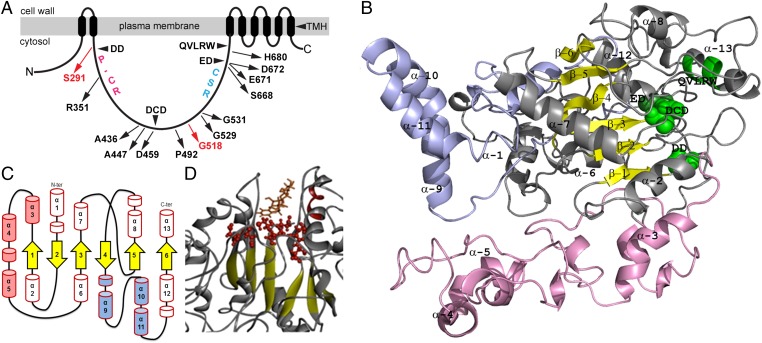
Plant CESA has a transmembrane region with eight predicted transmembrane helices (TMH) and a large (∼500 amino acids) cytosolic region. The cytosolic region of plant CESAs has four characteristic conserved motifs containing DD, DCD, ED, and QVLRW residues (3, 5) (Fig. 1A) that were predicted to be involved in substrate and/or acceptor binding, a plant-conserved region (P-CR) and a class-specific region (CSR). For GhCESA1 from Gossypium hirsutum (cotton), deletion of the first conserved region containing DD abolished UDP-glucose binding in vitro (6), and four missense mutations causing cellulose deficiency occur in the conserved DCD or ED residues of CESAs in the model plant Arabidopsis thaliana (called hereafter Arabidopsis) (7–9). A few amino acids (D, D, D, QxxRW) within the plant CESA conserved motifs are more broadly conserved and required for catalysis in other GT-2 enzymes such as hyaluronan and chitin synthases (10–12). In GT-2 enzymes a conserved DxD motif is usually part of a GT-A fold, as shown in solved structures of spore coat polysaccharide biosynthesis protein (SpsA) from Bacillus subtilis (13), chondroitin polymerase from Escherichia coli K4 (K4CP) (14) and most recently Rhodobacter sphaeroides cellulose synthase (BcsA) (15). Plant CESAs will likely have a similar fold due to conservation of the cellulose polymerization mechanism, but experimental evidence is lacking.
Fig. 1.
Predicted structure of the GhCESA cytosolic region. (A) Diagram of GhCESA1 showing eight predicted TMH and the large cytsolic loop between TMH2 and TMH3. Labels within the cytosolic loop indicate the approximate locations of the four conserved motifs; the P-CR region; the CSR region; and the analogous locations for published (black) and previously undescribed (red) missense mutations in Arabidopsis CESAs. (B) Snapshot of the Gh506 structure. The catalytic core is gray, the P-CR is pink, and the CSR is light blue. The catalytic core contains a β-sheet (yellow) with six strands: β-1 S287-S291; β-2 D253-S257; β-3 F454-D459; β-4 C532-N535; β-5 Y488-F491; and β-6 S686-C689. Green highlights DD, DCD, ED (directly behind DCD), and the QVLRW within α-13. The five α-helices that are part of the GT core are α-2 L267-A278; α-6 H433-V448; α-7 N466-D479; α-8 N508-K517; and α-13 S705-R725. (C) Diagram of the secondary structure showing six β-strands (yellow arrows) and 13 major α-helices (barrels) in three regions: catalytic core (red outlines); P-CR (pink fill); and CSR (blue fill). Possible additional shorter helical regions are indicated as unnumbered small barrels. (D) UDP-Glc (orange) docked into the catalytic site.
In contrast to bacteria, the plant CESA cytosolic region contains large insertions specific to plants only, namely the P-CR and the CSR (5, 6, 16). Although their exact functions are unknown, the CSR and P-CR are hypothesized to mediate aspects of cellulose synthesis unique to plants, such as the formation of rosette-like CSCs that move through the plasma membrane producing cellulose fibrils through the coupling of β-1,4-glucan polymerization and crystallization (1, 17, 18). However, no insight into the structure, folding, and putative role in CSC assembly of the plant-specific CESA regions has been reported.
To fill in the gaps in our understanding about the tertiary structure of plant CESAs, we generated a model of the 3D structure of 506 amino acids from a cytosolic region of cotton GhCESA1 (GenBank Accession P93155) (6), called hereafter the Gh506 structure. GhCESA1 is an apparent ortholog of AtCESA8 from Arabidopsis, and its gene is highly expressed during cotton fiber secondary wall thickening (6, 19). Structural coalignment of selected regions of BcsA, the recently solved bacterial cellulose synthase, (15) with the plant Gh506 model revealed numerous structural commonalities within the GT-2 domains despite poor sequence similarity over the cytosolic region. This result supports the veracity of the plant CESA model, given that BcsA was not used as homolog for Gh506 prediction. Moreover, the Gh506 structure reveals how plant-specific P-CR and CSR domains are interfaced with the GT domain and showed possibilities for how they may participate in CESA oligomerization to generate plant-specific CSCs.
