Abstract
The nucleoprotein (NP) of segmented negative-strand RNA viruses such as Orthomyxo-, Arena-, and Bunyaviruses coats the genomic viral RNA and together with the polymerase forms ribonucleoprotein particles (RNPs), which are both the template for replication and transcription and are packaged into new virions. Here we describe the crystal structure of La Crosse Orthobunyavirus NP both RNA free and a tetrameric form with single-stranded RNA bound. La Crosse Orthobunyavirus NP is a largely helical protein with a fold distinct from other bunyavirus genera NPs. It binds 11 RNA nucleotides in the positively charged groove between its two lobes, and hinged N- and C-terminal arms mediate oligomerization, allowing variable protein–protein interface geometry. Oligomerization and RNA binding are mediated by residues conserved in the Orthobunyavirus genus. In the twofold symmetric tetramer, 44 nucleotides bind in a closed ring with sharp bends at the NP–NP interfaces. The RNA is largely inaccessible within a continuous internal groove. Electron microscopy of RNPs released from virions shows them capable of forming a hierarchy of more or less compact irregular helical structures. We discuss how the planar, tetrameric NP–RNA structure might relate to a polar filament that upon supercoiling could be packaged into virions. This work gives insight into the RNA encapsidation and protection function of bunyavirus NP, but also highlights the need for dynamic rearrangements of the RNP to give the polymerase access to the template RNA.
Keywords: protein–RNA interactions, structural biology
The largest family of segmented negative-strand RNA viruses (sNSVs), Bunyaviridae, comprises more than 350 species belonging to five genera: Orthobunyavirus, Phlebovirus, Nairovirus, Hantavirus, and Tospovirus. Bunyaviruses generally infect mammals, except Tospoviruses that infect plants, and use arthropods as vectors for their spread, although Hantaviruses are rodent borne (1, 2). Several species can cause severe zoonotic diseases in humans such as La Crosse Orthobunyavirus (LACV, childhood encephalitis), Rift Valley Fever Phlebovirus (RVFV), and Crimean Congo Haemorrhagic Fever Nairovirus (CCHFV). As vector-borne diseases, their geographical occurrence is closely linked to the environment, and climate change or other ecological factors may lead to altered distribution or emergence of new viruses (3–5).
Bunyaviruses have a trisegmented negative-strand RNA genome. The L segment encodes for the RNA-dependent RNA polymerase or L protein, the M segment for glycoproteins and a nonstructural protein, and the S segment encodes the nucleoprotein (NP) and another nonstructural protein. The genome is always coated by multiple copies of NP forming ribonucleoprotein particles (RNPs), which also contain the viral polymerase. RNPs are the only form under which the genome is efficiently replicated and transcribed, and NP and L are necessary and sufficient to perform these functions. Like the polymerase, which catalyses RNA replication and transcription by cap-snatching (6), NP is multifunctional. In coating exclusively genomic or antigenomic viral RNA (but not viral mRNA or cellular RNA), it protects the viral genome from degradation, avoids formation of dsRNA between viral RNAs of opposite polarity, compacts the RNA into RNPs, and by interacting with the glycoprotein tails inside the virion membrane, actively promotes packaging into progeny virions (7, 8). Dynamic interactions of NP with the polymerase are also critical, allowing the polymerase controlled access to the RNA within template RNPs on the one hand and nucleating assembly of progeny RNPs only when the polymerase is functioning as a replicase.
NPs with essentially the same functions are found in all NSVs, yet recent progress in their structural characterization has shown them to be remarkably diverse in structure. Whereas those of nonsegmented NSVs (nsNSVs) are structurally homologous, each genus of sNSVs possesses an NP with a distinct fold (reviewed in ref. 9). Nevertheless, all characterized NPs are mainly helical proteins with a basic groove that binds single-stranded RNA (ssRNA) sequence independently. Protein–protein oligomerization occurs through interaction of N- (e.g., RVFV) and/or C-terminal extensions (e.g., rabies), internal protruding loops (e.g., influenza), or subdomains (e.g., CCHFV) that link one NP to the next (10–14). Diversity of NP is particularly apparent within the Bunyaviridae family, with each of the five genera having nonhomologous NP sequences. The only structural similarity noted so far is that CCHFV NP possesses a globular domain structurally similar to the RNA-binding domain of Lassa Arenavirus NP (13–15). It is thought that NPs must also exist in an auto-inhibited form to avoid RNA binding or oligomerization in the wrong biological context. This can occur by intra-, rather than intermolecular, binding of the flexible arms [e.g., RVFV (16), influenza (17)], rotation of an entire subdomain [e.g., CCHFV stalk domain (13, 14)], or chaperoning by another protein [e.g., the phosphoprotein in nsNSVs (18)].
To give further insight into the multifunctionality of sNSV NPs, we undertook a structural analysis of LACV NP, representative of the Orthobunyavirus genus for which no structure has yet been published. The crystal structure of LACV NP in the absence of RNA reveals a unique protein fold and flexibly linked N- and C-terminals arms that mediate oligomerization. The structure of a tetrameric complex of NP with ssRNA shows how each protomer sequesters 11 nucleotides (nts) of RNA. We compare these results with the structure of the NP–RNA complex from RVFV (19) and with an extensive mutagenesis analysis performed on the homologous Bunyamwera virus (BUNV) NP (20). Finally, based on electron microscopy of native LACV RNPs released from virions, we discuss how the LACV NP–RNA tetramer might relate to higher order structures observed in RNPs.
