Abstract
Malaria parasites are transmitted to humans by mosquitoes of the genus Anopheles, and these insects are the targets of innovative vector control programs. Proposed approaches include the use of genetic strategies based on transgenic mosquitoes to suppress or modify vector populations. Although substantial advances have been made in engineering resistant mosquito strains, limited efforts have been made in refining mosquito transgene expression, in particular attenuating the effects of insertions sites, which can result in variations in phenotypes and impacts on fitness due to the random integration of transposon constructs. A promising strategy to mitigate position effects is the identification of insulator or boundary DNA elements that could be used to isolate transgenes from the effects of their genomic environment. We applied quantitative approaches that show that exogenous insulator-like DNA derived from the Drosophila melanogaster gypsy retrotransposon can increase and stabilize transgene expression in transposon-mediated random insertions and recombinase-catalyzed, site-specific integrations in the malaria vector mosquito, Anopheles stephensi. These sequences can contribute to precise expression of transgenes in mosquitoes engineered for both basic and applied goals.
Keywords: transgenesis, transposase
The World Health Organization estimated the annual malaria burden in 2010 to be ∼216 million clinical cases with ∼655,000 deaths (1). Malaria parasites are transmitted to humans by mosquitoes of the genus Anopheles, and therefore, these insects are the targets of vector control programs. However, insecticide resistance challenges malaria eradication efforts and innovative approaches for control are needed where traditional methods no longer work (2). Proposed approaches include the use of genetic strategies based on transgenic mosquitoes to suppress (population reduction) or modify (population replacement) vector populations (3–5). Population replacement requires the introgression of a parasite-refractory or resistance gene into wild, malaria-susceptible mosquito populations, thereby interrupting transmission (6). Although substantial advances have been made in engineering resistant mosquito strains (7–18), limited efforts have focused on refining mosquito transgene expression, which can result in variations in phenotypes and impacts on fitness due to the random integration of transposable elements (19–21).
Transgene expression varies both among and within transgenic animal strains due primarily to position effects and position-effect variegation (PEV). Position effects arise as a direct consequence of the different transcriptional status of the integration site and/or the influence of nearby enhancers or repressors of gene expression (22). PEV results when expression is varied among siblings of a family with a single, common insertion that likely borders a euchromatic/heterochromatic boundary. Both types of variable expression can explain phenotypes observed in transgenic mosquito lines (7, 8, 23). These phenomena complicate efforts to identify and characterize transgenic lines in mosquitoes, especially Aedes aegypti, which has large amounts of interspersed heterochromatin throughout its genome (24). Perhaps the most promising strategy to mitigate position effects is the identification of insulator or boundary DNA elements that could be used to isolate transgenes from the effects of their genomic environment.
Insulators regulate interactions among promoter and enhancer elements and are able to organize independent gene regulatory domains to prevent inappropriate gene expression. Position effects can be minimized by flanking transgenes with insulators as they can block the effects of neighboring enhancers and silencers as well as encroaching heterochromatin (25). A number of different DNA sequences with insulating activity have been identified in both invertebrate and vertebrate species including scs/scs’, a portion of the gypsy retrotransposon from the fruit fly, Drosophila melanogaster, sites in the sea urchin histone H3 genes (sns), human Matrix Attachment Regions (MARs), the chicken β-globin genes (cHS4), the ribosomal RNA genes of Xenopus, the human T-cell receptor (TCR)-α/δ locus, and the CTCF factor (26–33). The Drosophila gypsy insulator [also known as the Su(Hw) insulator] phenotype results from a nucleotide sequence ∼350 base pairs (bp) in length derived from the DNA adjacent to the 3′-end of the 5′ long terminal repeat in the gypsy retrotransposon and a number of host-derived DNA binding proteins (34–36). The gypsy insulator complex is proposed to regulate gene expression by establishing higher order domains of chromatin structure and blocking the interference of nearby enhancers or repressors (37–39).
We developed quantitative approaches that show that Drosophila gypsy insulator DNA included in transgenes inserted into the genome of the malaria vector mosquito, Anopheles stephensi, can increase and stabilize transgene expression in transposon-mediated random insertions and recombinase-catalyzed, site-specific integrations. These sequences can contribute to precise expression of transgenes in mosquitoes engineered for both basic and applied goals.
Results
Design of gypsy-Insulated and -Uninsulated Vectors.
