Abstract
The partial purification of mouse mammary gland stem cells (MaSCs) using combinatorial cell surface markers (Lin−CD24+CD29hCD49fh) has improved our understanding of their role in normal development and breast tumorigenesis. Despite the significant improvement in MaSC enrichment, there is presently no methodology that adequately isolates pure MaSCs. Seeking new markers of MaSCs, we characterized the stem-like properties and expression signature of label-retaining cells from the mammary gland of mice expressing a controllable H2b-GFP transgene. In this system, the transgene expression can be repressed in a doxycycline-dependent fashion, allowing isolation of slowly dividing cells with retained nuclear GFP signal. Here, we show that H2b-GFPh cells reside within the predicted MaSC compartment and display greater mammary reconstitution unit frequency compared with H2b-GFPneg MaSCs. According to their transcriptome profile, H2b-GFPh MaSCs are enriched for pathways thought to play important roles in adult stem cells. We found Cd1d, a glycoprotein expressed on the surface of antigen-presenting cells, to be highly expressed by H2b-GFPh MaSCs, and isolation of Cd1d+ MaSCs further improved the mammary reconstitution unit enrichment frequency to nearly a single-cell level. Additionally, we functionally characterized a set of MaSC-enriched genes, discovering factors controlling MaSC survival. Collectively, our data provide tools for isolating a more precisely defined population of MaSCs and point to potentially critical factors for MaSC maintenance.
Keywords: FACS sorting, mammary gland transplant, shRNA screen
The murine mammary gland resembles, to some extent, the human mammary gland in development, milk production, and progression to carcinogenesis, making it an ideal system to develop methodologies and form hypotheses of relevance to women. The use of cell surface markers to isolate selected cell types from mice has greatly enhanced our understanding of development and our knowledge of molecular pathways and interactions that influence it. Mammary gland stem cells (MaSCs) have commanded attention because of not only their roles in the cycles of gland morphogenesis but also their potential contribution in tumor initiation. Full characterization of MaSCs, however, has been hampered by their scarcity. Enrichment of the MaSC compartment has, until now, been achieved by using a combination of cell surface markers (Lin−CD24+CD29hCD49fh) (1, 2). Thus far, these cells have been enriched to 1 MaSC per every 64 cells stained Lin−CD24+CD29h (1). This is sufficient to test for MaSC repopulation capacity and to some extent, roles in tumorigenesis, but this level of purity is less suitable for more complex molecular analyses that define MaSCs and their properties.
Additional characterization of MaSCs has been achieved using a transgenic mouse model expressing GFP under the control of the s-ship promoter (3). This gene is expressed in embryonic and hematopoietic stem cells but not differentiated cells (4). GFP+ cells in this mouse model were shown to reside at the tips of the terminal end buds, where MaSCs are believed to be located in these developing mammary gland structures (3, 5). Transplantation of the MaSC-enriched GFP+CD49fh cells improved the mammary reconstitution unit (MRU) frequency to 1/48 cells, an increase over the previous shown frequency for CD24+CD29hCD49fh cells. Although being very elegantly performed and enhancing our understanding of MaSC localization, studies with this mouse model did not achieve a greater enrichment for MaSCs using more conveniently accessible markers, such as cell surface proteins.
Given the limitations in accurately purifying MaSCs, we sought to devise a method better suited for identifying this population. Here, we describe the use of long-term label retention to increase the MRU frequency within MaSC-enriched CD24+CD29h cells. This approach, previously applied to the isolation of skin stem cells (6), enables the identification of slowly dividing cells, a characteristic of adult stem cells. To mark slowly dividing cells, expression of the H2b histone, linked to GFP, is regulated by a tetracycline responsive element (TRE) and a tet-controlled transcription activator (tTA) under the endogenous keratin K5 promoter (K5tTA-H2b-GFP). In the absence of tetracycline or its analog doxycycline (DOX), the tTA binds to TRE and activates transcription of H2b-GFP. Treatment with DOX prevents the tTA binding to TRE, and transcription of H2b-GFP is terminated (6). As the cell divides, newly synthesized, unlabeled H2b replaces the H2b-GFP; therefore, the more slowly dividing cells will retain GFP expression for an extended period.
