Abstract
Marine diatoms are important primary producers that thrive in diverse and dynamic environments. They do so, in theory, by sensing changing conditions and adapting their physiology accordingly. Using the model species Thalassiosira pseudonana, we conducted a detailed physiological and transcriptomic survey to measure the recurrent transcriptional changes that characterize typical diatom growth in batch culture. Roughly 40% of the transcriptome varied significantly and recurrently, reflecting large, reproducible cell-state transitions between four principal states: (i) “dawn,” following 12 h of darkness; (ii) “dusk,” following 12 h of light; (iii) exponential growth and nutrient repletion; and (iv) stationary phase and nutrient depletion. Increases in expression of thousands of genes at the end of the reoccurring dark periods (dawn), including those involved in photosynthesis (e.g., ribulose-1,5-bisphosphate carboxylase oxygenase genes rbcS and rbcL), imply large-scale anticipatory circadian mechanisms at the level of gene regulation. Repeated shifts in the transcript levels of hundreds of genes encoding sensory, signaling, and regulatory functions accompanied the four cell-state transitions, providing a preliminary map of the highly coordinated gene regulatory program under varying conditions. Several putative light sensing and signaling proteins were associated with recurrent diel transitions, suggesting that these genes may be involved in light-sensitive and circadian regulation of cell state. These results begin to explain, in comprehensive detail, how the diatom gene regulatory program operates under varying environmental conditions. Detailed knowledge of this dynamic molecular process will be invaluable for new hypothesis generation and the interpretation of genetic, environmental, and metatranscriptomic data from field studies.
Keywords: diurnal cycle, oceanography, photosynthesis, phytoplankton, gene expression
Diatoms are widespread throughout the oceans and freshwater ecosystems and have evolved to flourish under diverse and changing environments (1, 2). They are important for the uptake of CO2 and the production of organic carbon (3–5), as well as the cycling of macronutrients including nitrogen, silicon, and phosphorus (6). Marine phytoplankton account for nearly half of the primary production of organic carbon on the planet (7). Diatoms are estimated to account for up to 40% of marine productivity and up to 90% in highly productive nutrient-rich coastal regions (8, 9). Thus, it is important to study the effects of climate change, nutrient availability, and other stresses on marine diatoms, as these effects will propagate from single cells into marine ecosystems.
The biology and adaptation of diatoms, as in all organisms, critically depend on mechanisms of physiological regulation encoded in the genome. The availability of diatom genome sequences provides the opportunity for detailed examinations of the genetic and mechanistic bases of diatom ecophysiology and evolution (10, 11). The genetic repertoire of diatoms consists of more than 10,000 predicted genes, the majority of which bear no similarity to genes of known function. This novelty precludes a complete understanding of the physiology, regulation, and adaptation of this important class of organisms.
Measures of genome-wide expression patterns of the up- or down-regulation of diatom genes under both laboratory conditions and in the wild have implicated many genes in the responses and/or acclimation to environmental perturbations (12–16). In nature, diatoms must use sensory and regulatory mechanisms to carry out their physiological programs in the context of additional variations including recurring changes in nutrient conditions, pH, cell density, and diel light cycling. Our experiments were designed to identify genes responsible for general homeostasis under fluctuating conditions and expression patterns that are coherent across multiple environmental scenarios. We analyzed the recurring physiological and transcriptional changes that occur in the model diatom Thalassiosira pseudonana (10) as cells divide and transition from nutrient-replete to nutrient-depleted conditions. Physiological characteristics and genome-wide transcript levels were measured over the course of 5 d for axenic cultures on a 12:12 h dark:light cycle. Data collected from growth experiments at two CO2 levels, ambient (400 ppm) and elevated (800 ppm), were compared and then combined to identify core patterns of expression that are shared in common between current and future estimates of CO2 levels in the ocean (17–20).
Results and Discussion
Cultures of axenic T. pseudonana cells that were entrained to a 12:12 h light:dark cycle were grown for 5 d to survey the physiological and regulatory changes that occur over the growth curve. Cell density increased exponentially for the first 3 d, with the highest growth rates and the majority of cell divisions occurring during the light phase (Figs. S1 and S2). Light-dependent photosynthetic activity drove increases in pH and oxygen levels during the light period. Cellular fluorescent capacity (CFC) peaked early during the light phases and decreased to a minimum during the dark period. The cultures entered a stationary phase at a cell density of ∼3 × 106 cells/mL, concurrent with the exhaustion of nitrate in the media. This was accompanied by a decrease in photosynthesis, as evident in the decrease in the magnitude of pH and O2 fluctuations, as well as a decrease in photosynthetic capacity and cell division. Physiology and growth characteristics were similar under both CO2 regimes (Figs. S1 and S2).
