BIOCHEMISTRY Correction for “Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex,” by Toshiharu Tsurumura, Yayoi Tsumori, Hao Qiu, Masataka Oda, Jun Sakurai, Masahiro Nagahama, and Hideaki Tsuge, which appeared in issue 11, March 12, 2013, of Proc Natl Acad Sci USA (110:4267–4272; first published February 4, 2013; 10.1073/pnas.1217227110).
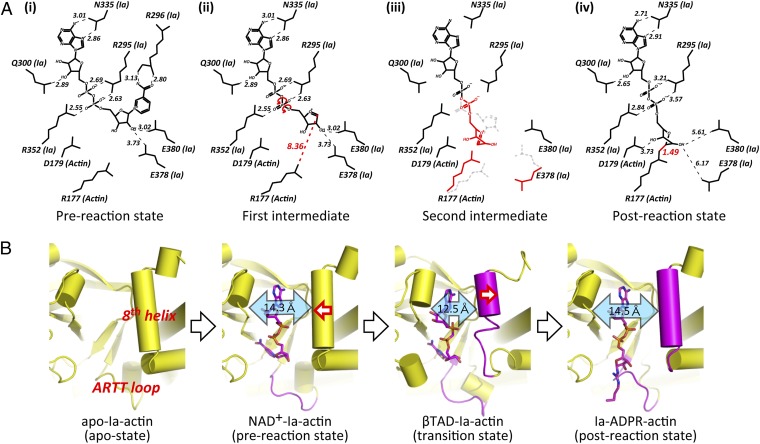
The authors note that Fig. 7 appeared incorrectly. There was a drawing error in the nicotinamide of NAD+ within Fig. 7A, and distance values were refined based on the last coordinate. The corrected figure and its legend appear below. This error does not affect the conclusions of the article.
Fig. 7.
Schematic illustrating the mechanism of ADP-ribosylation. (A) SN1 mechanism in Ia: (i) NAD+-Ia-Actin as the prereaction state; (ii) nicotinamide cleavage occurs via an SN1 reaction induced by an NMN ring-like structure and the first oxocarbenium cation intermediate is formed with a strained conformation; (iii) the second cationic intermediate is induced through alleviation of the strained conformation mainly by O3-NP and NP-NO5 rotation, and then NC1 of N-ribose nucleophilically attacks Arg177 of actin; (iv) Ia-ADPR-actin as the postreaction state. (B) Successive structures during ADP-ribosylation and the structure of each reaction: step 1 [apo-Ia-actin (apo-state)], step 2 [NAD+-Ia-actin (prereaction state)], step 3 [βTAD-Ia-actin (transition state)], and step 4 [Ia-ADPR-actin(postreaction state)].