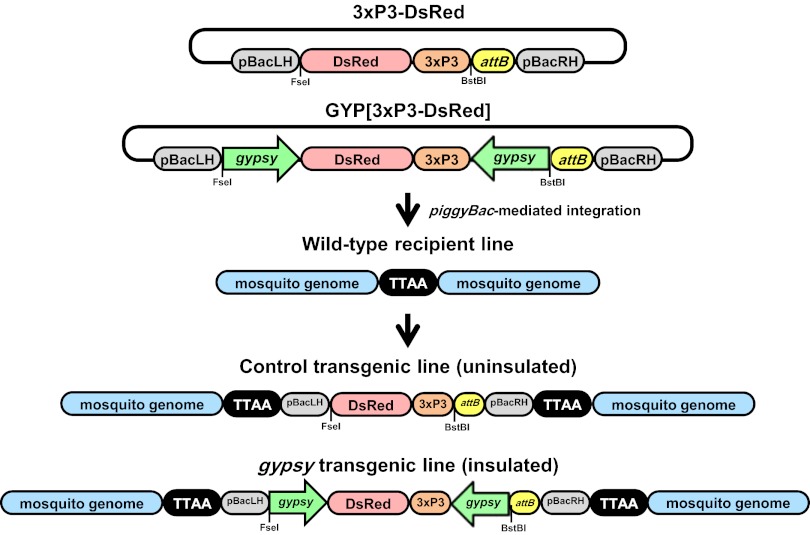
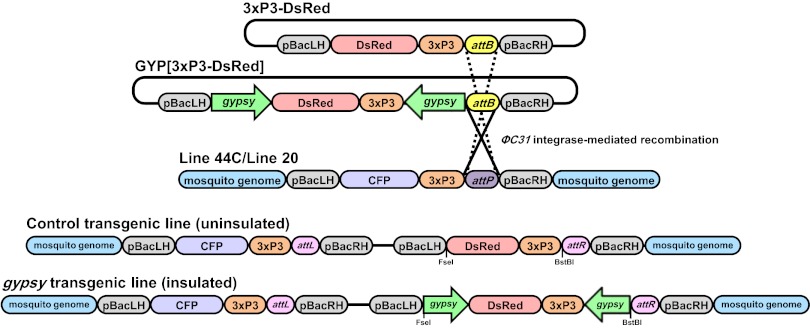
An import vector, GYP[3xP3-DsRed], containing gypsy insulator DNA derived from D. melanogaster was constructed to test effects on transgene expression in An. stephensi (Fig. 1) (GenBank Accession No. KC733875). GYP[3xP3-DsRed] expresses DsRed, a fluorescent marker protein distinguished easily from the cyan fluorescent protein (CFP) expressed by recipient-line mosquitoes (docking-site lines 44C and 20) (18, 40). The recipient lines were selected because they have single docking-site transgene insertions at different places in the genome and vary visually in the intensity and distribution of their CFP phenotype (Fig. S1). Line 20 was chosen as a “high”-intensity phenotype with brighter and more widely distributed fluorescence, and 44C was chosen as comparatively “low,” less bright, and with more tissue-specific expression. The gypsy DNA sequences are cloned as inverted repeats so as to flank the DsRed marker gene on both sides. The construct, 3xP3-DsRed, lacks the gypsy DNA and was used to generate control transgenic lines. The entire DsRed cassette is flanked in both vectors by the piggyBac right- and left-hand inverted terminal repeat (ITR) DNA to allow transposase-mediated random integration of the transgene into the genome. A φC31 attB sequence also is inserted into the left-hand piggyBac ITR to allow φC31 recombinase-catalyzed transgene insertion into the attP-containing docking-site lines (Fig. 2). The use of plasmids with identical structures for both random and site-directed integration experiments is expected to reduce some potential variation due to different construct architecture.
Fig. 1.
Schematic representation of gypsy-containing and control transgene random integration into the An. stephensi genome. The DsRed (DsRED) gene is included in the pUC18 vector (black line) along with piggyBac left- and right-hand (pBacLH; pBacRH) ITR DNA. DsRed expression (DsRED) is driven by the Pax3 promoter (3xP3). GYP[3xP3-DsRed] constructs have the DsRed cassette flanked with Drosophila gypsy insulator sequences (gypsy). An attB site (not relevant to these experiments) also is included. The piggyBac transposase mediates random integration of 3xP3-DsRed and GYP[3xP3-DsRed] into the wild-type mosquito genome at TTAA sites to produce uninsulated and insulated lines, respectively. Relative locations are shown for the recognition sites for the restriction endonucleases, BstBI and FseI.
Fig. 2.
Schematic representation of gypsy-containing and control site-specific integration in An. stephensi docking-site lines. The 3xP3-DsRed and GYP[3xP3-DsRed] plasmid structures are identical to Fig. 1. The docking-site lines (Line 44C/Line 20) have piggyBac LH and RH sequences joined to a CFP transformation marker driven by the 3xP3 promoter and a phage attachment site (attP) (18). The φC31 integrase catalyzes recombination of the attB and attP sites to generate right and left attachment sites (attR and attL) and results in integration of the 3xP3-DsRed (broken lines) or GYP[3xP3-DsRed] plasmids (solid lines). Relative locations are shown for the recognition sites for the restriction endonucleases, BstBI and FseI.
gypsy DNA Elevates and Stabilizes the Expression of DsRed in Transgenic Lines with piggyBac Transposase-Mediated Random Integrations.