Results
Purification and Characterization of Recombinant LACV NP.
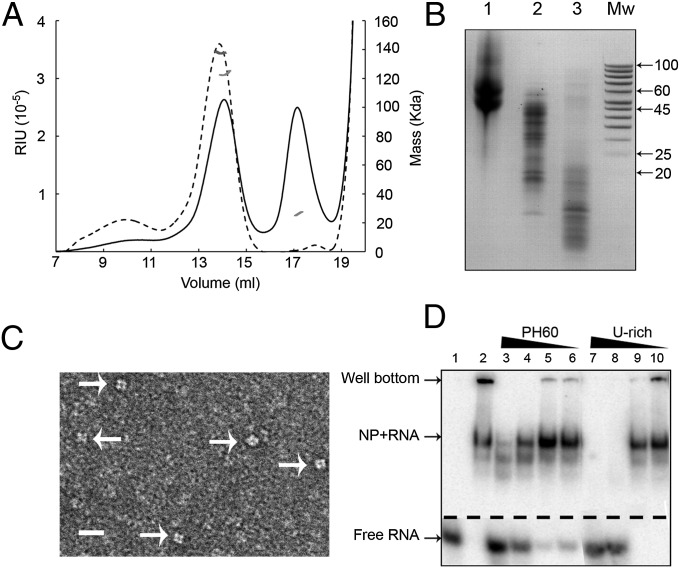
Full-length LACV NP was expressed in Escherichia coli and purified as a tetrameric complex associated with bacterial RNA in the range of 45–60 nts (Fig. 1 A and B), as visualized by electron microscopy (Fig. 1C). RNAs up to about 50 nts were protected from degradation by RNase A treatment (Fig. 1B). Similar results were previously found for the homologous BUNV NP (21, 22). Crystals of apo-NP (see below) could be grown from RNase-treated tetramers, but only in the presence of 0.2 M thiocyanate. Indeed adding thiocyanate to the lysis buffer destabilizes the tetramer, giving a mixture of both tetrameric and monomeric NP (Fig. 1A) and rendering longer RNAs more susceptible to degradation (Fig. 1B). Monomeric NP was further purified by heparin affinity chromatography, resulting in the complete removal of RNA. This allowed NP–RNA complexes with defined RNAs to be reconstituted (see below). Using competition experiments and two different RNAs (PH60 based on the LACV genomic panhandle and U-rich) (Materials and Methods), we confirmed that RNA binding is sequence independent, as expected (Fig. 1D).
Fig. 1.
Recombinant LACV NP characterization. (A) Gel filtration of NP tetramers purified from E. coli before and after treatment with RNase A and 0.4 M thiocyanate (discontinuous and continuous lines, respectively). Multiangle laser light scattering measurements for each peak are shown in dark and light gray lines, respectively. The mass observed before treatment is 131 KDa, corresponding to a tetramer with bound RNA. After treatment, there are two species of 126 KDa (tetramer with shorter RNA) and 26 KDa (monomer). (B) RNA content of tetramers purified from E. coli after various treatments. Lane 1, RNA isolated from tetramers directly purified from E. coli; lane 2, after treatment with RNase A; lane 3, after treatment with RNase A and 0.4 M thiocyanate. E. coli-derived RNAs of length 45–60 are preferentially bound to the tetramer. RNAs up to about 45 nts are protected from degradation, although there is some accessibility of RNase probably at the NP–NP interfaces. Thiocyanate destabilizes the tetramer, making more accessible the RNA that is not tightly bound to a single monomeric NP. (C) EM image of untreated tetramers negatively stained with 1% sodium silico-tungstate. The black line indicates 20 nm. Arrows point toward individual tetramers. (D) Electrophoresis mobility shift assay using native acrylamide gels of 32P–labeled PH60 viral RNA with cold competitor RNA incubated for 1 h at room temperature with monomeric NP. Lane 1, labeled PH60 only; lane 2, labeled PH60 only incubated with monomeric N; lanes 3–6, labeled PH60 with 12 µM and successive threefold dilutions of cold PH60; lanes 7–10, the same with cold U-rich RNA.
Structure of RNA-Free LACV NP.