Taking advantage of the high conservation between seed plant CESA sequences, we mapped Arabidopsis CESA missense mutations that alter cellular morphogenesis via effects on cellulose synthesis onto the Gh506 structure. The confluence of some of the point mutations allows us to propose the existence of previously unidentified functional nodes within CESA. These insights into structure/function relationships in plant CESAs may have importance for optimization of the properties of renewable biomass.
Results and Discussion
In Silico Predicted Structure of the GhCESA1 Cytosolic Region.
A rough initial model of 506 amino acids of the GhCESA1 cytosolic region (Fig. 1A, Fig. S1) was generated by the SAM-T08 server using 20 solved protein structures (Table S1). The template structures were selected via multipass Blastp search for putative homologs in the National Center for Biotechnology Information nonredundant protein database (Table S1, Fig. S2 A–C). Two of the top selected structures were from the bacterial protein templates of SpsA and K4CP that have been extensively used to examine the molecular basis for catalysis and substrate recognition of glycosyltransferases (13, 14). Note that the recently solved structure of BcsA was not included in the prediction of Gh506 as it was not available at the time. After refinement with molecular dynamics (MD) simulations, the Gh506 structure (Fig. 1B, Fig. S2E) had a Pro-SA Z score of -6.09 and an ERRAT2 quality factor of 86.9%, which is the percentage of the protein where the calculated conformational error falls below the 95% rejection limit. The overall quality of the Gh506 structure is consistent with solved structures of three other GT-2 enzymes obtained from crystallography with 2 Å resolution (Table S2, Fig. S3). The regions with conformation errors either have high local mobility or are deeply buried. Similar difficulties in full refinement arise for some regions within solved crystal structures (Fig. S3B).
The Gh506 structure contains 13 α-helices and 6 β-strands, which are organized into a β-sheet near the catalytic site where UDP-glucose binds, forming a GT-A domain with a canonical Rossmann fold (Fig. 1 C and D, Fig. S2, Table S3, Dataset S1) similar to bacterial GT-2 enzymes, such as SpsA and BcsA (13, 15, 20). In this core GT-2 domain, the structural elements include five core α-helices (α−2, -6, -7, -8, and -13) and the β-sheet (six β-strands) that helps to stabilize the catalytic residues (Fig. S2; Table S3). The catalytic pocket of the Gh506 structure contained the closely arranged conserved motifs. The QVLRW motif might interact with the newly polymerized cellulose with its tryptophan residue (21), and it is located in the center of α-13 above a pocket with linearly arranged DD, DCD, and ED motifs (Fig. 1 B and D), matching their proximal locations in early CESA diagrams (22). By analogy to BcsA, which contains a cocrystallized glucan chain and a UDP molecule, we can postulate the functions of the classical conserved motifs in plant CESA: (i) to coordinate UDP (D292 of DD, D459 and D461 of DCD, R713 of QVLRW); (ii) to act as the catalytic base (D672); and (iii) to stabilize the acceptor glucan (W714 of QVLRW). The catalytic site of the Gh506 structure was supported by docking UDP-glucose into its solvent-exposed catalytic pocket in proximity to DCD (Fig. 1D). Density Functional Theory calculations supported the coordination of UDP via a divalent cation interacting with D459 and D461 of the Gh506 structure (Fig. S2F), similar to other glycosyltransferases (23, 24). In addition, we identified three loops in the vicinity of the UDP-glucose binding site in the Gh506 structure that may control catalysis through modulation of local accessibility to key residues: first loop (1): T258–L267 is located at the end of β-2; second loop A294-F300 is just after DD and leading to α−3 of the PCR; and third loop Y421-H432 is between α−5 and α−6 (Fig. S2G). The function of these loops can be further explored in future experiments. Overall, the predicted Gh506 structure shows a highly conserved single active site for coordinating the donor and acceptor sugars for cellulose synthesis.