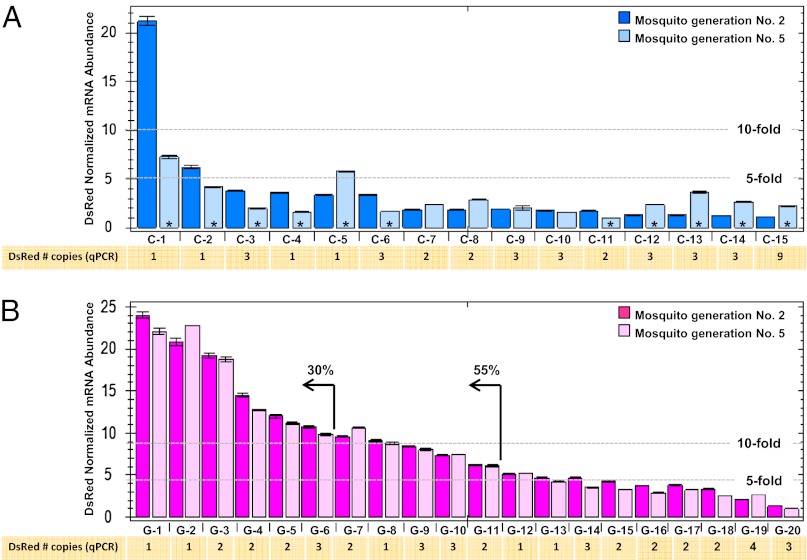
Hemizygous transgenic lines were generated by coinjecting separately control and gypsy-containing plasmids along with a piggyBac helper plasmid into wild-type An. stephensi embryos. The resulting lines have the transgene inserted in different places, and in some cases, in multiple copies throughout the genome (Fig. S2). Fifteen control and 20 gypsy lines were recovered, and the abundance of DsRed mRNA in individual mosquitoes was quantified by real-time PCR and normalized to the An. stephensi ribosomal S7 gene (Fig. 3). Individual mosquitoes were used to minimize variations in gene copy number that could arise from independent assortment in lines carrying multiple, unlinked insertions. Relative DsRed mRNA levels from two independent experiments (biological replicates) showed that 11 (55%) and six (30%) of the gypsy-containing transgenic lines had ≥5- or ≥10-fold higher, respectively, marker gene mRNA abundance compared with the S7 control. In contrast, only one of the 15 (6%) control lines had DsRed mRNA accumulation levels ≥10-fold. Furthermore, real-time PCR analyses of individual second-generation mosquitoes (G2, following establishment the transgenic lines) show that marker gene mRNA accumulation in these experiments is independent of DsRed genome copy number. All transgenic lines were outcrossed to wild-type animals at each generation, and therefore, the copy numbers represent multiple and distinct hemizygous insertions of the transgene in the genome. Finally, replicate biological experiments with G2 and G5 mosquitoes show that gypsy-containing transgenic lines are significantly more consistent (all P values >0.01) than controls (P values ranging from 0.0015 to 0.46) in the levels of DsRed mRNA accumulation among mosquitoes of the different generations (Tables S1 and S2). These results support the conclusion that the exogenously derived gypsy insulator-like DNA is functional in this mosquito and contributes to higher and more consistent transgene mRNA abundance.
Fig. 3.
DsRed normalized mRNA abundance and genome copy number from uninsulated and insulated transgenes inserted randomly into the An. stephensi genome. (A) Fifteen uninsulated, control transgenic lines (C-1 to C-15), and (B) 20 insulated transgenic lines (G-1 to G-20) were generated and analyzed for DsRed mRNA abundance levels. DsRed mRNA accumulation was quantified by qRT-PCR in two different mosquito generations, G2 (dark-color bars) and G5 (light-color bars). Statistically significant differences in mRNA abundance between generations are marked with an asterisk (*). The number of DsRed genes present in G2 mosquitoes was determined by qPCR and is indicated below each transgenic line. DsRed mRNA levels were quantified by qRT-PCT and normalized using ribosomal gene S7 transcript abundance as a reference.
gypsy DNA Modulates and Stabilizes the Expression of DsRed in Transgenic Lines with φC31-Mediated Site-Specific Integrations.