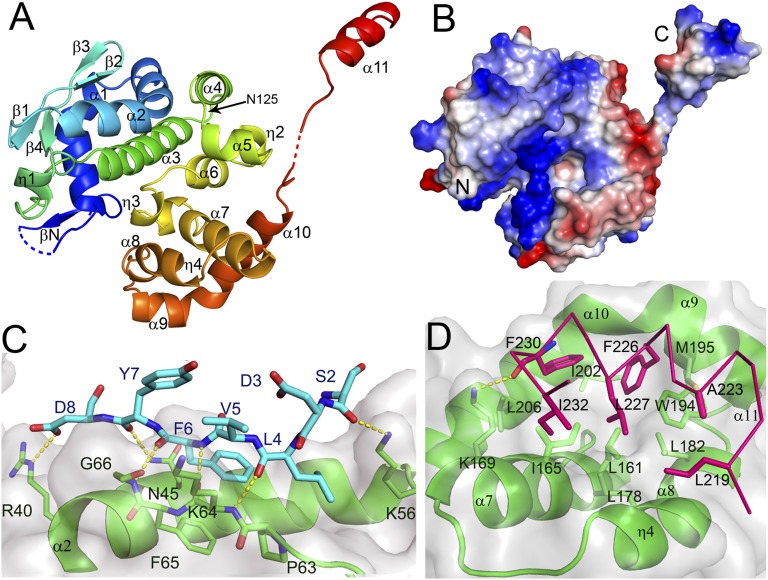
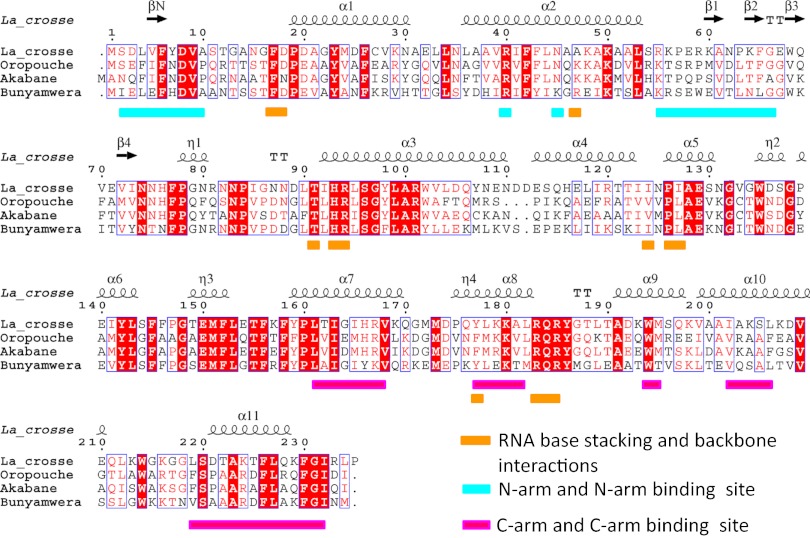
Crystals of RNA-free LACV NP grown from tetramers in the presence of thiocyanate diffracted to 1.8 Å resolution. The structure was solved by the single anomalous dispersion method using selenomethionine-labeled protein, resulting in a model with Rwork/Rfree of 0.182/0.206 (Table S1). LACV NP has 11 helical segments and one prominent beta-hairpin (Figs. 2A and 3). No significant overall structural homology with any other known NP was found using the PDBeFold server. There are two subdomains (residues 17–124 and 125–214, respectively), which pack together to form a single globular structure with a deep positively charged cleft between them (Fig. 2 A and B). N- and C-arms are linked to each extremity of the globular core by flexible, glycine-containing peptides of, respectively, eight (amino acids 9–16) and three residues (215–217), the hinges having poor or missing electron density. The N- and C-arms make intimate contacts with the core domain of neighboring molecules, forming a 3D network throughout the crystal (Fig. S1). The N-arm forms a short parallel β-strand (βN) with the external strand (β2, residues 64–66) of the β-hairpin of a neighboring molecule (Fig. S1). The interaction is reinforced by additional hydrophobic and polar contacts (Fig. 2C). The C-arm forms an amphipathic α-helix (α11) whose hydrophobic face meshes into a hydrophobic pocket on the core of a neighboring NP formed by residues from helices α7–10 (Fig. 2D). The strong complementarity of both N- and C-arm contacts to the body, and the high conservation in the Orthobunyavirus genus of the residues involved (Fig. 3), suggests that these interactions are responsible for multimerisation of NP in RNPs, as corroborated by the structure of the NP–RNA complex described below.
Fig. 2.
Structure of the RNA-free LACV NP. (A) Ribbon representation of LACV NP with rainbow coloring from N- (blue) to C-terminus (red). Disordered hinge regions are dotted. Indicated secondary structure elements are consistent with the sequence alignment in Fig. 3. (B) Electrostatic surface of the LACV NP, the positive and negative charges, respectively, being in blue and red. (C) The N-arm oligomerization interface. The N-arm (blue sticks) forms a β-strand (βN) parallel with β2 on the body of the next protein (green with transparent surface). The three intermolecular main-chain hydrogen bonds are shown as yellow dotted lines. Asp8 and Arg40 form a salt-bridge. Residues Leu4, Phe6, Pro63, and Phe65 form a hydrophobic cluster. Asn45 side-chain makes two hydrogen bonds to the main-chain of Tyr7. (D) The C-arm oligomerization interface. Hydrophobic residues on one side of the C-arm amphipathic helix α11, notably Leu219, Ala223, Phe226, Leu227, Phe230, and Ile232 (red sticks), bind to a hydrophobic pocket on the body of the next protein (green ribbons and sticks with transparent surface) formed by residues from helices α7–α10 (notably Leu161, Ile165, Leu178, Leu182, Trp194, Met195, Ile202, and Leu206).
Fig. 3.
Sequence alignment of representative Orthobunyavirus NPs. The secondary structure of LACV NP is shown above and key functional residues colored as indicated.
Structure of the NP–RNA Tetramer.
NP–RNA complexes were reconstituted from monomeric NP and ssRNA of 7, 9, 11, 41, and 45 nts length and extensive crystallization trials made. The best crystals were of a complex reconstituted with a 45 nts U-rich RNA and diffracted to 3.4 Å resolution. The structure was solved by molecular replacement, and a model comprising four NPs and 44 nts RNA in the asymmetric unit was refined to Rwork/Rfree of 0.205/0.260 (Table S1). Several different crystal forms were obtained with an 11 nts RNA but diffracted less well (Table S2).