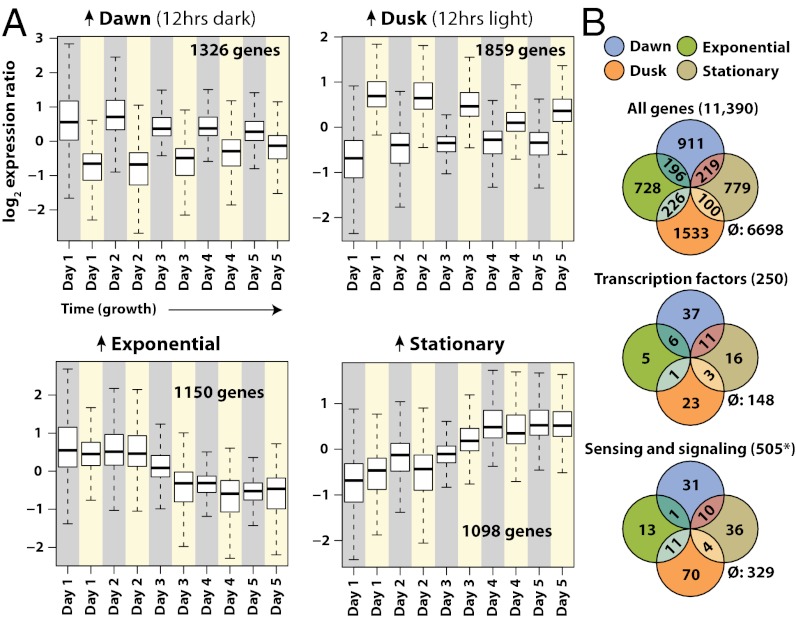
Whole-genome expression patterns were measured twice daily for 5 consecutive days: once at the end of the 12-h dark period (hence referred to as “dawn”) and once at the end of the 12-h light period (hence referred to as “dusk”). These sampling points represent distinctly different growth conditions for these obligate photoautotrophs, which must regulate their physiology and metabolism based on light and nutrient availability (21, 22). Gene-level expression changes for all corresponding time points were highly correlated for both CO2 regimes (P = 2.2 × 10−16, Fig. S3B). The expression time series were combined to detect the growth-dependent dynamics of expression that generalize over both conditions. The expression patterns displayed recurring diel transitions as well as a large co-occurring growth-phase–dependent effect over the 5-d growth curve. Expression of roughly 40% of T. pseudonana genes (two-factor ANOVA) was associated with four cellular states (Fig. 1A): (i) dawn, following 12 h of darkness (1,326 up-regulated genes), (ii) dusk, following 12 h of light (1,859 genes), (iii) exponential phase, nutrient-replete growth (1,150 genes), and (iv) stationary phase nutrient-limited culture (1,098 genes). The genes that were most differentially expressed between these states are shown in Fig. S3A and Table S1. In addition to broad changes in the core physiology of T. pseudonana under each of these conditions, the large repertoires of sensing, signaling, and regulatory genes in this organism were highly patterned and differentially expressed under conditions that usually fluctuate under typical growth conditions (Fig. 1B). The physiological and transcriptomic variations that occurred during these experiments resulted in a broad picture of the cell state transitions that prevail under these conditions (Fig. 2).
Fig. 1.
Four principal genome-wide expression patterns observed over the growth cycle. (A) The mean and quantiles of expression ratios for state-associated genes are shown over time during the course of the experiment. Gene groups consist of genes whose expression changes were significantly associated with dark:light or exponential:stationary transitions by ANOVA at P < 0.01. Dawn-associated genes were significantly associated with the dark:light transition and higher in expression at the end of the 12-h dark periods (gray backgrounds). Dusk-associated genes were significantly associated with the dark:light transition and higher in expression at the end of the 12-h dark light (yellow backgrounds). Exponential and stationary genes were significantly associated with the exponential:stationary transition. Genes labeled exponential were higher in expression during the exponential phase (up to day 3). Genes labeled stationary were higher in expression during the stationary phase (following day 3). (B) Euler diagrams illustrate the numbers of genes associated with each condition. Genes for which associations were not detected are indicated by the ∅ symbol. *Sensing and signaling genes include all photoreceptors and genes with the following InterPro domains: IPR000719 (protein kinase), IPR001019 (G protein alpha subunit), IPR001632 (G protein beta subunit), IPR001789 (response regulator receiver), IPR003018 (GAF), IPR000337 (GPCR, family 3), IPR000276 (GPCR, rhodopsin-like), IPR002182 (NB-ARC), IPR001806 (Small GTPase), IPR002073 (cyclic nucleotide phosphodiesterase), IPR001054 (guanylyl cyclase), IPR000014 (PAS), IPR001680 (WD40), IPR001611 (Leucine-rich repeat (LRR)). WD40 and LRR domains (110 and 81 genes) are multifunctional and may not all represent signaling genes.
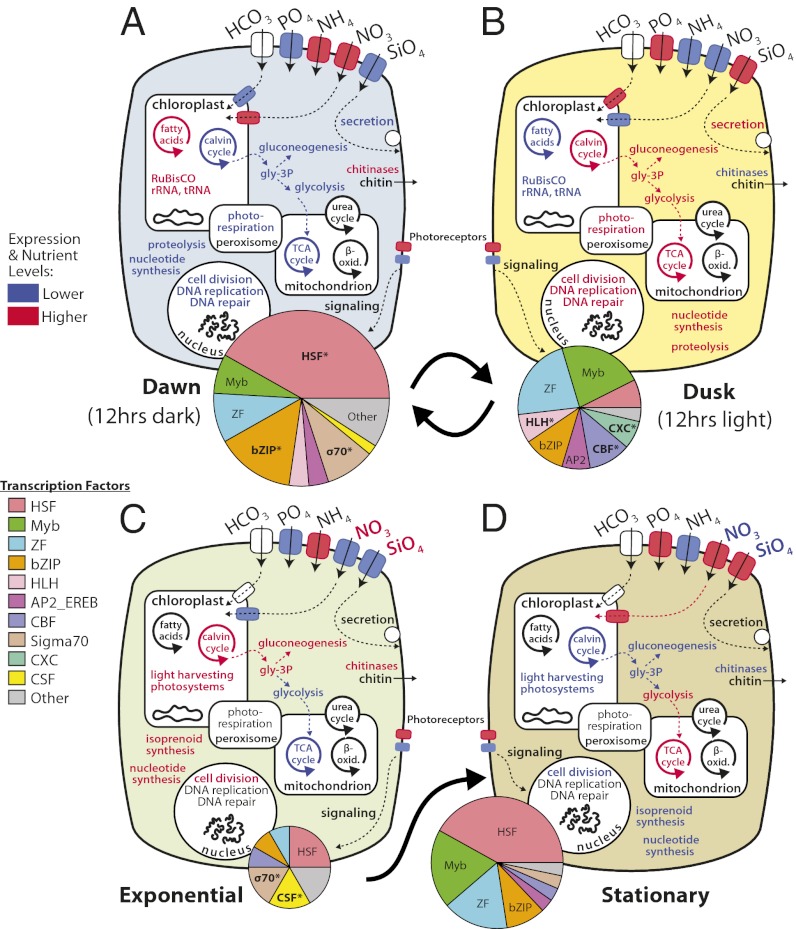
Fig. 2.