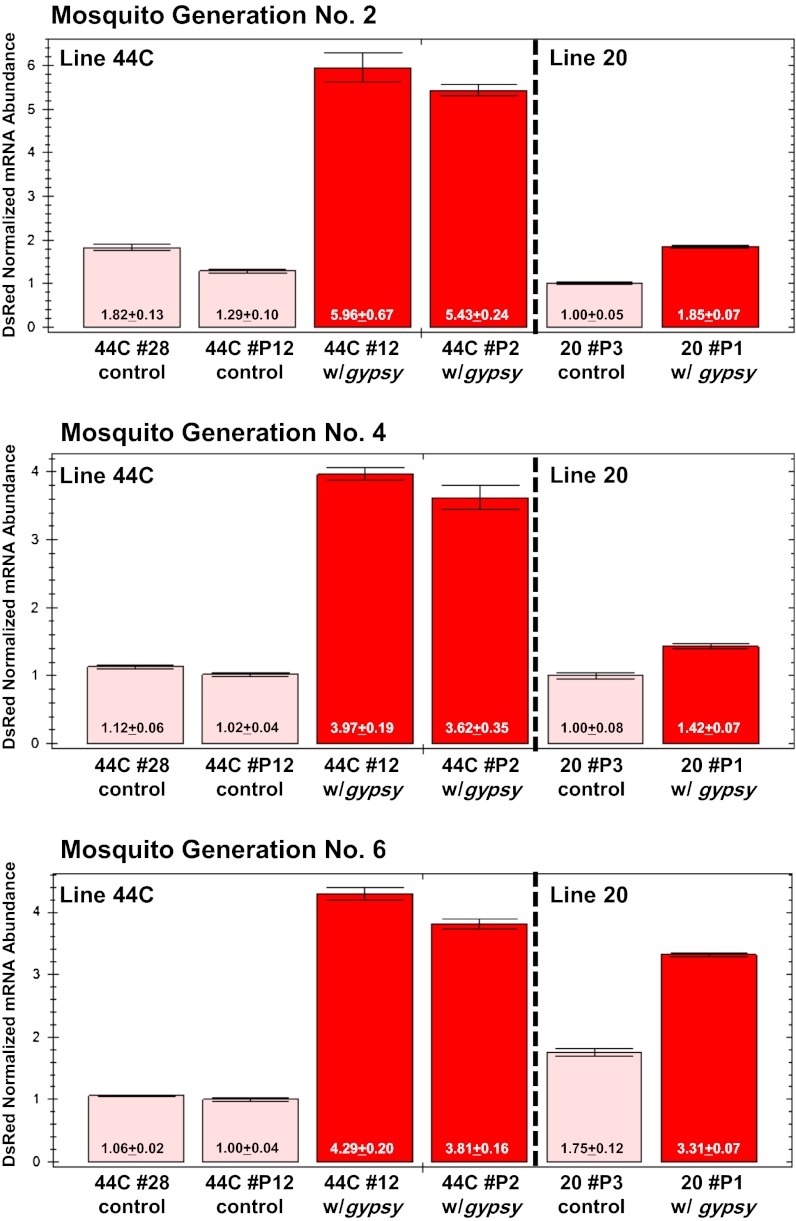
Mosquito embryos from docking lines 44C and 20 containing attP sites for φC31 integrase (18, 40) were coinjected with control and gypsy-containing plasmids and the integrase mRNA. Two lines each were derived independently in docking line 44C (44C #12 and 44C #P2 contain GYP[3xP3-DsRed] and 44C #28 and 44C #P12 carry 3xP3-DsRed). Pooled mRNA samples from 10 individuals were prepared for these experiments. No statistical differences were seen in the normalized abundance of DsRed mRNA between each gypsy-containing corresponding pair, although the values in 44C #12 were consistently 9–12% higher than 44C #P2 (Fig. 4; Tables S3–S5). The range of variation, 6–41%, was larger between the independently derived control lines (44C #28 and 44C #P12), but the low absolute values distort these differences. The low variation among the three experimental replicate reactions of each sample support the conclusion that the procedures used were consistent throughout the experiments. In contrast, GYP[3xP3-DsRed] showed a significant threefold increase in DsRed mRNA abundance compared with the 3xP3-DsRed control lines. However, DsRed mRNA abundance was increased only ∼1.8-fold in docking line 20 when GYP[3xP3-DsRed] (line 20 #P1) was compared with its corresponding control (line 20 #P3). Direct comparisons of the values of DsRed mRNA abundance among the biological replicates of G2, G4, and G6 mosquitoes of both docking lines are complicated likely by environmental and epigenetic variation. However, the DsRed-normalized mRNA abundance in gypsy-containing lines was consistently higher than controls. These results support the conclusion that gypsy DNA can have a positive and stable effect on single-copy transgene expression.
Fig. 4.