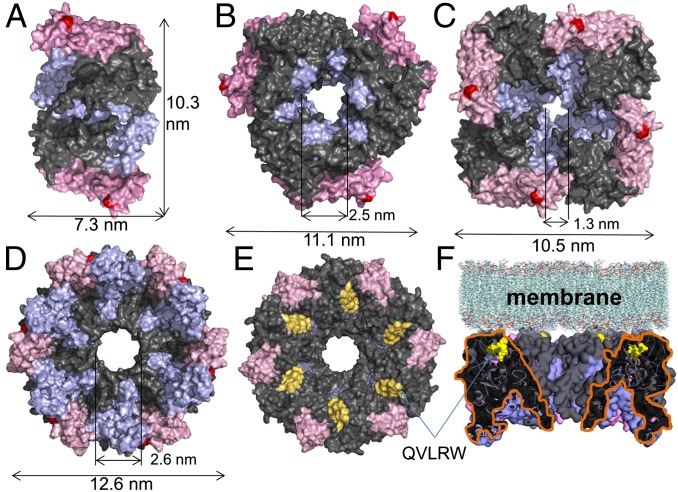
Regions unique to plant CESAs, the P-CR and CSR domains, extend away from the GT-domain of Gh506 toward the cytosol where they may feasibly regulate other aspects of plant CESA function including the assembly of rosette CSCs (Fig. 1B). The relatively high structural independence of these plant-specific regions was indicated by cross-correlated atomic fluctuations (Fig. S4). Based on the Gh506 structure, we propose that these regions partake in the oligomerization of CESAs to form the rosette CSCs that are found in land plants and their close relatives. To examine possible roles of the CSR and P-CR in assembly of CESA homo-oligomers, we used the Rosetta Symmetry docking protocol to show possible dimers, trimers, tetramers, and hexamers of the Gh506 structure (Fig. 2, Fig. S5). The assemblies show that the CSR and P-CR regions are located at the interfaces of the monomers, supporting the possibility that these regions may help to stabilize CESA assembly through noncovalent interactions. Interestingly, the CSR region is more important for assemblies of dimers and trimers whereas both the CSR and P-CR participate in assemblies of tetramers and hexamers. Future computational and laboratory experiments can be designed to test how the N-terminal zinc finger region, which is also unique to plant CESAs but not included in the Gh506 structure, may help to modulate CESA assembly through dimerization as shown previously for GhCESA1 (25).
Fig. 2.
Possible oligomeric assemblies of the Gh506 cytosolic structure under (A) C2, (B) C3, (C) C4, and (D) C6 crystallization symmetries. The catalytic region is gray, the CSR is light blue, the P-CR is pink, QVLRW is yellow, and the site of fra6 mutations is red. (D) Bottom, (E) top, and (F) side view of the hexameric Gh506 assembly.
No known missense mutation exists in the CSR, but one does occur in the P-CR: Atcesa8R362K (fra6), which was reported to cause reduced cellulose content when homozygous in Arabidopsis. However, no phenotype resulted from overexpression of the mutant gene in wild-type plants (26). In our tetrameric model, the fra6 mutation is located at the surface, which potentially could affect the assembly of oligomers into rosette CSCs (Fig. 2C). In our hexameric assembly, fra6 is located at the interface between the CESA monomers, which could disrupt the assembly of oligomers (Fig. 2D). This result suggests that the affected arginine residue may be important for CESA oligomerization within the rosette CSC.
Comparison Between Bacterial and Plant Cellulose Synthases.
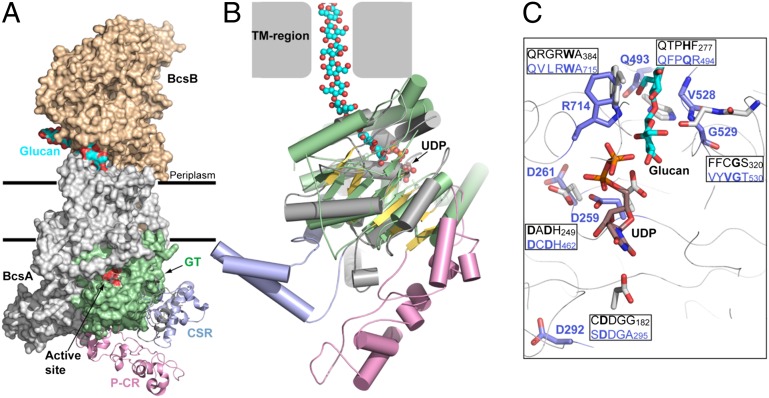
The inherent differences in CSC formation and resultant cellulose fibril properties between bacteria and plant CESAs must arise from differences in their protein sequences and, thus, structures. For example, a sequence comparison of the cytosolic region responsible for cellulose synthesis between bacterial BcsA (276 amino acids from GenBank Accesssion Q3J125) and plant GhCESA1 (506 amino acids) shows 17.5% identity, 26.1% similarity, and 49.9% gaps. The plant CESA cytosolic region is longer, mostly due to the presence of P-CR and CSR insertions specific to plants (5, 6, 16). Even with omission of the P-CR and CSR regions, the coaligned sequences of the edited bacterial and plant cytosolic regions (240 or 259 residues, respectively) showed 28% identity, 44% similarity, and 10% gaps. However, a structural alignment between BcsA (solved at 3.25 Å resolution; PDB ID 4HG6) and Gh506 resulted in a 3.9 Å rmsd overall (Fig. 3A, Fig. S6).
Fig. 3.