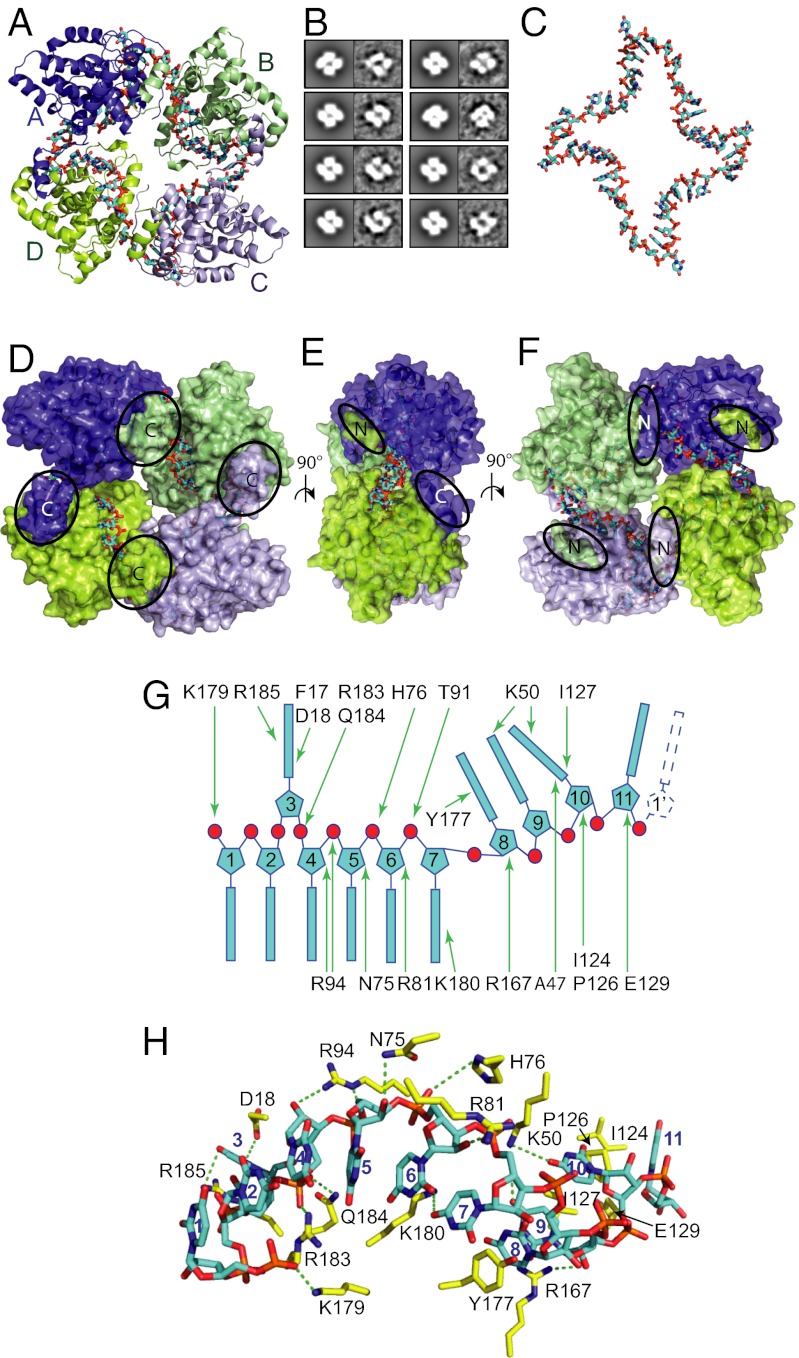
The structure shows a tetrameric NP–RNA complex that is a dimer of dimers with only a twofold rather than a fourfold symmetry (Fig. 4A). The 2D projections of this structure are consistent with the EM class averages of tetrameric NP bound to bacterial RNA (Figs. 1C and 4B). The intermolecular interactions that each N- and C-arm makes with the core of its neighbors in the tetramer are the same as observed in the apo-NP crystals, although the hinges bend the arms in different directions (discussed below), forming a closed tetramer rather than an extended filament. Inside the tetramer, the positively charged groove forms a continuous path into which 44 nts RNA was modeled as poly(U) (denoted 5′ U1–U44 3′), the bases, although not their identity, being clearly resolved in sharpened maps (Fig. S2A). The RNA thus forms a closed ring, which is strongly bent at each NP–NP interface (Fig. 4C). Each NP binds 11 nts, close to the 12 nts estimated for BUNV from gel analysis of RNase A digestion products (22). The RNA is largely inaccessible in the tetramer (Fig. 4 D–F), with about 1,400 Å2 of RNA surface buried per subunit [calculated using the protein interfaces, surfaces, and assemblies (PISA) server, www.ebi.ac.uk/msd-srv/prot_int/pistart.html], consistent with its protection from RNase treatment (Fig. 1B).
Fig. 4.