We were able to improve the MaSC enrichment by isolating GFP-retaining cells after a long-term inhibition of transgene expression. We refer to these cells as H2b-GFPh MaSCs (CD24+CD29hH2b-GFPh). Comparisons between expression profiles of all mammary gland cell types suggested that H2b-GFPh MaSCs differentially expressed several genes involved in pathways previously described as playing roles in other adult stem cells. Additional analysis of the H2b-GFPh MaSC expression signature led to the identification of a cell surface marker that, combined with conventional markers, resulted in the isolation of an MaSC population with an elevated proportion of MRUs. In addition, we performed a focused shRNA screen, targeting genes that were differentially expressed in our newly characterized MaSC-enriched cell population, revealing potential regulators of mammary gland biogenesis. Overall, this work improves our ability to purify MaSCs and provides valuable insights into their role in mammary gland development and perhaps, even tumor initiation.
Results
H2b-GFP Label-Retaining Cells Enrich for MaSCs.
To better enrich for the MaSC population, we assessed the feasibility of using mammary gland label-retaining cells to select for MaSCs, given that a slower division rate is an excepted characteristic of adult stem cells. We adopted a system wherein expression of the H2b histone, linked to GFP, is regulated by a TRE and a tTA under the endogenous keratin K5 promoter K5tTA-H2b-GFP (a gift from Elaine Fuchs, Rockefeller University, New York, NY). Keratin K5 is expressed in cells of the basal compartment, the region considered to be home to MaSCs (7). This system displays some advantages over the previous gene reporter-based methods used to isolate MaSCs, because it takes advantage of one of the more general properties of stem cells: their relative quiescence. In support of the use of this mouse model, there were previous hints that MaSC-enriched CD24+CD29h cells display BrdU label-retaining properties (1), although label-retaining populations were not functionally characterized.
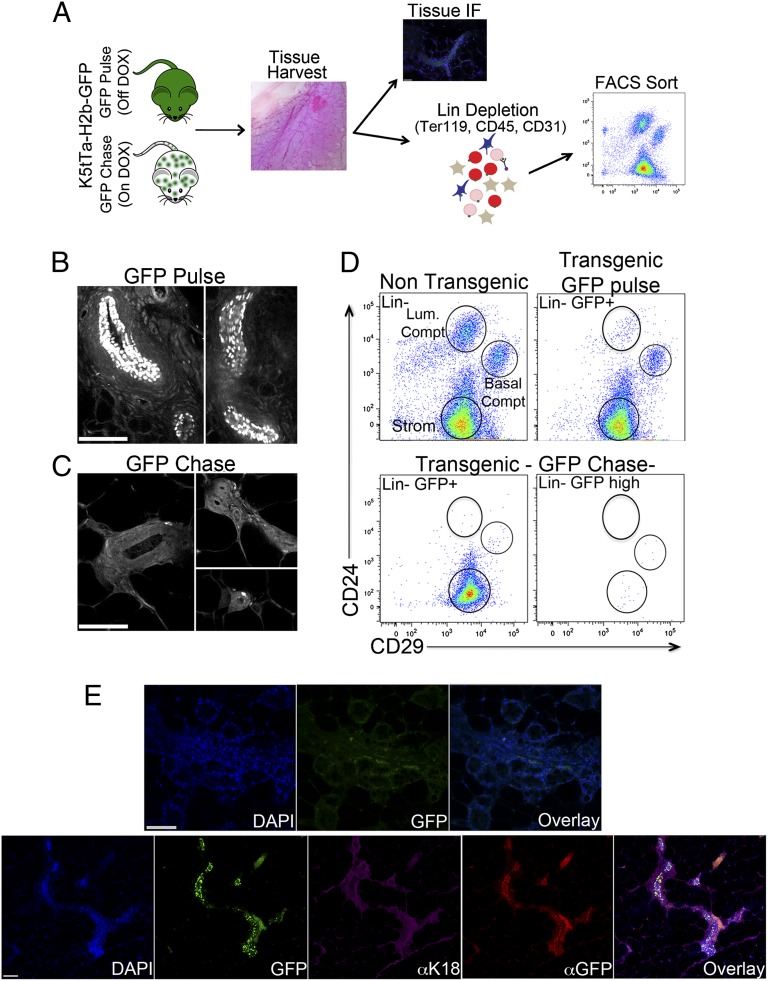
Initial experiments using the H2b-GFP mice assessed the expression and distribution of GFP-positive cells in the adult mammary gland (Fig. 1A). Histological sections revealed the presence of several GFP+ cells located within structures resembling the mammary gland ductal epithelium (Fig. 1B and Fig. S1A, Upper). Treatment of H2b-GFP mice with DOX over a 12-wk period, thus ceasing transcription of H2b-GFP transgene, dramatically reduced the number of cells expressing GFP. Notably, those cells that remained GFP+ were located at the tips of the terminal end buds. These distinct sites in the ductal epithelium are the areas currently believed to be resident by MaSCs (8) (Fig. 1C and Fig. S1A, Lower).