Comparison of the Gh506 with the bacterial cellulose synthase. (A) Surface representation of the Rs cellulose synthase (PDB ID 4HG6) superimposed with the Gh506 structure at the GT-domain (colored green). (B) Superimposition of the BcsA GT-domain with the Gh506 by secondary structure matching. The BcsA GT-domain is colored green, Gh506 GT-domain is gray and yellow, P-CR and CSR domains are colored pink and light blue, and UDP and glucan are shown as spheres. (C) Conserved sequence motifs that form the binding site for UDP and the acceptor glucan are compared between Gh506 (blue letters) and RsBcsA (black letters) with depicted residues in bold and colored as blue (Gh506) and gray (BcsA), and UDP (rust) and glucan (cyan).
The bacterial BcsA GT-domain adopts a GT-A fold consisting of a mixed seven-stranded β-sheet surrounded by seven α-helices (15). Our Gh506 model aligns well with the BcsA GT-domain, particularly with its central β-sheet and four of the surrounding α-helices (Fig. 3 A and B), and the Gh506 structure shares five of seven α-helices and six of seven β−strands as found in the GT-A fold of BcsA. Other structural features that are likely to function similarly in plant CESAs and BcsA are noted in Table S3. Fig. 3C shows that the invariant DD, DCD, and QVLRW motifs of our Gh506 model align well with the bacterial cellulose synthase structure. This comparison importantly confirms a high degree of structural similarity between the catalytic sites of eukaryotic and prokaryotic cellulose-synthesizing proteins, which indicates a conserved mechanism of cellulose polymerization. Moreover, the structural alignment orients the P-CR and CSR domains of Gh506 toward the cytosol (Fig. 3A).
Genetic Mutations Demonstrate Functional Nodes Within Plant CESA Structure.
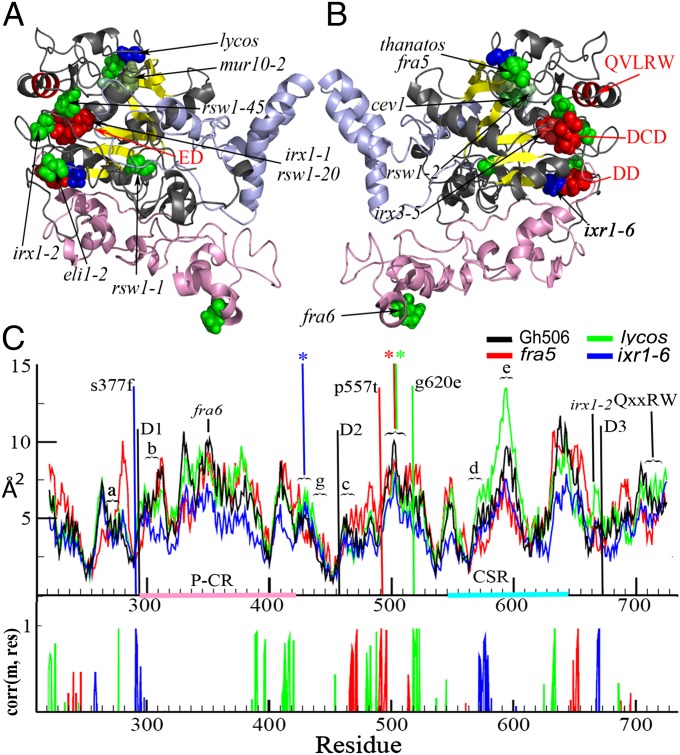
The relatively high sequence conservation between seed plant CESAs (Fig. S1) allowed Arabidopsis CESA missense mutations to be mapped onto the analogous residue in the Gh506 structure (Fig. 4, Table S3; see Table S3 for nomenclature of Arabidopsis CESA missense mutations). Based on the primary sequence, several previously identified Arabidopsis CESA missense mutations coincide with the conserved ED motif [Atcesa1E779K (rsw1-45), Atcesa8 D683N (irx1-1), and Atcesa1D780N (rsw1-20)] or the first D in the DCD motif (Atcesa7D524N) (7–9). However, other missense mutations are dispersed throughout the cytosolic region of CESAs at locations with no known function. Interestingly, in our Gh506 model, the mutated residues primarily converged in a spatially discrete cluster around the catalytic site even though the residues were dispersed throughout the sequence (Fig. 4). A plausible interpretation for this result is that the catalytic region retains an overarching tertiary structure across plant CESAs and that most of the currently known missense mutations that lead to reduced cellulose content cluster around this core domain.
Fig. 4.
Previously known (green) and previously undescribed (blue) Arabidopsis CESAs mutations mapped onto the Gh506 structure. (A and B) Two sides of Gh506 are shown with DD, DCD, ED, and QVLRW motifs in red. The equivalent GhCESA1 amino acid positions are: R351 (fra6); A436 (eli1-2); A447 (rsw1-1); D459 (irx3-5); P492 (fra5 and thanatos); G529 (rsw1-2); G531 (cev1) ; S668 (irx1-2); E671 (rsw1-45); D672 (irx1-1 and rsw1-20); and H680 (mur10-2). (C) Cross correlation of atomic fluctuations over simulation trajectories by residue. The peaks shown had at least 97% correlation, indicating distant effects of the mutations analogous to ixr1-6 (blue), lycos (green), and fra5 (red).