RNA binding mode in tetrameric LACV NP. (A) Cartoon representation of LACV NP tetramer in complex with 44 nts RNA shown as cyan and orange sticks. A pseudo-twofold symmetry axis into the figure makes monomers A (dark blue) and C (light blue) equivalent as well as B (green) and D (lemon). (B) Correspondence between 2D projections of LACV NP–RNA tetramer crystal structure (Left, filtered to 20 Å) and EM class averages of LACV tetramers purified from E. coli and bound to bacterial RNA (Right). Class averages corresponding to top and tilted views (up to 30°) are displayed; side views are not observed. (C) RNA conformation within LACV NP–RNA tetramer showing S-shaped backbone conformation bound to each monomer and sharp kinks at the NP–NP interfaces. The top and bottom bends at the A–B and C–D interfaces are less sharp than the right and left bends at the D–A and B–C interfaces (Fig. S3A). Whereas the RNA backbone continuity across the NP–NP interfaces is clear between U11–U12, U22–U23, and U33–U34, the electron density for U43–U44 and the connectivity to U1 is less good, consistent with the fact that the RNA actually has 45 nts, one more than necessary to occupy all four NPs. (D–F) Orthogonal views of a surface representation of the NP–RNA tetrameric complex showing that the RNA is largely buried within interior grooves with only partial solvent access to mainly the backbone. The N- and C-arms are highlighted. (G) Schematic of protein–RNA interactions, highlighting the stacking arrangement of the bases. (H) Stick representation of NP–RNA interactions with the 11 nts RNA shown in cyan and interacting protein residues in yellow. Putative hydrogen bonds are shown as green dotted lines.
The 11 nts segment of RNA bound to each NP has the same conformation (Fig. 4 G and H), except for the 5′ nucleotide, which differs for the two nonequivalent conformations, according to the variable orientation of the N-arm (see below and Fig. S3A). Bases U11′–U1–U2 and U4–U7 are stacked consecutively on each other in an A-form–like conformation, whereas base U3 is flipped out into a separate pocket and stacks between Arg185 and the Phe17–Asp18 peptide plane. Tyr177 intercalates between bases U7 and U8 and forms a triple stack with bases U8 and U9, resulting in an S-shaped backbone that maintains the RNA in contact with the protein groove. Base U10 is again flipped out to be sandwiched between Ile127 and Ala47. Pro126/Ile124 stacks on one side of base U11, the other side stacking on base U1′, which begins the next 11 mer. Apart from these stacking interactions, key contacts to the RNA backbone are made by genus-conserved residues Arg183 and Gln184 to the phosphate of U4 (denoted U4P), Arg94 to U5P, His76 to U6P and U7P, and Thr91 to U7P. In addition, Thr12 and Phe17 main-chain, and Asp18, Lys50, Arg167, and Lys180 side-chains are in a position to make hydrogen bond interactions with bases, although it is likely that any base contacts adapt to the RNA sequence. Remarkably, there are no major conformational changes in the protein core upon RNA binding (root-mean-square-deviation of Cα positions ∼0.6 Å for residues 18–216), although residues around the intercalating Tyr177 are shifted by up to 2 Å.
From Closed Tetramer to Extended Helical Structure.
The tetramer has almost exact twofold symmetry with chains A/C and B/D being pairwise equivalent (Fig. 4A). Each protomer interacts in head-to-tail fashion with two neighbors; for instance, the N- and C-arms of chain A bind, respectively, to the cores of chain B and chain D, resulting in burial of about 1,880 Å2 at each NP–NP interface. The major difference between nonequivalent protomers is the conformation of the flexible hinges that place the N- and C-arms in different positions (Fig. 5A), while conserving the same arm–core contacts as in the apo-structure. In the A/C protomers, the N-arm folds back toward the core and all residues can be modeled (although residues 9–15 have weak density), whereas for the B/D protomers, the N-arm is extended away from the core and the hinge residues 9–15 lack electron density (Fig. 5A). Concerning the helical C-arm, in the A/C protomers it extends away from the core, whereas for the B/D protomers it orientates back toward the core (Fig. 5A). The different arm conformations affect the RNA conformation and protein–RNA interactions at the NP–NP interfaces, showing clearly the coupling between the two (Fig. S3A).
Fig. 5.
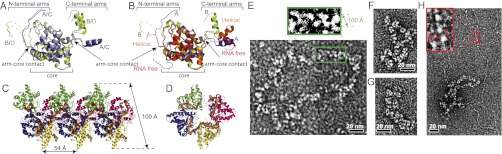
The hinged N- and C-arms enables flexible packaging of the genome. (A) Superimposition of the four monomers from the NP–RNA tetramer colored as in Fig. 4 showing the different positions of the N- and C-arms from the A/C and B/D pairs. (B) Superposition of LACV NP monomers from the RNA-free (red), tetrameric (chain A, blue and B, green), and P41 helical (orange) forms. The N- and C-arms of the helical form are respectively more similarly orientate to the B N-arm and A C-arm. The RNA-free arms (which result in an extended network in the crystal) are both distinct from other observed conformations. (C) Molecular packing of NP in a 41 helix as observed in the P41 crystal form, with the width and pitch of the helix indicated. RNA is modeled as an orange tube on the internal surface of the helix. Successive 11 nt RNA segments were positioned by superposing on the core of each helix protomer the B subunit from the tetramer together with its bound 11 nts of RNA. Without making any adjustments, the near continuity of the RNA segments shows that the RNA binding mode observed in the tetramer is compatible with a much longer RNA binding within an extended helix. (D) Near end view of 41 helical NP–RNA model showing resemblance of projection to the tetramer (Fig. 4A). (E) LACV RNPs from disrupted virions examined by electron microscopy after negative staining with 2% uranyl-acetate. The size of the RNP shown is ∼160 × 75 nm. The RNPs are flexible but in some areas display regions with apparent helical characteristics as highlighted in green and shown in close-up. (F and G) Purified LACV RNPs displaying partial (F) or complete (G) supercoiling. In G the RNP dimensions are ∼60 nm long × 18 nm wide. (H) RNPs are partially unwound after incubation at 37 °C and show long, thin necklaces representing individual NPs bound to RNA. A close-up view of three NPs bound to RNA with interprotein distance of ∼50 Å.