DsRed mRNA abundance in uninsulated and insulated site-specific transgenic lines. Docking lines 44C and 20 (left and right, respectively, in each panel) were used to generate DsRed site-specific integrations. DsRed mRNA abundance levels were compared among uninsulated (44C #28, 44C #P12, 20 #P3) and insulated (44C #12, 44C #P2, 20 #P1) lines, over three mosquito generations (2, 4, 6). Average values for each line (numbers within each histogram) were derived from triplicate technical replicates and the bars represent the SD in each set of samples. DsRed mRNA levels were quantified by qRT-PCT and normalized using ribosomal gene S7 transcript abundance as a reference.
gypsy DNA Affects Expression of a Marker Gene Outside the Insulated Region.
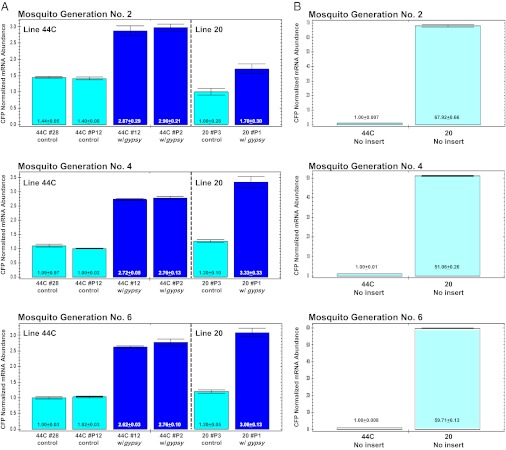
gypsy sequences have insulator activity not only on genes surrounded by them, but also on those outside but nearby (41). The CFP marker gene is flanked on only one side by gypsy DNA (Fig. 2). Comparisons were made of the abundance of CFP mRNA among the various combinations of docking-site lines (Fig. 5; Tables S6–S8). Line 20 has on average an ∼60-fold higher basal level of CFP mRNA abundance than line 44C, and this is consistent with the subjective interpretation that it has a high-intensity CFP fluorescence phenotype. Recombinase-mediated insertions of both GYP[3xP3-DsRed] and 3xP3-DsRed constructs in line 20 resulted in a significant 20–60-fold reduction in CFP mRNA abundance. Furthermore, CFP expression in line 20 was only ∼1.7-fold higher in the insulated line (20 #P1) than the control line (20 #P3). In contrast, CFP mRNA showed a significant ≥2-fold higher abundance in docking line 44C with gypsy DNA sequences compared with either the uninsulated control lines (44C #28 and 44C #P12) or the original docking line without any additional sequence insertions. Normalized CFP mRNA levels were similar among the three mosquito generations (G2, G4, and G6) for both docking lines. These results support the conclusions that there are some proximity effects of gypsy insulator-like DNA in transgenic lines but that the effect, although stable, can be positive (line 44C) or negative (line 20).
Fig. 5.
CFP abundance from uninsulated/insulated docking-site lines. CFP expression levels were derived and are presented as in Fig. 4. (A) Comparisons of uninsulated (44C #28, 44C #P12, 20 #P3) and insulated (44C #12, 44C #P2, 20 #P1) docking lines 44C and 20 (Right and Left, respectively) over three mosquito generations (2, 4, 6). (B) Comparisons of CFP transcript expression levels in docking-site lines 44C and 20 over three mosquito generations (2, 4, 6). Please note the differences in the scale of the y-axes in A and B.
Orthologous Genes Encoding gypsy Insulator Complex Proteins Are Found in Mosquito Species.
The conceptual translation products of the D. melanogaster gypsy complex genes with demonstrated insulator function, Su(Hw), Mod(mdg4)2.2, and CP190, were used in reciprocal Blast analyses to identify putative orthologous genes in the mosquitoes An. stephensi, An. gambiae, Ae. aegypti, and Culex quinquefasciatus. The highest scoring matches were screened visually for the conservation of the functionally characterized binding domains, and a putative ortholog for each gene was found (Table S9). Annotations were returned for only the putative C. quinquefasciatus genes, and these were described as a gonadotropin-inducible transcription factor, a modifier of mdg4, and a microtubule-binding protein for Su(Hw), Mod(mdg4)2.2, and CP190, respectively. With the possible exception of the Su(Hw) ortholog, these annotations are consistent with the demonstrated function of the corresponding D. melanogaster proteins.
Reciprocal Blast analyses using the amino acid sequences encoded by the An. gambiae putative gypsy complex orthologous genes returned the same genes discovered with the fruit fly query, and the relative similarities of the proteins support the established phylogeny of the species investigated. The two Anopheles sets of genes were more similar to one another than the Aedes and Culex genes, and as seen previously, the D. melanogaster sequences had the lowest similarity.