Schematic of physiological and transcriptional states of T. pseudonana during diurnal growth. Pathways and functions associated with each state are highlighted. (A) Dawn following 12 h of darkness. (B) Dusk following 12 h of light. (C) Exponential growth in replete nutrients. (D) Stationary phase and nutrient depletion. Those pathways highlighted in red contain genes significantly up-regulated, those in blue contain genes significantly down-regulated, and those in black do not display conditional associations. The association of putative transcription factors (TFs) with each state is depicted in pie charts whose sizes are proportional to the total number of associations detected. TFs that were significantly enriched under a condition are marked with an asterisk.
Diel Cycling of Core Physiological Functions.
Day and night length, maximum irradiance, and spectral composition are important variables in the diel modulation of photosynthesis and metabolic processes in phytoplankton (23–25). The expression of 3,185 genes fluctuated significantly and recurrently between the light and dark periods (Fig. 1A). After 12 h of light (dusk), the most highly expressed genes included those that encode enzymes for cell division, DNA replication and repair, carbon metabolism, and oxidative phosphorylation (Fig. S3A). Genes involved in cell membrane, cell wall biogenesis (actin, dynein, tubulin, protein transport protein Sec61-1 and -2, and several vacuolar ATPases) were also up-regulated during the light period (Fig. S4) as expected during periods of cell division when the silicified cell wall (frustule) is generated to reencase daughter cells (26). Significantly higher transcript levels for superoxide dismutases and peroxidases under light (Fig. S5A), followed by the up-regulation of genes for DNA repair, the mitigation of oxidative stress, and five of six putative metacaspases (Fig. S5B) (27, 28) by the end of the light period suggest active mechanisms to ameliorate light- and growth-related stresses on the cell.
After 12 h of darkness (dawn), the expression of light-associated genes had strongly decreased in accordance with the absence of light and the decrease in growth rates and cell division (Fig. S3 and Table S1). Genes whose transcript levels were higher after 12 h of darkness were enriched for those encoding ribosomal biogenesis, aminoacyl-tRNAs, and key photosynthetic enzymes (Fig. S3C and Table S2). Genes for key photosynthetic enzymes [ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO), aldose-1-epimerase, and transketolase] and protein synthesis were up-regulated in apparent anticipation of the impending light phase (Fig. S6), in agreement with prior findings for genes encoding photosynthetic functions in algae (29–32). This supports the theory that diatom gene regulation becomes entrained to anticipate the diurnal cycle such that the up-regulation of photosynthetic genes precedes the onset of light (33, 34).
Transition from Exponential Growth to Nutrient-Depleted Stationary Phase.
In the natural environment, diatoms commonly experience growth limitation due to depletion of nitrogen or silicon. In these experiments, the transition from nutrient-replete exponential growth to a nutrient-depleted stationary phase was accompanied by changes in the expression of 2,248 genes. The 1,150 genes expressed at higher levels during exponential growth were predominantly associated with photosynthesis, anabolism, and cell division, which are required for growth and increased biomass (Fig. 2C). Genes encoding fucoxanthin chlorophyll binding proteins (n = 30) were the most differentially expressed class of genes between exponential and stationary phases, decreasing in expression an average of 5.8-fold (P = 6 × 10−12, Fig. S3A). This has been previously observed for select fucoxanthin genes (33). The 1,098 genes that were more highly expressed during the stationary phase are related to nutrient assimilation and respiratory and catabolic pathways characteristic of nutrient limitation and starvation (Fig. S3). Nitrate and silicic acid transporters (n = 6) increased in expression an average of 3.5-fold following depletion of both nutrients during the stationary phase (P = 2 × 10−12, Fig. S7), in agreement with prior studies (12). Despite the lack of added ammonium to the media, ammonium transporters (n = 4) were expressed (1.5-fold) during the exponential phase of growth (P = 2 × 10−5), perhaps due to an engrained regulatory mechanism to uptake this preferred nutrient during growth (35–37).
Growth Phase and Nutrient Status Affect Diel Regulation.
In addition to broadly altering physiology and gene expression, the exponential-to-stationary transition also altered diel expression patterns of many genes. A decrease in the magnitude of diel expression changes occurred during the nutrient-depleted stationary phase (Fig. 1A), and the expression patterns of roughly 500 genes were statistically associated with the interactions between the light:dark and exponential:stationary factors: the extent to which the expression of these genes correlated with light status depended on the phase of growth (Fig. S8 and Dataset S1). Most of these genes ceased to fluctuate in dark vs. light expression during the stationary phase (Fig. S8A). This could be explained by the concomitant decreases in photosynthesis, cellular photosynthetic capacity, carbon fixation, and cell division. Self-shading due to increased cell density may also have influenced photosynthesis and gene expression. In contrast, a small number of genes exhibited diel fluctuations only during the stationary phase, including hypothetical proteins and a dawn-associated putative pyruvate carboxylase (11075) that may be indicative of gluconeogenesis or lipid biosynthesis (Fig. S8B). Roughly 100 genes, including genes likely involved in carbohydrate metabolism (11212, 27850, 269248, and 1972) and transcriptional regulation (268788), appeared to switch from dawn to dusk expression (or vice versa) following the transition to stationary phase (Fig. S8C). Overall, genes that were affected by interacting light and growth phase effects included many photoreceptors, transcription factors, and signaling proteins, as well as a putative C-terminal jumonji (JmjC)-like signal-sensing chromatin state regulator (22122). This underlines the importance of decoding the roles of sensing and regulatory genes in circadian regulation.