Fig. 1.
H2b-GFP label-retaining cells represent a population of MaSCs. (A) Experimental scheme. Mammary glands were harvested from K5tTa-H2b-GFP transgenic mice either off (GFP pulse) or on DOX diet (GFP chase) and further processed for immunological staining or single-cell suspension FACS sorting. (B and C) Tissue histology H2b-GFP+ cells distribution. Mammary glands from transgenic mice off and on DOX diet were harvested, defatted, embedded in agarose, and imaged with two-photon microscopy. (B) Mice off DOX diet (GFP pulse) showing a broad distribution of GFP+ cells in mammary gland ductal structures. (C) After a 12-wk DOX chase, H2b-GFP+ label-retaining cells became restricted to the edges of the ductal structures. (D) Flow cytometry profile of H2B-GFP+ cells. Upper Left shows the profile of a lineage-depleted (CD45−, Ter119−, and CD31−) nontransgenic mammary gland according to CD24 and CD29 staining and highlights the three cell compartments: luminal (CD24hCD29+; comprising luminal progenitor cells, luminal alveolar cells, and luminal ductal cells); basal (CD24+CD29h; comprising myoepithelial progenitor cells, myoepithelial differentiated cells, and MaSCs); and stromal. Total GFP+ cells from H2b-GFP transgenic mice off DOX diet (GFP pulse mice; Upper Right) displayed a similar cellular compartmental distribution with fewer luminal-type cells. The CD24CD29 cell profile of H2b-GFP+ cells from GFP chase mice (on DOX) were analyzed using two strategies to define GFP-expressing cells. Lower Left displays CD24CD29 staining of total H2b-GFP+ cells, whereas Lower Right shows the CD24CD29 staining of H2b-GFPh cells. The focus on GFPh cells, the most label-retaining cells, drastically decreased the cellular content of all mammary gland compartments and retained a greater proportion of cells inside of the basal compartment, potentially representing MaSCs. (E) Histological analysis of mammary gland H2b-GFPh MaSCs outgrowths. Cleared fat pads from prepubescent female mice were injected with either total H2b-GFP− MaSCs (CD24+CD29hGFP− cells) or H2b-GFPh MaSCs (CD24+CD29hGFPhcells), harvested 12 wk after transplantation, embedded in agarose, stained with antibodies, and imaged on a Zeiss 710 LSM (Zeiss) confocal microscope. Images display outgrowths of two distinct glands injected with H2b-GFPh MaSCs.