Discussion
This study supports the conclusion that exogenously derived gypsy insulator DNA can function to modulate and stabilize transgene expression in the vector mosquito, An. stephensi. gypsy insulators were characterized first in D. melanogaster associated with the eponymous retrotransposon, where it was shown that a number of specific proteins interacted with and bound conserved DNA domains in the mobile element and others distributed throughout the fruit fly genome (42–44). We interpret our results to indicate that a functionally similar group of proteins exist in An. stephensi and that they have the ability to reproduce the insulating phenotype using introduced DNA binding domains. This similarity extends to the ability to modulate gene expression both positively and negatively and the imposition of polar effects on transcriptional units near but not flanked by the insulator DNA (41). This similarity also supports the hypothesis that the functional requirements for these insulator-like elements may have been conserved evolutionarily over the >250 million years that these taxa are estimated to have diverged (45, 46).
Although the DNA components of the insulating elements were found first associated with the gypsy retrotransposon, it was shown later that there are independent gypsy-like and functional endogenous sequences in the D. melanogaster genome (47). However, there is debate as to whether the gypsy DNA sequences represent components of a canonical insulating system. While there are gypsy-like retrotransposons in mosquitoes, the existing data support a hypothesis that the target DNA and corresponding interacting proteins most likely originated in ancestral genomes and were acquired later by the mobile element (47). This hypothesis receives additional, although indirect, support by the discovery of putative Su(Hw), Mod(mdg4)2.2, and CP190 orthologous genes in the four mosquito species. Furthermore, a gypsy-like DNA insulator system is likely to not be the only one functioning in mosquito genomes. Putative CTCF homologous genes were identified and characterized in Ae. aegypti and An. gambiae (48). These genes encode polypeptides with primary amino acid similarity to vertebrate CTCFs, despite ≥500 million years of divergence.
The results from the piggy-Bac–mediated random insertions of gypsy-containing and control plasmids support the inclusion of gypsy DNA in routine mosquito transformation experiments. While there are published experiments in which this DNA was included in mosquito transgenes, no efforts were made to do a rigorous quantitative analysis of their impact and there is no previous evidence that they function in mosquitoes (49). The results we obtained show that transgene expression can be enhanced, leading to greater mRNA abundance, and that this effect is reproducible across a number of generations. Analyses of these or similar lines are needed over a much longer term to determine if the gypsy insulator prolongs the mRNA abundance phenotypes. A noninsulated transgene in Ae. aegypti experienced silencing following long-term culture (G17) (50), and it is not clear if insulator elements would have mitigated these effects or if the silencing phenotype extends to mosquitoes of the genus Anopheles.
An interesting observation from the piggy-Bac–mediated insertions is the lack of correlation of insert copy number with marker gene mRNA abundance levels. piggy-Bac often inserts into multiple and distinct sites of the An. stephensi genome during transgenesis experiments (11, 18, 40). As such, many transgenic lines are actually populations of mosquitoes consisting of individuals with different qualitative and quantitative transgene representations generated by insertion-site effects, linkage, independent assortment, and hemi- or homozygosity. These differences may account for some of the variation in the phenotypes of many of the transgenes reported in the literature (51, 52) and for the observed variation in the biological replicates done here. These data support a hypothesis that the major qualitative and quantitative effects of a transgene result likely from only one of the multiple insertions. While it is reasonable to expect an increase in mRNA abundance and transgene function when an individual locus is made homozygous, adding another copy at another location in the genome does not necessarily add to this.
The results of the site-specific transgene integrations reinforce strongly the impact of insertion sites on gene function within the added context of a single-copy gene. The two docking-site lines were selected for these experiments because they had visually detectable differences in the intensity and distribution of their respective CFP marker gene phenotypes; line 20 mosquitoes had a much “brighter” and more widely distributed expression pattern under fluorescence microscopy. The discovery that docking line 20 had ∼60-fold higher normalized CFP mRNA abundance than line 44C confirms this subjective observation. Both the visual phenotypes and quantitative evaluations of mRNA abundance are direct demonstrations of the differential impact of genome location on the function of an identical gene. However, previous studies of transgene insertions in these two docking sites show that line 20 is not as permissive as line 44C for expression from different integrated promoters (18). Both components of a transgene construct comprising dual antiparasite effector genes were expressed well in line 44C but only one of the two promoters functioned in line 20.
gypsy insulator DNA had a measurable and significant effect on transgene mRNA abundance in both docking-site lines. The net impact on insulator-flanked DsRed-expressing transgenes was positive for insertions in line 44C. The insulating effect was not as strong for insertions into line 20, although it was positive through all three biological replicates. In contrast, the effects of insertions on CFP mRNA abundance varied between the two docking-site lines. There was a large and significant reduction in CFP mRNA abundance in line 20 as a consequence of the integration of either gypsy-containing or control transgenes, while control insertions had little effect in line 44C. Interestingly, gypsy DNA increased CFP mRNA abundance in line 44C and reduced the negative impact of an insertion in line 20. These results support a general model that the insulator-like sequences mitigated the effects of a suppressor in the region of the 44C genome insertion site, while in line 20 they impact the influence of a strong enhancer. The results from line 20 are counterintuitive, as it might be predicted that the insulated construct would suppress even more the insertion-site influences. However, the mechanisms of insulator activity in mosquitoes are unknown, and further experiments are needed to explain this observation.