Coordination of Cellular State Transitions by Sensing, Signaling, and Regulatory Systems.
The ability of diatoms to adjust their physiology in response to the diurnal cycle and growth phase fundamentally depends on sensing, signaling, and regulatory mechanisms (38, 39). The expression levels of 41% of known transcription factors, 35% of all potential signaling domains, and 31% of genes involved in chromatin state dynamics were up-regulated under one or more of the four identified growth states (Fig. 1B). The physiological changes and differential expression of over 4,000 genes that accompanied day:night and exponential:stationary transitions were presumably conducted in part by the modulation of these genes. Whereas the precise flow of information through multiple levels of regulation cannot be elucidated based on these data alone, the conditional coassociation of genes belonging to these classes begins to explain how and which genes are receiving, transmitting, and executing the gene regulatory program over changing conditions.
Light Sensing and Signaling Mechanisms.
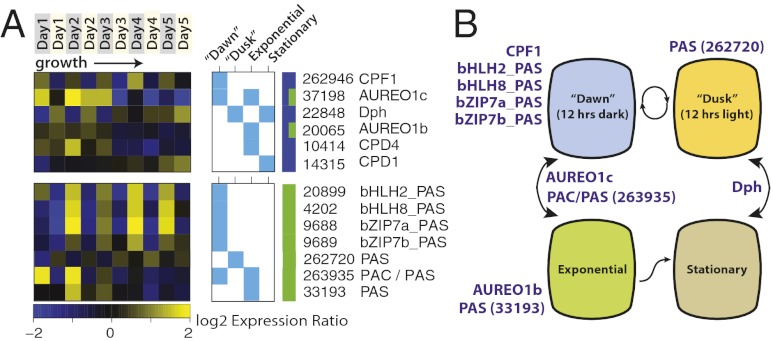
Photoreceptors modulate, at a transcriptional level, a wide array of different light-driven processes. These include photomorphogenesis and the entrainment of the circadian clock in higher plants (40). Diatoms contain multiple photoreceptors to sense and respond to light signals of varying and specific spectral properties (38, 39). In T. pseudonana, one (far) red-light photoreceptor (diatom phytochrome DPh), four blue-light cryptochrome/photolyase family proteins (CPF1–4), and four aureochrome-like blue-light–regulated transcription factors have been identified (38, 39, 41, 42). Aureochrome AUREO1a (33340) was recently shown to control blue-light–induced initiation of diatom cell division by activating the diatom-specific cyclin 2 (dsCYC2) promoter in Phaeodactylum tricornutum (43). In our experiments, CPF1 (262946), AUREO1c (37198), and DPh (22848) were all diurnally expressed, with consistently higher transcript levels for CPF1 at dawn and consistently higher transcript levels for DPh at dusk. In contrast, the CPD photolyases (CPD; DNA repair), CPF2-4, AUREO1a, and AUREO2 (33407) were not detectably associated with the light:dark cycle (Fig. S9A). CPF1 is the only putative photoreceptor for which the diel pattern of expression cycled consistently throughout the experiment, regardless of nutrient status or cell density (Fig. 3A). This is in contrast to AUREO1c and DPh, for which diel changes in expression were higher during the exponential and stationary phases, respectively. CPF1 has also been shown to exhibit an endogenous and entrained circadian rhythm upon transition to constant light conditions in Ostreococcus tauri (44). This suggests that CPF1 may be involved in a diurnally entrained circadian mechanism that is independent of other factors affecting cell status. AUREO1c may putatively exert a regulatory function by binding and activating promoter regions of exponential growth phase-related target genes using its basic region/leucine zipper (bZIP) domain, possibly in conjunction with a secondary bZIP protein, in a manner similar to the AUREO1a activation of dsCYC2 promoter in P. tricornutum (43). The function of the (far-) red light-sensitive DPh receptor in diatoms remains enigmatic, because red light is selectively attenuated with depth in the ocean (45), but in higher plants phytochromes have been shown to play a variety of roles, including the entrainment of the circadian clock, photomorphogenesis, and shade avoidance, among others (40).
Fig. 3.
Genes with putative light-sensing and signaling functions differentially expressed between dark:light and exponential:stationary conditions. (A) The expression patterns of light-sensing and signaling genes during diurnal growth. Genes are in rows and growth time points are in columns. Yellow and blue cells show higher and lower expression ratios during the experiment. Only genes with gene/condition associations by ANOVA at P < 0.01 or higher are shown (aqua boxes). See Fig. S9 for expression of all known photoreceptors. The multicolored bar indicates the following classes of genes: blue, putative photoreceptors and photolyases and green, PAS/PAC domain-containing genes. (B) Model for putative light-sensitive regulation of cell states by photoreceptors. Cellular states (interior labels) are hypothetically driven by the sensory and regulatory activities of differentially expressed photoreceptors (purple text).
Despite the abundance of high-energy light in the ocean (violet, blue, and green), sequences encoding phototropins (blue light) and rhodopsins (green light) are lacking in diatom genomes currently sequenced, with the exception of bacterial-like (proteo) rhodopsins found in open-ocean Pseudonitzschia granii and Fragilariopsis cylindrus (16). We examined the T. pseudonana genome for novel putative light-sensitive regulators by looking for genes containing light-sensing and regulatory domains. Aureochromes, for example, consist of a flavin-binding light-oxygen-voltage (LOV) domain in conjunction with a bZIP domain (41). The discovery of aureochromes in stramenopiles may suggest that these organisms have evolved unique and alternative ways to control (blue) light-regulated processes. To identify additional putative light-responsive regulators, we examined the expression profiles for genes containing signal-sensing Per-ARNT-Sim (PAS) and/or C-terminal PAS (PAC) domains in conjunction with transcription factor domains (Fig. S10). Two transcripts with PAS domains and a basic helix–loop–helix (bHLH) DNA-binding domain (20899 and 4202) and two transcripts with PAS and bZIP domains (9688 and 9689) all showed strong diel oscillations regardless of growth phase or nutrient status (Fig. 3). This is similar to the expression pattern of CPF1, suggesting that these genes may be involved in maintaining light-regulated circadian rhythms. One PAS-domain containing gene (262720) was expressed similarly to DPh, with higher expression levels at dusk, and two PAS genes (263935 and 33193) were expressed specifically during exponential growth. These genes, especially the bZIP and bHLH-containing sequences, are thus implicated as light-sensitive stramenopile-specific photoreceptors that could regulate gene expression levels by directly binding to the diatom DNA.