The net result of the compensating alternation of two NP orientations is a quasi-planar tetramer rather than an extended chain structure (Fig. 4A). However, the arm flexibility allows this structure to be sheared, as observed in a C2 crystal form of the NP–RNA complex, cocrystallized with 11 nts RNA, which exhibits a slightly different tetramer, although the same architectural principles apply (Fig. S3 B and C). However, neither of these tetramers are likely to be the building blocks of wild-type RNPs, which might be expected to have some kind of, perhaps irregular, helical structure to be able to compact and encapsidate very long RNA molecules. Interestingly, in a P41 crystal form of LACV NP, grown from thiocyanate and RNase-treated tetramers (Fig. 1A, solid line and Fig. 2B, third lane), a polar, helical arrangement of linked NPs is observed, which could give some insight into wild-type RNP structure. Despite diffracting only to 7.4 Å resolution, the structure could unambiguously be solved by molecular replacement and shows two NPs in the asymmetric unit but no RNA (Fig. S2B). One of these generates a continuous 41 helix throughout the crystal with a pitch of 54 Å and width of about 100 Å (Fig. 5 C and D). Interestingly, in the crystallographic 41 helix, the N-arm is orientated roughly as observed in the A/C protomer of the tetramer and the C-arm as in the B/D protomers, this combination allowing formation of an extended, regular helix, rather than a closed tetramer (Fig. 5B). Note that the disposition of the arms that forms the network in the RNA-free crystal form is different again (Fig. 5B). Using the crystallographically observed 41 helix as a template, we constructed a corresponding helical model of an NP–RNA filament by superposing a monomer with 11 nts RNA bound from the tetramer on the helix protomer to position the RNA within the helix. This shows that the 11 nts segments can join up to form a continuous helical arrangement of the RNA with 44 nts per 54 Å pitch (Fig. 5 C and D). As shown next, electron microscopy gives an indication that such a helical structure may exist in wild-type RNPs released from LACV virions, although because of arm flexibility, we do not expect exact correspondence with the 41 helix observed, perhaps fortuitously, in a particular crystal form.
EM of Native LACV RNPs Reveals a Structural Hierarchy.
To gain insights into the native RNP structure, we analyzed RNPs released from wild-type LACV virions by electron microscopy after disruption of their membrane with mild detergent. Negatively stained EM images reveal the presence of circular RNPs of different sizes (Fig. S4 A and B) as previously observed (23). The RNPs are highly flexible, but display in some areas an order compatible with a helical structure (Fig. 5E). The width (100–110 Å) and pitch (around 50 Å) of these regions are comparable with the structural characteristics of the LACV NP 41 helix described above (Fig. 5C), but the flexibility precludes more quantitative analysis of this higher order structure. Other RNP segments appear as “pearl necklaces” with a width of around 35 Å and monomer separation of about 50 Å that likely represent discrete NP “daisy chains” bound to RNA (Fig. S4A).
To investigate further, we purified RNPs by ultracentrifugation and analyzed the effect of pH, salt concentration, and temperature on their structure. Buffer exchange from 30% (wt/vol) CsCl to 200 mM NaCl induced partial or complete supercoiling of RNPs (Fig. 5 F and G and Fig. S3 C and D). Heating-purified RNPs in 200 mM NaCl for 1 h at 37 °C favored RNP unwinding into very long, thin filaments containing discrete NPs coating RNA, the distance between NPs being again around 50 Å (Fig. 5H). These filaments often emerge from more condensed regions (Fig. S4 E–G). These observations of different levels of structure within virion-released RNPs suggest that such RNPs are more or less partially unraveled versions of a perhaps more compact state existing inside the virion but that the NP–NP interactions responsible for higher order structure are relatively weak and sensitive to conditions.
Discussion
Correspondence with Mutational Studies.
Whereas no mutagenesis results are available on LACV NP, several studies have been made on NP from the prototypical Orthobunyavirus BUNV. Since BUNV and LACV NP are 42% identical (63% similar) (Fig. 3), it is thus expected that results obtained on one should be applicable to the other. Note that LACV NP numbering is the same as that of BUNV up to 111, then are shifted by one as LACV has a single residue insertion. The first attempt to map functional domains of BUNV NP showed that both the N-terminal and C-terminal extremities were important for multimerisation (24), in clear agreement with the structural results presented here. Subsequently, an exhaustive mutational analysis was performed by making point mutants at 110 different positions on BUNV NP, including 40 at absolutely conserved sites in the Orthobunyavirus genus (20). Many of the conserved residues whose mutation had severe effects in BUNV are either buried in the hydrophobic core of the protein (e.g., Phe26, Leu118, Trp135, Tyr142, Phe145, Tyr159, and Trp214, LACV numbering) or involved in NP–NP interactions (e.g., Leu4, Phe6, Leu161, Val168, Leu178, Trp194, Phe226, Leu227, Phe230, and Ile232). However, several residues observed to make direct contacts to the RNA in the LACV NP–RNA structure also led to nonrescuable virus, notably Arg94, Pro126, Tyr177, and Lys180. Arg94 interacts with U5P and Tyr177 intercalates between bases U7 and U8 and is critical in shaping the RNA conformation to maintain contact with the protein (see above). Another BUNV mutant, Lys50A, was moderately attenuated consistent with its perhaps less critical role in base contacts. In another study on BUNV NP, in which RNA binding was directly assayed, the mutant Arg94A was found to eliminate RNA binding, and Arg40A (which makes a salt-bridge with Asp8, stabilizing the position of the N-arm) (Fig. 2C) and Lys50A were significantly deficient in RNA binding, again consistent with our results (25). Thus, the LACV NP–RNA structure explains extremely well the available BUNV mutagenesis data, and also demonstrates that our results can most likely be generalized to all Orthobunyavirus NPs.