To compliment this observation, under the hypothesis that mammary gland label-retaining cells comprise a population of potential MaSCs, we investigated the correlation between GFP retention and expression of previously defined MaSC-enriched cell surface markers, CD24 and CD29. Using FACS analysis, we were able to subdivide the mammary gland (after depletion of endothelial and hematopoietic cells as shown in Fig. S1B) into three distinct cell compartments: luminal (CD24hCD29+), occupied by luminal cells; basal (CD24+CD29h), occupied by myoepithelial cells and MaSCs; and stromal (CD24−CD29+) (1) (Fig. 1D, Upper Left). The majority of GFP+ cells from a transgenic H2b-GFP mouse off DOX could be categorized into either basal or stromal compartments, with far fewer GFP+ cells occupying the luminal compartment (Fig. 1D, Upper Right and Fig. S1C, Left). After a 12-wk DOX chase, the overall proportion of GFP+ cells decreased by more than one-half, and the presence of a GFP+ luminal compartment was all but eliminated (Fig. 1D, Lower Left and Fig. S1C, Center). Focusing on GFP intensity (a measure that directly relates to the rate of cell division), selection of only the brightest GFP+ cells (GFPh) resulted in a greater proportion remaining in the CD24+CD29h basal compartment, whereas the stromal compartment was significantly reduced after GFPdim cells were removed (Fig. 1D, Lower Right and Fig. S1C, Right). This result suggests that the most label-retaining cells reside within the basal compartment and may represent the MaSCs population.
The benefit of using GFP to test for label retention, as opposed to BrdU, is that its detection does not require fixation and staining. We were then able to test the biological differences, using mammary gland transplants, between GFPh cells (H2b-GFPh MaSCs) and GFP− cells (H2b-GFP− MaSCs) within the MaSC-enriched compartment. Transplantation assays are a fundamental criterion to evaluate stemness and have been used previously for several tissues, including the mammary gland (1, 2, 9). For these experiments, the inguinal glands were removed from the endogenous tissue of prepubescent females before injection of donor cells. Donor cells were harvested from mammary glands of H2b-GFP mice after a 12-wk DOX chase, dissociated, lineage-depleted, and sorted according to GFP intensity (Fig. S1D). Cells (GFPh and GFP−) were then injected, and outgrowths from donor cells were compared (by visualization of GFP+ epithelium) 12 wk posttransplantation. Given that the recipient animals are not treated with DOX, all cells derived from the donor mice will resume expression of the H2b-GFP transgene and give rise to GFP+ outgrows. MRU frequency was estimated according to the previously described algorithm (10). Transplantation of 500 H2b-GFPh MaSCs (n = 5) gave rise to GFP+ epithelium in all injected glands. This ability to reconstitute was still retained when only 50 cells were transplanted (Fig. 1E). In contrast, only one-half of the glands injected with 500 H2b-GFP− MaSCs displayed fluorescent outgrowths, decreasing to just 29% with injection of 50 cells (Dataset S1). These results represent an increase in the estimated frequency of MRUs from 1/70 cells, when MaSC selection was performed using CD24+CD29h alone (1), to 1/33 cells, with restriction to H2b-GFPh cells to further define MaSCs. Comparatively, the MRU frequency among H2b-GFP− MaSCs was estimated to be 1/149 (Dataset S1). Colony-forming ability was also twofold greater for H2b-GFPh MaSCs when 500 of these cells were seeded in a Matrigel (BD Bioscience) and cultured for 7 d (Fig. S1E).
Considered together, these data suggest that mammary gland H2b-GFPh label-retaining cells represent a subset, if not an entire population, of the MaSCs. Our experiments using a repressible H2b-GFP transgene have built on previous knowledge regarding the label-retaining properties of stem cells in the mammary gland and confirmed that MaSC CD24+CD29h cells reside mainly within the H2b-GFPh label-retaining cell population. In addition to these experiments, we also found that hormone-dependent activation of MaSC proliferation and differentiation, triggered by one complete cycle of pregnancy and involution in transgenic H2b-GFP mice treated with DOX, completely depleted GFP+ cells, validating that H2b-GFPh cells truly represent a population of slowly dividing cells rather than being a transgenic artifact.
It has been proposed that MaSCs comprise less than 5% of the total basal compartment. Our findings support this notion given that we find label-retaining H2b-GFPh cells to account for ∼0.2% of the total CD24+CD29h population (Fig. S1D, Upper Right). We also compared the distribution of H2b-GFPh–retaining cells with expression of a recently identified marker for myoepithelial progenitor-like cells, CD61. This marker was expressed by most of the H2b-GFPdim population, whereas virtually all H2b-GFPh cells were negative for CD61 staining, suggesting perhaps a unique mammary gland cell differentiation pattern, where H2b-GFPh label-retaining cells might occupy the top of hierarchy.