In addition, the location of missense mutations in the Gh506 structure provided insights about putative functionally important nodes within CESA. For example, the analog of the Atcesa7H734Y (mur10-2) mutation (27) located after TED in α−12 makes contact with β-5 and β-6, as well as the site of the Atcesa1G631S (rsw1-2) mutation (28) even though these mutated histidine and glycine are separated by ∼150 residues in the sequence. The Atcesa7H734Y plants have dwarfed shoots and cellulose-deficient xylem secondary walls (27). The Atcesa1G631S mutant seedlings have ∼75% less crystalline cellulose and swollen organs (28). The mapped sites of the Atcesa1G631S and Atcesa3G617E (cev1) mutations are separated by one amino acid, and Atcesa3G617E mutant plants were dwarfs with radial cell swelling and cellulose deficiency compared with wild type (29). The Atcesa1G631S and Atcesa3G617E mutation sites lie at the end of β-4 in a VYVGTG motif, which structurally aligns with the FFCGS motif of BcsA in the core GT domain. The perturbation of a β-sheet structure may affect the structure of the catalytic site and substrate binding. In BcsA, FFCGS binds the terminal dissaccharide of the glucan acceptor on the opposite side compared with QRGRW (15). Therefore, the Atcesa7H734Y, Atcesa1G631S, and Atcesa3G617E mutation sites may represent a functional node that controls the acceptor glucan placement or conformation within the active site (Fig. 4).
To further investigate the effect of mutations, we mapped and cloned two previously undescribed missense mutations to regions of interest within Arabidopsis CESAs. The first of these, Atcesa3S377F, confers resistance to the cellulose synthesis inhibitor, isoxaben, and the mutant Arabidopsis plants showed reduced growth and a lower relative crystallinity although the cellulose content was not statistically different from in the control (Table 1). According to our Gh506 model, the analogous affected residue, S291 in GhCESA1, resides within a water-accessible pocket at the end of β-1 and before the A294-F300 loop containing DD (Fig. S2H). Interestingly, this mutation disturbs the crystallization process with little impact on cellulose catalysis despite proximity to the conserved DD residue. To explore how this mutation disrupts CESA structure, MD simulations were performed on the mutated model CESA. In Fig. 4C, each peak represents a highly motile residue that is often solvent accessible whereas the valleys are largely populated by buried amino acids. For example, mapped peaks are the analogs of Atcesa8R362K (fra6) located in the P-CR and AtcesaS679L (irx1-2) three residues below the conserved ED motif. This analysis showed that S291 in GhCESA1 is tightly coupled to the conserved ED motif by the short T258–L267 loop and to residues S572-R580 in α-9 within the CSR. As previously explained based on analogy to BcsA, the ED residues are likely to affect catalysis directly as well as interact with glucose when it is bound to UDP. Disturbance of glucose positioning in the active site could affect glucan chain conformation and/or the rate of catalysis, which could affect cellulose crystallization. Possible effects arising through coupling to the CSR are not easily defined given the unknown function of this domain.
Table 1.
Plant phenotypes for previously undescribed CESA mutations
| Allele/genotype | Dark-grown hypocotyl length, % wt | Height, mature stem, cm (SE) | Cellulose content, mature stem, % wt | RCI, mature stem (SE) |
| Atcesa3S377F (ixr1-6) LER background | 45.6* | 20.8 (0.5)* | 87.5 | 32.8 (4.7)** |
| Atcesa1G620E (lycos) Col-0 background | 100 | 13.4 (0.5)* | 61.7* | 41.9 (1.0)*** |
| Wild type (LER) | 100 | 30.7 (3.1) | 100 | 48.4 (1.1) |
| Wild type (Col-0) | 100 | 39.1 (0.8) | 100 | 49.2 (2.1) |
Significantly different compared with wild-type (LER or Col-0) as determined by t test: *P < 0.001; **P = 0.009; ***P < 0.01. RCI, Relative crystallinity index.
In general, a single point mutation of a key residue may affect CESA function in multiple ways. For example, in the Gh506 structure, the S291 residue that is analogous to Atcesa3S377F contacts L442 within core α−6 (Fig. S2H), and MD simulations of mutated Ghcesa1S291F revealed a larger distance between these residues compared with wild type (Table S4). Notably, the analog of the A residue in the temperature-sensitive Atcesa1A549V (rsw1-1) mutation is at the base of core α−6 (Fig. S2H). Arabidopsis Atcesa1A549V mutants grown at the restrictive temperature showed severely impaired crystalline cellulose deposition and seedling growth (30), effects that are similar to the Atcesa3S377F mutation. The analog of the Atcesa3A522V (eli1-2) mutation, which also caused similar phenotypes (31, 32), is 10 residues away from the Atcesa1A549V mutation site at the other end of core α−6 within a HKKAGA motif (Fig. S2H) that is coaligned with the HAKAGN motif of BcsA. In BcsA, the A225 and K226 residues of HAKAGN lie on the other side of the pocket that may accommodate glucose when bound to UDP (15). Taken together, these results suggest that core α−6, predicted to be in the interior of CESA behind β-1–3, may help to control the positioning of the donor glucose in the catalytic site, which in turn may modulate the organization of glucan chains into crystalline cellulose fibrils.