Membrane-Associated Signal Transduction.
Additional signaling and regulation likely occurs in diatoms through membrane-associated second-messenger and two-component signaling (TCS) systems. Two-component signaling, which is commonly used by bacteria and other organisms to sense environmental cues such as pH, temperature, and nutrient status, involves a membrane-bound histidine kinase and a cytosolic response regulator (RR). The T. pseudonana genome encodes 194 putative protein kinases, of which 8 contain putative transmembrane regions (TMs), as well as 4 histidine kinases without predicted TMs (38). Other genes that are potentially involved in environmental perception and signaling include those bearing similarity to membrane-bound G-protein coupled receptors (GPCRs), leucine-rich repeat (LRR)-containing genes, and recently described nucleotide-binding NB-ARC domains (46). Genes from each of these classes varied significantly in expression between dark:light and exponential:stationary conditions (Fig. S9E). Additionally, one of the most highly expressed genes at dawn (268270) encodes a PATCHED domain, which is found in the transmembrane receptors of hedgehog signaling pathways in vertebrates and insects.
Chromatin State Regulation Pervades Normal Growth.
DNA methylases and histone modifiers epigenetically control chromatin state and gene silencing (47). T. pseudonana possesses a large set of genes encoding chromatin regulatory functions and may use extensive chromatin state regulation (48). Diel cycling of expression for these genes suggests that chromatin state regulation and gene silencing is important during the diurnal cycle (Fig. S11). Of four genes resembling a C5 cytosine-specific DNA methylase (C5-DMase), one (21517) was more highly expressed at dawn, along with lysine acetyltransferases (22580 and 9040), a methyltransferase (264323), and a jumonji-like protein (22122) that may be a signal-dependent histone modifier. The expression of histones H2A and H2B was consistently higher at dusk, related perhaps to DNA replication and cell division during the light period. Various lysine deacetylases (16405, 16384, and 269060) and a demethylase (1836) were coexpressed with these histones and likely regulate the formation of nucleosomes. During the nutrient-depleted stationary phase, two lysine demethylases (1863 and 22122), two lysine deacetylases (16405 and 15819), and one acetyltransferase (37928) were up-regulated. These genes may be involved in the shift to decreased cell division, nutrient scavenging, and quiescence. The cycling expression of these genes suggests that chromatin state regulation is pervasive in diatoms even under normal conditions.
Forty Percent of Transcription Factors Are Differentially Expressed During Typical Growth Transitions.
Expression changes in transcription factors (TFs) are known to influence downstream gene regulation. Increases or decreases in the expression of TFs under specific conditions implicate their function in the acclimation or response of the organism to that condition. The T. pseudonana genome contains an estimated 250 genes encoding transcription factors, with the largest number of genes bearing similarity to heat-shock factor (HSF) and “myeloblastosis” family (Myb) domains (94 and 37, respectively) (49). One hundred and two (102) TFs were associated with one or more growth states (Fig. 1B and Fig. S10A). Diel fluctuations were more prevalent for transcription factors than the shift from exponential to stationary phase (71 vs. 42 TFs), and the majority of associations involved diel fluctuations with higher expression at dawn (54/102 TFs). Twenty-three HSFs were significantly enriched among dawn-associated TFs (P = 0.008, hypergeometric), suggesting that heat shock factor activity may be important for diatoms to cope with nighttime conditions and/or the transition to light. Genes bearing similarity to σ70 (49) were also enriched in dawn-associated TFs (P = 0.006) and for those associated with the exponential phase (P = 0.016). In contrast, helix-loop-helix (HLH), CCAAT-binding factor (CBF), and cysteine-rich (CXC-type) family TFs were enriched among dusk-associated TFs, implicating the functions of these factors in daytime-associated functions and the transition to darkness. Four putative transcription factors (269238, 6958, 25911, and 9375), three of which are putative heat-shock factors, were among the top twenty most differentially expressed known genes during the stationary phase (Fig. S3A). Thus these may be important for the response and acclimation to nutrient limitation. Expression levels for many TFs differed significantly over both day:night and exponential:stationary transitions (Fig. S10B). The recurrent differential expression of almost half the transcription factor repertoire in T. pseudonana indicates a high degree of complexity in transcriptional regulation during typical growth conditions.
Core Transcriptomic States and Regulatory Relationships Generalize over Multiple Independent Experiments.
The large, recurrent changes in the transcriptome during our growth experiments establish the core functional and regulatory modules associated with recurring changes in growth conditions (Fig. 2). To determine the extent to which these expression modules reflect fundamental properties of the T. pseudonana gene regulatory network, we examined the coexpression of genes across an integrated dataset that included published microarray data from several additional T. pseudonana growth conditions [high pH, iron, nitrogen and silicon depletion, low temperature (12), iron stress (15), benzopyrene exposure (50), and recovery from silicon limitation (51)]. The genes identified in our experiment were highly correlated (P < 0.01) across 40 microarray conditions, reconstituting the four principal expression states that we observed (Fig. S12A). Data from these orthogonal experiments begin to subordinate large groups of coexpressed genes into submodules that are theoretically enriched in functionally related and coregulated genes (Fig. S12A and Dataset S2).