H2b-GFP Cells Display a Stem Cell-Like Expression Signature.
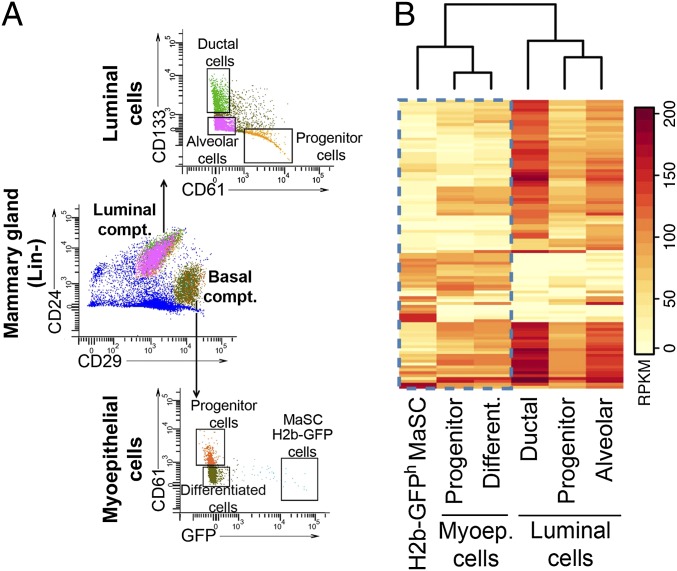
Having established that H2b-GFPh MaSCs have reconstitution properties, we next sought to determine where these cells fall in the mammary differentiation hierarchy with regard to their gene expression patterns. Using a combination of cell surface markers (1, 11), six distinct cell types were isolated by FACS to a purity of >90%: H2b-GFP MaSCs (Lin−CD24+CD29hH2b-GFPhCD61−), myoepithelial progenitor-like cells (Lin-CD24+CD29hH2b-GFP−/lCD61+), myoepithelial differentiated cells (Lin−CD24+CD29hH2b-GFP−CD61−), luminal progenitor cells (Lin−CD24hCD29+CD61+CD133−), luminal ductal cells (Lin−CD24hCD29+CD61−CD133+), and luminal alveolar cells (Lin−CD24hCD29+CD61− CD133−) (Fig. 2A). The myoepithelial progenitor-like cells were defined by expression of CD61 as a positive cell surface marker and their positioning as the second most label-retaining cell population.
Fig. 2.
H2b-GFPh MaSCs display a stem-like expression signature. (A) Sorting strategy. We used a combination of four cell surface markers in addition to H2b-GFP expression to segregate the lineage-depleted mammary gland cells into six distinct cell types: H2b-GFPh MaSCs (Lin−CD24+CD29hH2b-GFPhCD61−), myoepithelial progenitors cells (Lin-CD24+CD29hH2b-GFP−CD61+), myoepithelial differentiated cells (Lin−CD24+CD29hH2b-GFP−CD61−), luminal progenitor cells (Lin−CD24hCD29lCD61+CD133−), luminal ductal cells (Lin−CD24hCD29lCD61−CD133+), and luminal alveolar cells (Lin−CD24hCD29lCD61− CD133−). For each library, two biological replicates were analyzed. (B) Mammary gland differential expression heat map. Clustering of RPKM profiles for the 100 genes with highest variance across all samples. Two main cell clusters were generated according to the expression patterns of analyzed genes: luminal- (progenitor, alveolar, and ductal cells) and basal-type cells (H2b-GFPh MaSCs, progenitors, and differentiated cells). Note that H2b-GFPh MaSCs cluster with other basal compartment cells but have an expression signature distinct from the other two cell types in this cluster.