The differences in mRNA abundance levels for the corresponding, independently derived pairs of experimental and control strains generated from line 44C were heritable over multiple generations in the three biological replicate experiments. Since the same gene is inserted at the same locus in each of these pairs of lines, these data support a hypothesis that one or more modifier genes or alternate alleles that either stimulate or repress transgene expression at a low level segregated or assorted independently in one of the isolates. Furthermore, carrying out biological replicates over multiple generations mimics conditions that might prevail during the establishment and maintenance of any transgenic mosquito line for basic or applied studies. The small variations in normalized mRNA abundance values among the three replicates most likely arise from extrinsic and environmental variables. We expect this variation to be negligible as specific insulated strains are produced and standardized for practical applications. As with any biological agent, lines adapted for disease control programs will need to be monitored for continued efficacy.
We expect future studies of gypsy-insulating DNA in mosquitoes to reveal the extent to which this specific system represents a fundamental property of genome organization, at least in insects, and how it can be used in both basic and applied studies of mosquito gene expression. Toward these ends, we have initiated analyses of the mosquito orthologous genes encoding the protein components of the insulating-like system and are searching for endogenous gypsy-like sites in mosquitoes.
Materials and Methods
Transposon- and recombinase-mediated transgenesis technologies were used to produce experimental and control strains of An. stephensi. Experimental and control transgenes were identical except for the inclusion of gypsy element insulator target DNA in the former. Gene copy numbers were verified using Southern blot analyses and gene amplification.
Standardized quantitative gene amplification methods and statistical analyses were used to determine the mRNA abundance of a marker gene (encoding DsRed) flanked or not by gypsy DNA and compare variation among samples, respectively. Both technical and biological replicates were performed with the latter conducted over successive generations. The mRNA abundance also was determined for a second marker gene (CFP) with or without gypsy DNA on one side only.
Bioinformatic approaches were used to identify putative orthologous genes encoding the protein components of the gypsy insulator complex in four mosquito species and evaluate their relative similarity. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Karen Chow, Stephanie Mattingly, and Aniko Fazekas for mosquito husbandry. R.C.-L. was supported by a postdoctoral fellowship from UC MEXUS and CONACYT (UCM49318) and National Institutes of Health National Institute of Allergy and Infectious Diseases (AI29746) to A.A.J.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KC733875).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304722110/-/DCSupplemental.
References
- 1.WHO World Malaria Report. 2011. www.who.int/malaria/world_malaria_report_2011/en/
- 2.Alonso PL, et al. A research agenda for malaria eradication: vector control. PLoS Med. 2011;8(1):e1000406. doi: 10.1371/journal.pmed.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James AA. Gene drive systems in mosquitoes: Rules of the road. Trends Parasitol. 2005;21(2):64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Marshall JM, Taylor CE. Malaria control with transgenic mosquitoes. PLoS Med. 2009;6(2):e20. doi: 10.1371/journal.pmed.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terenius O, Marinotti O, Sieglaff D, James AA. Molecular genetic manipulation of vector mosquitoes. Cell Host Microbe. 2008;4(5):417–423. doi: 10.1016/j.chom.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerntsen BT, James AA, Christensen BM. Genetics of mosquito vector competence. Microbiol Mol Biol Rev. 2000;64(1):115–137. doi: 10.1128/mmbr.64.1.115-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jasinskiene N, et al. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci USA. 1998;95(7):3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coates CJ, Jasinskiene N, Pott GB, James AA. Promoter-directed expression of recombinant fire-fly luciferase in the salivary glands of Hermes-transformed Aedes aegypti. Gene. 1999;226(2):317–325. doi: 10.1016/s0378-1119(98)00557-5. [DOI] [PubMed] [Google Scholar]
- 9.Catteruccia F, et al. Toward Anopheles transformation: Minos element activity in anopheline cells and embryos. Proc Natl Acad Sci USA. 2000;97(5):2157–2162. doi: 10.1073/pnas.040568397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm] Insect Biochem Mol Biol. 2001;31(12):1137–1143. doi: 10.1016/s0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- 11.Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito Anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. J Biol Chem. 2002;277(11):8759–8762. doi: 10.1074/jbc.C100766200. [DOI] [PubMed] [Google Scholar]
- 12.Grossman GL, et al. Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol Biol. 2001;10(6):597–604. doi: 10.1046/j.0962-1075.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 13.Perera OP, Harrell RA, II, Handler AM. Germ-line transformation of the South American malaria vector, Anopheles albimanus, with a piggyBac/EGFP transposon vector is routine and highly efficient. Insect Mol Biol. 2002;11(4):291–297. doi: 10.1046/j.1365-2583.2002.00336.x. [DOI] [PubMed] [Google Scholar]
- 14.Lycett GJ, Kafatos FC, Loukeris TG. Conditional expression in the malaria mosquito Anopheles stephensi with Tet-On and Tet-Off systems. Genetics. 2004;167(4):1781–1790. doi: 10.1534/genetics.104.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W, et al. Ectopic expression of a cecropin transgene in the human malaria vector mosquito Anopheles gambiae (Diptera: Culicidae): Effects on susceptibility to Plasmodium. J Med Entomol. 2004;41(3):447–455. doi: 10.1603/0022-2585-41.3.447. [DOI] [PubMed] [Google Scholar]