A second previously undescribed Arabidopsis missense mutation, Atcesa1G620E, conferred resistance to the cellulose synthesis inhibitor quinoxyphen and also caused reductions in stem height, relative crystallinity, and cellulose content (Table 1). Its analogous residue in GhCESA1, G518, helped to support the functional importance of the solvent-accessible P492-G518 loop that lies between β-4 and β-5 and behind α-13/QVLRW in the Gh506 structure (Fig. S2I). The G518 residue is predicted to sit adjacent to the P492 residue, which is analogous to the site of the Atcesa7P557T (fra5) and Atcesa3P578S (thanatos) mutations (26, 33). The P492 and G518 residues appear to act as hinge points for the loop between them. The range of motion for this loop was established from the MD simulation trajectory (Table S4). The tip of the loop contains three aspartic acid residues (Fig. S2I), and it is able to contact the QVLRW motif and may potentially modulate its interaction with the newly forming β-1,4-glucan chain. Thus, changing the dynamic behavior of the loop by mutations at its base may adversely affect QVLRW interaction with the cellulose product.
Several computational experiments showed putative effects of altering the predicted hinge points of the P492-G518 loop through: (i) substitution at P492 of threonine (analogous to Atcesa7P557T) or serine (analogous to AtcesA3P578S); and (ii) substitution of glutamic acid at G518 (analogous to Atcesa1G620E). The dynamic behavior of the three D residues at the tip of the loop was reduced for mutant E518 compared with wild-type G518 in MD simulations based on the Gh506 structure (Table S4). Reasonably, the larger glutamic acid residue could cause constrained movement, steric clash, and changed hydrophobicity of the solvent-exposed loop. However, the expected reduced local rigidity for mutant T492 compared with wild type was not observed (Table S4), possibly due to the effects of intermittent hydrogen bonding interactions between T492 and Y688 of β-6 observed in the MD simulations. This hydrogen bonding interaction may serve to stabilize the mutant T492-G518 loop (Fig. S7). Similarly, E518 showed intermittent hydrogen bonding with the adjacent L517 residue.
Mutations at these putative hinge points also had widely distributed effects on the Gh506 structure as determined through correlations of residue fluctuations. The position of mutant E518 strongly couples to atomic motions in β-3, β-6, and part of the P-CR and CSR regions. The mutant T492 position couples to β-2, β-5, β-6, and F696 near the QVLRW motif. Therefore, both of these mutations are likely to perturb the β-sheet, which can affect catalysis and substrate binding (34). Previous modeling of 185 amino acids with the HMMSTR/Rosetta Server suggested that the catalytic domain structure was altered by Atcesa3P578S (thanatos) mutation (35). However, we could not reproduce this result with de novo modeling using the SAM-T08 server for the 506 amino acid-long GhCESA1 cytosolic region containing the analogous mutation.
Commonalities in the Atcesa7P557T (fra5) mutation and the Atcesa3P578S (thanatos) mutation can now be explained through effects on the same functional loop. Both are semidominant: Atcesa3P578S causes reduced primary wall cellulose synthesis (35) and Atcesa7P557T causes reduced cellulose content of fiber cells (26). Both mutations exert dominant negative effects when overexpressed in wild type, which has been described only for these two Arabidopsis CESA missense mutations (26, 35). Therefore, the mutant proteins must compete effectively for entry into the rosette CSC, which is logical given the location of the analogous residues near the catalytic region of the Gh506 structure. Together, the data presented here illustrate the utility of the predicted tertiary structure of the GhCESA1 cytosolic region to provide insight into mechanisms of cellulose polymerization in plants, help systematize data on CESA missense mutations, and illuminate possible new structure/function relationships that are broadly conserved among plant CESAs.
Overall, we were able to predict a complex 3D structure of plant cellulose synthase from Gossypium hirsutum using a molecular modeling approach. Our model is in close agreement with the core region of the recently solved structure of the bacterial BcsA cellulose synthase (15) despite substantial differences in the plant and bacterial sequences. Given that BcsA was not used as structural homolog for model prediction, this structural convergence supports a conserved mechanism for cellulose polymerization. The clustering of most Arabidopsis missense mutations around the structurally conserved catalytic site further supports the similarity of the cellulose catalytic mechanism across Kingdoms. Moreover, unique regions to plant CESAs, the CSR and P-CR, were revealed to fold into distinguishable subdomains within the cytosolic region, and these regions can be explored further for how they potentially control the assembly of plant CSCs, other regulatory aspects of plant cellulose synthesis, and, consequently, the unique material properties of plant cellulose.