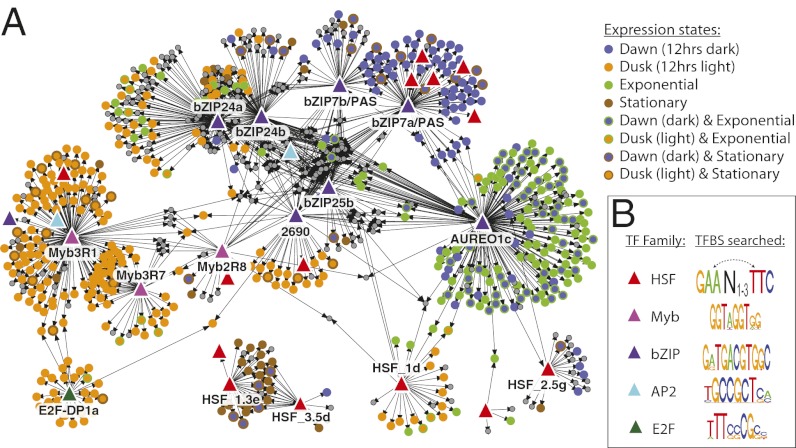
Many genes involved in sensing, signaling, gene regulation, and chromatin dynamics were exclusively correlated with particular functional clusters (Fig. S12B). To assess the potential mechanistic relationships by which transcription factors in T. pseudonana regulate the transcription of other genes, we combined coexpression data obtained from all available microarrays with the occurrence of potential transcription factor-DNA binding sites (TFBSs) in the upstream promoter regions of all genes. We identified the putative diatom TFBS using conserved profiles from orthologous TF families (HSF, Myb, bZIP, AP2, and E2F) found in higher plants (P < 1 × 10−4). Assuming that diatom TFs can specifically bind these plant-derived motifs and that binding may alter target gene transcription, we generated a set of potential transcriptional activation pathways between several TFs and many significantly coexpressed genes (P < 0.05; Fig. 4). This partial gene regulatory network model (Dataset S3) provides a number of new hypotheses about the potential direct regulators of tightly controlled physiological and metabolic pathways. For example, the majority of putative target genes clustering with the dawn/exponentially expressed photoreceptor AUREO1c (Fig. 3A) are involved in photosynthesis and sugar metabolism (Dataset S4). Key regulatory roles are also implied for the putative light-responsive transcription factor bZIP7a/PAS, which potentially regulates several other transcriptional regulators, and bZIP24b, which potentially regulates nucleosome and chromatin organization. In combination with photosensory patterns observed over the day/night cycle (Fig. 3), these inferences identify the most likely regulators of diel transitions in diatoms. These findings can be used to design new experiments to dissect this process, as well as to develop comprehensive models of physiological regulation in diatoms.
Fig. 4.
Inferred gene regulatory relationships. Shown are potential transcriptional activation pathways that were consistent with all published T. pseudonana microarray data and known conserved transcription factor-DNA binding sequence motifs for five major TF families. (A) Transcription factors (TFs, colored triangles) are proposed as candidate regulators of target genes (circles) if: (i) the TF and its target genes were highly correlated in expression changes over multiple array experiments (bootstrap P < 0.025); (ii) the upstream region of each target gene contains at least one potential DNA binding site that matches a conserved DNA-binding motif for the TF family (motif P < 1 × 10−4); and (iii) the target genes are enriched in putative TF-DNA binding sites, compared with all genes (hypergeometric P < 0.05). Gray circles represent target genes that were not statistically associated with diel states. (B) Putative DNA-binding motifs for conserved TF families in higher plants. Dashed arrows denote possible inversions in the heat-shock element for this family of transcription factors. Functional enrichment analysis of TF-gene clusters is provided in Dataset S4.
Conclusion
We identified the core physiological and transcriptomic transitions associated with diatom growth under conditions that shift between dark and light periods and from nutrient-replete to -depleted conditions. Thousands of genes of both known and unknown function fluctuated in expression during these experiments and were associated with markedly different conditions and cell states. T. pseudonana appears to be broadly entrained to anticipate the diurnal cycle at the level of gene expression, as previously suggested (29, 30). The shift from exponential growth to nutrient-depleted stationary phase attenuated many diel patterns of expression. A highly robust, flexible, and multifactorial gene regulatory program was apparent that involves large portions of the available regulatory repertoire under typical growth conditions. This sensing, signaling, and regulatory system, which includes several previously uncharacterized putative photoreceptors and signaling proteins, presumably detects environmental conditions and conducts large changes in cell physiology accordingly. The association of nearly half of T. pseudonana genes with environment-dependent regulatory modes partly explains how the diatom gene regulatory program operates under typically varying conditions. Together with studies conducted in related diatoms (52), this represents a broad model of diatom gene regulation against which to compare unique diatom responses to specific conditions and environments, which will be valuable for new hypothesis generation and the interpretation of genetic, environmental, and metatranscriptomic field studies.
Materials and Methods
Growth and Sampling.
Axenic cultures of T. pseudonana were grown in enriched artificial seawater (ESAW) medium modified with reduced levels of nitrate (80 µM), silicic acid (110 µM), and phosphate (20 µM). Before the experiment, the diatoms were acclimated to constant 20 °C temperature and ambient CO2 under a 12:12 h dark:light diurnal cycle at 125 µmol photons⋅m−2⋅s−1. The light quality and quantity remained constant throughout the light period. The cultures were continuously equilibrated at ambient (400 ppm) or elevated (800 ppm) CO2 by bubbling of mixed gasses (2 L/min) and stirring (50 rpm) in a 10-L glass bioreactor system (BioFlow). Eight-liter bioreactors were inoculated with 5 × 104 cells/mL of acclimated, axenic T. pseudonana and grown for 1 wk on a 12:12 h dark:light cycle. pH was measured continuously and calibrated by spectrophotometric measurements every 2 h. CFC was measured using the selective inhibitor of photosystem II, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) (53). Nutrients (NO3, NH4, PO3, and Si) were sampled twice daily.