Structure of RNPs in Bunyavirus Virions.
Cryo-EM images of La Crosse virions show that the particles are spherical but heterogeneous in size, ranging from 80 to 110 nm in diameter, the spikes and membrane accounting for about 28 nm of this value (26). RNPs were not directly visualized in the EM images. However, a viable LAC virion needs to package at least three distinct RNAs totaling 12,489 nts (L 6980, M 4526, and S 983 nts). Fully unraveled LACV RNPs are extremely long and thin (Fig. 5H and Fig. S3) and clearly must undergo compaction to be packaged. The 41 helical model discussed above, in which there are 44 nts per 54 Å, would result in RNPs of linear length 860, 555, and 121 nm, for the L, M, and S segments, respectively. Taking into account the LACV RNPs are in fact circularised by the complementary 5′ and 3′ ends forming a panhandle (which is most likely bound to the polymerase) (23, 27), this would correspond to idealized circles of diameter 274, 176, and 38 nm. Although the contour length, estimated at 420 nm, of the irregular closed RNP shown in Fig. 5E is somewhat smaller than the size calculated for an M segment, it does suggest that RNPs are organized in this kind of way. Indeed in previous work, very similar circular RNPs with three modal contour lengths of 700, 510, and 200 nm and width 10–12 nm were observed (23). However, it is still possible that a higher order structure, perhaps of the supercoiled form observed in Fig. 5 F and G, is required to reach the compaction required to fit within a virion of internal diameter ∼80 nm. Supercoiling of RNPs is best characterized in the case of influenza virus where rod-shaped RNPs of different lengths are clearly visible inside virions (28, 29). Similarly, cryo-electron tomography has enabled clear visualization of rod-like RNPs within Tula (30) and Hantaan (31) viruses, both of the Hantavirus genus. In Hantaan virus, parallel rod-like RNPs ∼10 nm in diameter and up to 100 nm long are observed, the median diameter of the virions being 132 nm (31). On the other hand, RNPs are much less distinctly seen in the interior of two Phleboviruses, Rift Valley Fever (8, 32) and Uukuniemi (33). However, it must be borne in mind that these viruses have distinct N proteins from Orthobunyaviruses, and so these results are not directly transferable to LACV.
Comparison of LACV with Other sNSV NP–RNA Complexes.
Recently crystal structures were published of several closed NP–RNA oligomers (tetramer, pentamer, and hexamer) of RVFV from the Phlebovirus genus (19). This followed earlier work on the structure of the RNA-free RVFV NP in auto-inhibited, monomeric (16), and hexameric forms (10). RVFV and LACV N proteins are of similar size (respectively, 245 and 235 residues), but have quite distinct folds. Nevertheless there are several similarities in the architectural principles by which both proteins form NP–RNA complexes, even though the details differ (Fig. S3 C and D). The similarities are the use of arms to mediate oligomer formation, the flexible hinge giving the ability to form assemblies with different geometry; the internal sequestration of the RNA in a positively charged groove within the core of the protein with mainly backbone contacts; and the sharp turn of the RNA at the protein–protein interfaces with coupling of RNA conformation to hinge conformation. The main differences are that RVFV NP has only one long N-arm that mediates protein–protein interactions rather than an N- and C-arm as in LACV NP; RVFV tetramer, pentamer, and hexamer have, respectively, four-, five- or sixfold symmetry (or close to), whereas the LACV tetramer only has twofold symmetry; and RVFV NP sequesters 7 nts per protomer compared with 11 nts for LACV. The only other known sNSV NP–RNA structure, the N-terminal domain of Lassa virus NP with 6 nts RNA, also shows the RNA largely sequestered in a positively charged groove (34). For LACV, based on different crystal structures and EM images of flexible but fairly compact RNPs released from virions, we discussed above a possible supercoiled helical model for the RNP. For RVFV, EM images of RNPs isolated from virally infected cells show a much more unraveled structure, which was interpreted as a flexible chain of monomers (19), and this is compatible with the lack of obvious discreet, rod-like RNP structures within Rift Valley Fever or Uukuniemi virions (8, 32, 33).
Conclusion
The structural studies reported here give unique insight into how Orthobunyavirus NP fulfils its protective, encapsidation function, but leave open a number of intriguing questions. Firstly, how budding virions select three complementary RNPs for packaging is unknown, especially if the bases, whose pairing between different segments is thought to play a role in influenza virus packaging (35), are inaccessible. Secondly, as with the case of RVFV, the observed NP–RNA structure is clearly incompatible with transcription or replication of the genomic RNA within RNPs without polymerase-induced disruption of the tight protein–RNA interactions. For Lassa virus NP, there is some evidence of a gating mechanism for RNA binding that could be manipulated by the polymerase (34). For LACV, since the RNA binding to the core does not involve significant conformational changes, we imagine that polymerase-induced changes in arm conformation, which as we have shown is coupled to RNA conformation, are most likely used to temporarily disrupt RNA binding. Indeed it is possible that the same kind of interaction of the NP arms with L protein, perhaps also involving specific RNA sequence recognition (36), enables recruitment of auto-inhibited NP into a conformation able to lock onto nascent genome replicates during de novo RNP assembly.