Methods and Materials
Simulations and Modeling.
We used secondary structure prediction tool PSI-PRED (36) to isolate the cytosolic region of GhCESA1 (P93155) (Fig. S1). Almost the entire region was modeled (506 amino acids; Q220–R725) beginning just after second transmembrane helix. Successful de novo structure modeling is predicated on an accurate energy function, an efficient search method, and selection of appropriate models from the ensemble. Heuristic Hidden Markov Model (HMM) protein structure prediction approaches, along with fragment-based assembly algorithms such as ROSETTA (www.rosettacommons.org/), have proven to be most successful (37, 38). However, due to the intensive computational time required, successful de novo folding with ROSETTA has generally been limited to 100–150 amino acids (39, 40). To overcome this limitation, we used the protein structure prediction server of SAM-T08 (41) to generate an initial homology model. The SAM-T08 server relies on the construction of HMM and multiple sequence alignments to generate structural homologs for parts of the target structure. The initial fragmented structure was manually refined and subjected to a series of MD simulations to explore the conformational space and develop the final modeled structure. After preliminary structural quality checks, the modeled structure was analyzed comprehensively using the Protein Structure Validation Software suite (PSVS) (42) and ERRAT (43). The UDP-glucose was docked into the structure with the help of Density Functional Theory carried out in Gaussian 03 (44) with the B3LYP/6–311+G(d,p) method and D residues constrained. Mutants based on the Gh506 structure were generated using the TLEAP tool of Amber 11 (45), subjected to MD simulations, and the resultant structural flexibility was assessed using cross correlation analysis (46). Putative homomeric assemblies of the Gh506 structure were generated using the symmetric docking protocol of Rosetta (47). Additional details are available in the supplemental methods (S1 Materials and Methods).
Previously Undescribed Mutations in Arabidopsis CESAs and Phenotypes of Mutant Plants.
Approximately forty-five thousand A. thaliana ecotype Landsberg (LER) and Columbia-0 (Col-0) seeds were mutagenized by ethyl methane sulfonate (EMS) by immersing seeds in a solution of 0.3% EMS (M1) for 16 h, extensively washed with distilled water (12 h), and sown into soil to generate M2 seeds. M2 seed were surface sterilized, and 1 million M2 seeds were plated on 0.5× strength Murashige and Skoog agar plates supplemented with 20 nM isoxaben (LER screen) or 5 μM quinoxyphen (Col-0 screen). Seed were stored at 4 °C for 4 d to synchronize germination and then exposed to 100 µE/m2/s white light at room temperature until seeds germinated and cotyledons had expanded. Resistant mutants grew above the surface of the agar whereas nonresistant plants did not. Resistant plants from the M2 generation were retested in the M3 generation to confirm heritability of the resistance phenotype. The previously undescribed isoxaben resistant (ixr) allele in AtCESA3 discussed here was named ixr1-6, and the previously undescribed quinoxyphen resistance allele in AtCESA1 discussed here was named lycos. For clarity, the mutants are referred to in the text as Atcesa3S377F and Atcesa1G620E, respectively. Methods for assessing phenotypes were as described previously (1).
Supplementary Material
Acknowledgments
Work by L.S., J.D.K., C.H.H., and Y.G.Y. was supported as part of The Center for LignoCellulose Structure and Formation, Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Office of Basic Energy Science under Award DE-SC0001090. Work by S.D. was supported by National Science Foundation Award IOS- 0922947. Work by J.Z. was support by National Institutes of Health Grant 1R01GM101001 and start-up funds from the University of Virginia School of Medicine. Work by D.B. was supported by the National Science and Engineering Research Council of Canada (NSERC).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301027110/-/DCSupplemental.