RNA Extraction, Labeling, and Hybridization.
Cells (∼3 × 107) were harvested at the end of the dark periods (dawn) and at the end of the light periods (dusk) by filtration (30–200 mL) and then flash frozen. Total RNA was extracted using the mirVANA kit from Invitrogen (total RNA extraction protocol). Transcribed mRNA was amplified and labeled using the Agilent Quick-Amp Labeling kit (two color). Agilent oligonucleotide array slides were hybridized with equal amounts of oppositely labeled sample and internal reference RNA and scanned using an Agilent two-color array scanner. Gene-specific arrays were obtained from Agilent Technologies. At least three probes were designed to match the 3′ end of each of the 11,390 Thaps3.0 nuclear gene models (10), as well as 180 chloroplast genes (54).
Microarray Data Processing and Gene Expression Analysis.
Two-color expression data were normalized using the limma package in R (55). Expression ratios were calculated relative to the mean within-experiment expression level for each probe to remove batch effects between experiments. A two-factor ANOVA (exponential:stationary, dark:light) was performed on the data using the MeV program (56), with significance assigned at an alpha level of 0.01 based on 1,000 permutations. Exponential samples were those collected before the light period of day 3 (∼80 h); stationary samples were those collected afterward. This grouping of samples coincided with the decline in specific growth rate of the freely growing cultures to a value below 1/d (Fig. S2). Dawn samples were collected in the dark after 12 h of constant darkness. Dusk samples were collected in the light after 12 h of constant light. Gene functional enrichment within these groups was analyzed using a binomial test with Benjamini–Hochberg correction in the Gitools program (57). All microarray data are available under GEO accession GSE45252.
Transcriptional Regulatory Network Inference.
A model for potential gene regulation by transcription factors on target genes was inferred for each transcription factor in five major families (49). Transcription factors were proposed as candidate regulators for target genes if all of the following were true: (i) the TF and its target genes were highly correlated in expression changes over multiple published array experiments (bootstrap P < 0.025), (ii) the upstream region of each target gene (−400 to +50 bp) contains at least one potential DNA binding site that matches a conserved DNA-binding motif for the TF family (motif P < 1 × 10−4), and (iii) the upstream regions of highly correlated target genes were enriched in putative TF-DNA binding sites, compared with all genes (hypergeometric P < 0.05). Putative DNA-binding motifs for conserved TF families were assumed from DNA sequence specificity profiles that have been experimentally determined for orthologous transcription factor families in plants (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank G. Miller Hennon and the University of Washington for assistance with chemical assays, as well as P. Troisch and B. Marzolf for microarray assistance. This work was funded by the National Science Foundation OCE-0928561 (to M.V.O. and N.S.B.) and OCE-0927238 (to E.V.A.). J.A. is a Gordon and Betty Moore Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE45252).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300962110/-/DCSupplemental.
References
- 1.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305(5682):354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 2.Armbrust EV. The life of diatoms in the world’s oceans. Nature. 2009;459(7244):185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- 3.Brzezinski MA, et al. A switch from Si(OH)4 to NO3- depletion in the glacial Southern Ocean. Geophys Res Lett. 2002;29(12):5. [Google Scholar]
- 4.Martin P, et al. Export and mesopelagic particle flux during a North Atlantic spring diatom bloom. Deep Sea Res Part I Oceanogr Res Pap. 2011;58:338–349. [Google Scholar]
- 5.Alkire MB, et al. Estimates of net community production and export using high-resolution, Lagrangian measurements of O2, NO3−, and POC through the evolution of a spring diatom bloom in the North Atlantic. Deep Sea Res Part I Oceanogr Res Pap. 2012;64:157–174. [Google Scholar]
- 6.Falkowski P, Raven J. Aquatic Photosynthesis. Princeton, NJ: Princeton Univ Press; 2007. [Google Scholar]
- 7.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 8.Nelson D, Treguer P, Brzezinski M, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles. 1995;9:359–372. [Google Scholar]
- 9.Raven J, Falkowski P. Oceanic sinks for atmospheric CO2. Plant Cell Environ. 1999;22:741–755. [Google Scholar]
- 10.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306(5693):79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 11.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456(7219):239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 12.Mock T, et al. Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc Natl Acad Sci USA. 2008;105(5):1579–1584. doi: 10.1073/pnas.0707946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen AE, et al. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc Natl Acad Sci USA. 2008;105(30):10438–10443. doi: 10.1073/pnas.0711370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maheswari U, et al. Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol. 2010;11(8):R85. doi: 10.1186/gb-2010-11-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thamatrakoln K, Korenovska O, Niheu AK, Bidle KD. Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ Microbiol. 2012;14(1):67–81. doi: 10.1111/j.1462-2920.2011.02468.x. [DOI] [PubMed] [Google Scholar]
- 16.Marchetti A, et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA. 2012;109(6):E317–E325. doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldeira K, Wickett ME. Oceanography: Anthropogenic carbon and ocean pH. Nature. 2003;425(6956):365. doi: 10.1038/425365a. [DOI] [PubMed] [Google Scholar]
- 18.Feely RA, et al. Impact of anthropogenic CO2 on the CaCO3 system in the oceans. Science. 2004;305(5682):362–366. doi: 10.1126/science.1097329. [DOI] [PubMed] [Google Scholar]
- 19.Orr JC, et al. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature. 2005;437(7059):681–686. doi: 10.1038/nature04095. [DOI] [PubMed] [Google Scholar]
- 20.Iglesias-Rodriguez MD, et al. Phytoplankton calcification in a high-CO2 world. Science. 2008;320(5874):336–340. doi: 10.1126/science.1154122. [DOI] [PubMed] [Google Scholar]