Materials and Methods
Full methods are given in SI Materials and Methods. Recombinant LACV NP was expressed in E. coli and purified in tetrameric form with bacterial RNA or monomeric RNA-free form after treatment with RNase A and sodium thiocyanate. Crystals were grown of RNA-free NP or reconstituted tetramers with either 11 or 45 nts synthetic RNA. Data collection was performed at the European Synchrotron Radiation Facility in Grenoble, France. The apo-NP structure was solved with a selenomethionine derivative and other structures by molecular replacement. Crystallographic data are in Table S1 and electron density for RNA in Fig. S2. RNPs released from LACV virions by mild detergent treatment or further purified by gradient ultracentrifugation were examined by negative stain electron microscopy.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility (ESRF)–European Molecular Biology Laboratory Joint Structural Biology Group for access to ESRF beamlines, the Partnership for Structural Biology for access to structural biology instrumentation, Jörg Schmidt and Julien Perard for technical assistance, and Rob Ruigrok and Dan Kolakofsky for critically reading the manuscript. H.M. holds a long-term European Molecular Biology Organization fellowship (ALTF2011-413), S.C. an Agence Nationale de Recherche (ANR) grant (ANR-GUI-AAP-04), and F.W. is supported by the Sonderforschungsbereich (SFB593) of the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4BGP (NP alone) and 4BHH (NP-RNA)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302298110/-/DCSupplemental.
References
- 1.Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol. 2011;92(Pt 11):2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- 2.Guu TS, Zheng W, Tao YJ. Bunyavirus: structure and replication. Adv Exp Med Biol. 2012;726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 3.Elliott RM. Bunyaviruses and climate change. Clin Microbiol Infect. 2009;15(6):510–517. doi: 10.1111/j.1469-0691.2009.02849.x. [DOI] [PubMed] [Google Scholar]
- 4.Maltezou HC, Papa A. Crimean-Congo hemorrhagic fever: Risk for emergence of new endemic foci in Europe? Travel Med Infect Dis. 2010;8(3):139–143. doi: 10.1016/j.tmaid.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Yu XJ, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364(16):1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6(9):e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol. 2007;81(7):3198–3205. doi: 10.1128/JVI.02655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huiskonen JT, Overby AK, Weber F, Grünewald K. Electron cryo-microscopy and single-particle averaging of Rift Valley fever virus: Evidence for GN-GC glycoprotein heterodimers. J Virol. 2009;83(8):3762–3769. doi: 10.1128/JVI.02483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Ferron F, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7(5):e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 12.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444(7122):1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 13.Carter SD, et al. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol. 2012;86(20):10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: Superhelical homo-oligomers and the role of caspase-3 cleavage. J Virol. 2012;86(22):12294–12303. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, et al. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci USA. 2012;109(13):5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2010;107(26):11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenavas S, et al. Monomeric nucleoprotein of influenza A virus. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavrakis M, et al. Rabies virus chaperone: Identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology. 2006;349(2):422–429. doi: 10.1016/j.virol.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 19.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci USA. 2012;109(47):19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J Virol. 2009;83(21):11307–11317. doi: 10.1128/JVI.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers JW, Zhou Q, Green TJ, Barr JN, Luo M. Purification, crystallization and preliminary X-ray crystallographic analysis of the nucleocapsid protein of Bunyamwera virus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 4):361–364. doi: 10.1107/S1744309106006397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohl BP, Barr JN. Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus. RNA. 2009;15(3):391–399. doi: 10.1261/rna.1367209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. J Virol. 1976;20(3):664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard VH, Kohl A, Osborne JC, McLees A, Elliott RM. Homotypic interaction of Bunyamwera virus nucleocapsid protein. J Virol. 2005;79(20):13166–13172. doi: 10.1128/JVI.79.20.13166-13172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter CT, Bento DF, Alonso AG, Barr JN. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J Gen Virol. 2011;92(Pt 1):80–84. doi: 10.1099/vir.0.024240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talmon Y, et al. Electron microscopy of vitrified-hydrated La Crosse virus. J Virol. 1987;61(7):2319–2321. doi: 10.1128/jvi.61.7.2319-2321.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63(1):122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439(7075):490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 29.Arranz R, et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338(6114):1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 30.Huiskonen JT, et al. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J Virol. 2010;84(10):4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battisti AJ, et al. Structural studies of Hantaan virus. J Virol. 2011;85(2):835–841. doi: 10.1128/JVI.01847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman MB, Freiberg AN, Holbrook MR, Watowich SJ. Single-particle cryo-electron microscopy of Rift Valley fever virus. Virology. 2009;387(1):11–15. doi: 10.1016/j.virol.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overby AK, Pettersson RF, Grünewald K, Huiskonen JT. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc Natl Acad Sci USA. 2008;105(7):2375–2379. doi: 10.1073/pnas.0708738105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hastie KM, et al. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci USA. 2011;108(48):19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J Gen Virol. 2010;91(Pt 2):313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 36.Osborne JC, Elliott RM. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J Virol. 2000;74(21):9946–9952. doi: 10.1128/jvi.74.21.9946-9952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.