References
- 1.Harris DM, et al. Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA. 2012;109(11):4098–4103. doi: 10.1073/pnas.1200352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama Y. Structure and properties of the cellulose microfi bril. J Wood Sci. 2009;55:241–249. [Google Scholar]
- 5.Roberts E, Roberts AW. A cellulose synthase (Cesa) gene from the red alga Porphyra yezoensis (Rhodophyta) J Phycol. 2009;45:203–212. doi: 10.1111/j.1529-8817.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- 6.Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93(22):12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12(12):2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beeckman T, et al. Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 2002;130(4):1883–1893. doi: 10.1104/pp.102.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang YK, et al. Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana. Plant J. 2010;64(4):679–686. doi: 10.1111/j.1365-313X.2010.04362.x. [DOI] [PubMed] [Google Scholar]
- 10.Saxena IM, Brown RM. Identification of cellulose synthase(s) in higher plants: Sequence analysis of processive beta-glycosyltransferases with the common motif 'D, D, D35Q(R,Q)XRW'. Cellulose. 1997;4:33–49. [Google Scholar]
- 11.Yoshida M, Itano N, Yamada Y, Kimata K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J Biol Chem. 2000;275(1):497–506. doi: 10.1074/jbc.275.1.497. [DOI] [PubMed] [Google Scholar]
- 12.Nagahashi S, et al. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J Biol Chem. 1995;270(23):13961–13967. doi: 10.1074/jbc.270.23.13961. [DOI] [PubMed] [Google Scholar]
- 13.Charnock SJ, Davies GJ. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38(20):6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 14.Sobhany M, Kakuta Y, Sugiura N, Kimata K, Negishi M. The chondroitin polymerase K4CP and the molecular mechanism of selective bindings of donor substrates to two active sites. J Biol Chem. 2008;283(47):32328–32333. doi: 10.1074/jbc.M804332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan JLW, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493(7431):181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpita NC. Update on mechanisms of plant cell wall biosynthesis: How plants make cellulose and other (1->4)-β-D-glycans. Plant Physiol. 2011;155(1):171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerriero G, Fugelstad J, Bulone V. What do we really know about cellulose biosynthesis in higher plants? J Integr Plant Biol. 2010;52(2):161–175. doi: 10.1111/j.1744-7909.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 18.Diotallevi F, Mulder B. The cellulose synthase complex: A polymerization driven supramolecular motor. Biophys J. 2007;92(8):2666–2673. doi: 10.1529/biophysj.106.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betancur L, et al. Phylogenetically distinct cellulose synthase genes support secondary wall thickening in arabidopsis shoot trichomes and cotton fiber. J Integr Plant Biol. 2010;52(2):205–220. doi: 10.1111/j.1744-7909.2010.00934.x. [DOI] [PubMed] [Google Scholar]
- 20.Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16(2):29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 21.Saxena IM, Brown RM., Jr Cellulose biosynthesis: Current views and evolving concepts. Ann Bot (Lond) 2005;96(1):9–21. doi: 10.1093/aob/mci155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delmer DP. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto K, Madej T, Bryant SH, Panchenko AR. Functional states of homooligomers: Insights from the evolution of glycosyltransferases. J Mol Biol. 2010;399(1):196–206. doi: 10.1016/j.jmb.2010.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiggins CAR, Munro S. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA. 1998;95(14):7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA. 2002;99(17):11109–11114. doi: 10.1073/pnas.162077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong RQ, Morrison WH, 3rd, Freshour GD, Hahn MG, Ye ZH. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 2003;132(2):786–795. doi: 10.1104/pp.102.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosca S, et al. Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 2006;142(4):1353–1363. doi: 10.1104/pp.106.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C. Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol. 2002;156(6):1003–1013. doi: 10.1083/jcb.200111093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14(7):1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arioli T, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279(5351):717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 31.Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003;34(3):351–362. doi: 10.1046/j.1365-313x.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- 32.Pysh L, Alexander N, Swatzyna L, Harbert R. Four alleles of AtCESA3 form an allelic series with respect to root phenotype in Arabidopsis thaliana. Physiol Plant. 2012;144(4):369–381. doi: 10.1111/j.1399-3054.2012.01575.x. [DOI] [PubMed] [Google Scholar]
- 33.Daras G, et al. Thanatos mutation in CesA3 gene exhibits a nonconditional semidominant-negative phenotype on Arabidopsis primary cell wall formation. FEBS J. 2008;275(Suppl S1):361. [Google Scholar]
- 34.Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147(7):1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daras G, et al. The thanatos mutation in Arabidopsis thaliana cellulose synthase 3 (AtCesA3) has a dominant-negative effect on cellulose synthesis and plant growth. New Phytol. 2009;184(1):114–126. doi: 10.1111/j.1469-8137.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- 36.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 37.Fleishman SJ, et al. Rosetta in CAPRI rounds 13-19. Proteins. 2010;78(15):3212–3218. doi: 10.1002/prot.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Wu S, Zhang Y. Ab initio protein structure prediction. In: Rigden DJ, editor. From Protein Structure to Function with Bioinformatics. London: Springer; 2009. Chap 1, pp 1-26. [Google Scholar]
- 40.Drew K, et al. The Proteome Folding Project: Proteome-scale prediction of structure and function. Genome Res. 2011;21(11):1981–1994. doi: 10.1101/gr.121475.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karplus K. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 2009;37(Web Server issue):W492-7. doi: 10.1093/nar/gkp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66(4):778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 43.Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frisch MJ, et al. Gaussian 03. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 45.Case DA, et al. Amber 11. 2010. (Univ of California, San Francisco) [Google Scholar]
- 46.Kormos BL, Baranger AM, Beveridge DL. A study of collective atomic fluctuations and cooperativity in the U1A-RNA complex based on molecular dynamics simulations. J Struct Biol. 2007;157(3):500–513. doi: 10.1016/j.jsb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.André I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci USA. 2007;104(45):17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.