- 16.Rodrigues FG, Oliveira SB, Rocha BC, Moreira LA. Germline transformation of Aedes fluviatilis (Diptera:Culicidae) with the piggyBac transposable element. Mem Inst Oswaldo Cruz. 2006;101(7):755–757. doi: 10.1590/s0074-02762006000700008. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida S, Watanabe H. Robust salivary gland-specific transgene expression in Anopheles stephensi mosquito. Insect Mol Biol. 2006;15(4):403–410. doi: 10.1111/j.1365-2583.2006.00645.x. [DOI] [PubMed] [Google Scholar]
- 18.Isaacs AT, et al. Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proc Natl Acad Sci USA. 2012;109(28):E1922–E1930. doi: 10.1073/pnas.1207738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces VG, Geyer PK. Interactions of retrotransposons with the host genome: The case of the gypsy element of Drosophila. Trends Genet. 1991;7(3):86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- 20.Brookfield JF. The ecology of the genome-mobile DNA elements and their hosts. Nat Rev Genet. 2005;6(2):128–136. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- 21.Pereira V, Enard D, Eyre-Walker A. The effect of transposable element insertions on gene expression evolution in rodents. PLoS ONE. 2009;4(2):e4321. doi: 10.1371/journal.pone.0004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 23.Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 1998;95(7):3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudson DL, et al. The Biology of Disease Vectors. Fort Collins, CO: Univ Press of Colorado; 1996. pp. 175–214. [Google Scholar]
- 25.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291(5503):447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 26.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185(2):341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 27.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6(10):1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 28.Palla F, Melfi R, Anello L, Di Bernardo M, Spinelli G. Enhancer blocking activity located near the 3′ end of the sea urchin early H2A histone gene. Proc Natl Acad Sci USA. 1997;94(6):2272–2277. doi: 10.1073/pnas.94.6.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namciu SJ, Blochlinger KB, Fournier RE. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol Cell Biol. 1998;18(4):2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74(3):505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 31.Robinett CC, O’Connor A, Dunaway M. The repeat organizer, a specialized insulator element within the intergenic spacer of the Xenopus rRNA genes. Mol Cell Biol. 1997;17(5):2866–2875. doi: 10.1128/mcb.17.5.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong XP, Krangel MS. An enhancer-blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc Natl Acad Sci USA. 1997;94(10):5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon H, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6(2):165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162(4):565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6(5):1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 36.Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92(4):511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 37.Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7(9):703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 38.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23(11):1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amenya DA, et al. Comparative fitness assessment of Anopheles stephensi transgenic lines receptive to site-specific integration. Insect Mol Biol. 2010;19(2):263–269. doi: 10.1111/j.1365-2583.2009.00986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.She W, et al. The gypsy insulator of Drosophila melanogaster, together with its binding protein suppressor of Hairy-wing, facilitate high and precise expression of transgenes in Arabidopsis thaliana. Genetics. 2010;185(4):1141–1150. doi: 10.1534/genetics.110.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20(10):2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16(5):737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Parnell TJ, et al. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol. 2006;26(16):5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19(5):748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 46.Schoborg TA, Labrador M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J Mol Evol. 2010;70(1):74–84. doi: 10.1007/s00239-009-9310-x. [DOI] [PubMed] [Google Scholar]
- 47.Adryan B, et al. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 2007;8(8):R167. doi: 10.1186/gb-2007-8-8-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray CE, Coates CJ. Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae. BMC Mol Biol. 2005;6:16. doi: 10.1186/1471-2199-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynd A, Lycett GJ. Development of the bi-partite Gal4-UAS system in the African malaria mosquito, Anopheles gambiae. PLoS ONE. 2012;7(2):e31552. doi: 10.1371/journal.pone.0031552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franz AWE, et al. Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol Biol. 2009;18(5):661–672. doi: 10.1111/j.1365-2583.2009.00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacs AT, et al. Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi. PLoS Pathog. 2011;7(4):e1002017. doi: 10.1371/journal.ppat.1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corby-Harris V, et al. Activation of Akt signaling reduces the prevalence and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PLoS Pathog. 2010;6(7):e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.