- 21.Parker M, Armbrust E. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2005;41:1142–1153. [Google Scholar]
- 22.Bender SJ, Parker MS, Armbrust EV. Coupled effects of light and nitrogen source on the urea cycle and nitrogen metabolism over a diel cycle in the marine diatom Thalassiosira pseudonana. Protist. 2012;163(2):232–251. doi: 10.1016/j.protis.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Prézelin BB. Diel periodicity in phytoplankton productivity. Hydrobiologia. 1992;238:1–35. [Google Scholar]
- 24.Dodd AN, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318(5857):1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 25.Monnier A, et al. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics. 2010;11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Jezequel V, Hildebrand M, Brzezinski MA. Silicon metabolism in diatoms: Implications for growth. J Phycol. 2000;36:821–840. [Google Scholar]
- 27.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat Rev Microbiol. 2004;2(8):643–655. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 28.Bidle KD, Bender SJ. Iron starvation and culture age activate metacaspases and programmed cell death in the marine diatom Thalassiosira pseudonana. Eukaryot Cell. 2008;7(2):223–236. doi: 10.1128/EC.00296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doran E, Cattolico RA. Photoregulation of chloroplast gene transcription in the chromophytic alga Heterosigma carterae. Plant Physiol. 1997;115(2):773–781. doi: 10.1104/pp.115.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granum E, Roberts K, Raven JA, Leegood RC. Primary carbon and nitrogen metabolic gene expression in the diatom Thalassiosira pseudonana (Bacillariophyceae): Diel periodicity and effects of inorganic carbon and nitrogen. J Phycol. 2009;45:1083–1092. doi: 10.1111/j.1529-8817.2009.00728.x. [DOI] [PubMed] [Google Scholar]
- 31.Wyman M. Diel rhythms in ribulose-1,5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.) Appl Environ Microbiol. 1999;65(8):3651–3659. doi: 10.1128/aem.65.8.3651-3659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul JH, Kang JB, Tabita FR. Diel patterns of regulation of rbcL transcription in a cyanobacterium and a prymnesiophyte. Mar Biotechnol (NY) 2000;2(5):429–436. doi: 10.1007/s101260000016. [DOI] [PubMed] [Google Scholar]
- 33.Leblanc C, Falciatore A, Watanabe M, Bowler C. Semi-quantitative RT-PCR analysis of photoregulated gene expression in marine diatoms. Plant Mol Biol. 1999;40(6):1031–1044. doi: 10.1023/a:1006256300969. [DOI] [PubMed] [Google Scholar]
- 34.Siaut M, et al. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406(1-2):23–35. doi: 10.1016/j.gene.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Goldman J, McCarthy J. Steady-state growth and ammonium uptake of a fast-growing marine diatom. Limnol Oceanogr. 1978;23:695–703. [Google Scholar]
- 36.Lomas MW, Glibert PM. Interactions between NH4+ and NO3- uptake and assimilation: Comparison of diatoms and dinoflagellates at several growth temperatures. Mar Biol. 1999;133:541–551. [Google Scholar]
- 37.Hildebrand M. Cloning and functional characterization of ammonium transporters from the marine diatom Cylindrotheca fusiformis (Bacillariophyceae) J Phycol. 2005;41:105–113. [Google Scholar]
- 38.Montsant A, et al. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana. J Phycol. 2007;43:585–604. [Google Scholar]
- 39.Depauw FA, Rogato A, Ribera d’Alcalá M, Falciatore A. Exploring the molecular basis of responses to light in marine diatoms. J Exp Bot. 2012;63(4):1575–1591. doi: 10.1093/jxb/ers005. [DOI] [PubMed] [Google Scholar]
- 40.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi F, et al. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci USA. 2007;104(49):19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coesel S, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10(6):655–661. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huysman MJJ, et al. 2013. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25(1):215–228.
- 44.Heijde M, et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 2010;33(10):1614–1626. doi: 10.1111/j.1365-3040.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 45.Levine JS, MacNichol EF. Color vision in fishes. Sci Am. 1982;246:140–149. [Google Scholar]
- 46.Zambounis A, Elias M, Sterck L, Maumus F, Gachon CMM. Highly dynamic exon shuffling in candidate pathogen receptors ... what if brown algae were capable of adaptive immunity? Mol Biol Evol. 2012;29(4):1263–1276. doi: 10.1093/molbev/msr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 48.Maumus F, Rabinowicz P, Bowler C, Rivarola M. Stemming epigenetics in marine stramenopiles. Curr Genomics. 2011;12(5):357–370. doi: 10.2174/138920211796429727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rayko E, Maumus F, Maheswari U, Jabbari K, Bowler C. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 2010;188(1):52–66. doi: 10.1111/j.1469-8137.2010.03371.x. [DOI] [PubMed] [Google Scholar]
- 50.Carvalho RN, Bopp SK, Lettieri T. Transcriptomics responses in marine diatom Thalassiosira pseudonana exposed to the polycyclic aromatic hydrocarbon benzo[a]pyrene. PLoS ONE. 2011;6(11):e26985. doi: 10.1371/journal.pone.0026985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shrestha RP, et al. Whole transcriptome analysis of the silicon response of the diatom Thalassiosira pseudonana. BMC Genomics. 2012;13:499. doi: 10.1186/1471-2164-13-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM. 2013. Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol 161(2):1034–1048.
- 53.Vincent WF. Mechanisms of rapid photosynthetic adaptation in natural phytoplankton communities. II. Changes in photochemical capacity as measured by DCMU-induced chlorophyll fluorescence. J Phycol. 1980;16:568–577. [Google Scholar]
- 54.Oudot-Le Secq M-P, et al. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol Genet Genomics. 2007;277(4):427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- 55.Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23(20):2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 56.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 57.Perez-Llamas C, Lopez-Bigas N. Gitools: Analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 2011;6(5):e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.