Hierarchical clustering of combined RNAseq replicates split mammary gland cells into two main branches: the basal compartment, comprising myoepithelial progenitor cells, myoepithelial differentiated cells, and H2b-GFP MaSCs, and the luminal compartment, with luminal progenitor cells and differentiated cells (Fig. 2B). As predicted by prior characterization of MaSCs (1), we found the expression profile of H2b-GFP MaSCs to be more closely related to the expression profile of myoepithelial cells than luminal cells; however, H2b-GFP MaSCs were still an outgroup compared with other cells in this cluster. Analysis over all mammary gland cell types yielded several hundred genes differentially expressed among all cell types (Fig. 2B), spanning diverse gene ontology groups and pathways (Dataset S2). More specifically, genes differentially expressed in H2b-GFP MaSCs were enriched in G protein-coupled receptors and pathways involving Wnt/B-catenin signaling, areas previously described to play fundamental roles in other adult stem cells (12). Differential expression patterns were confirmed on four genes by performing quantitative RT-PCR on H2b-GFP MaSCs (n = 4 individually sorted samples) and myoepithelial progenitor cells (n = 3 individually sorted samples) (Fig. S2). mRNA for the Cd24 and Cd29 genes was quantified as control, because all myoepithelial cells displayed similar levels of expression for these genes.
These results further confirmed that mammary cell types could be differentiated based on their gene expression profiles, allowing us to use these profiles to select cell type-specific genetic identifiers.
Additional Cell Surface Marker to Improve MaSCs Purification.
Because of limitations on the ability to purify MaSCs to homogeneity based on currently used cell surface markers, we searched for new surface markers that might identify MaSCs using the RNAseq data. We first generated a list of ∼500 genes that encode for cell surface markers according to their gene ontology term function (e.g., basolateral membrane, cell surface, membrane protein, or basement membrane). This list was further reduced to genes with high expression levels for the MaSC H2b-GFP cells. Five candidate cell surface proteins came out of this analysis (Fig. S3A): CD1d, a glycoprotein expressed on the surface of various mouse and human antigen-presenting cells (13); Cd59a, a regulator of the membrane attack complex (14); CD22, a regulatory lectin involved in repressing hyperactivation of the immune system (15); CD93, a C-type lectin involved in cell–cell adhesion processes (16); and CD74, an HLA class II protein, part of the major histocompatibility complex (17). Antibodies against CD1d, CD59a, and CD22 positively stained a distinct population of cells contained within the Lin-MaSC CD24+CD29h cells (Fig. 3A), whereas antibodies against the proteins, CD93 and CD74, failed to stain any mammary gland cells. We further tested CD1d MaSCs (CD24+CD29hCD1d+), CD59a MaSCs (CD24+CD29hCD59ah), and CD22 MaSCs (CD24+CD29hCD22+) for their ability to grow colonies in Matrigel culture. Two populations, CD1d MaSCs and CD59ah MaSCs [representing 1% and 4%, respectively, of the total MaSC (CD24+CD29h) population], displayed an approximately twofold increase in colony-forming ability compared with the total MaSCs population (Fig. S3B). However, we found CD1d MaSCs to have a greater colony-forming ability compared with CD59ah MaSCs, with one-half as many cells needed to produce the same number of colonies (200 and 500 cells, respectively, seeded on Matrigel). Additional analysis showed that all CD1d+ cells from the MaSC-enriched CD24+CD29h population were also CD59ah, whereas the remaining majority of CD59a+ cells from the MaSC-enriched CD24+CD29h population was negative for CD1d expression (Fig. 3B). Based on the enhanced colony-forming abilities of CD1d MaSCs over CD59ah MaSCs and the overlap of the two markers within the CD1d+ populations, we decided to pursue the experiments using CD1d as an MaSC marker.
Fig. 3.
Cd1d is an additional cell surface marker for purification of MaSCs. (A) FACS analysis of MaSC cell surface markers. Total MaSCs (CD24+CD29h cells) were additionally segregated according to the expression of the cell surface markers CD1d, CD59ah, or CD22. (B) CD1d is expressed by a subset of CD59ah cells. Lin− mammary gland cells were stained with antibodies against CD24, CD29, CD1d, and CD59a and further analyzed on an LSRII Cell Analyzer (BD Bioscience). The entire basal compartment (CD24+CD29h) was selected and analyzed according to CD1d and CD59a expression. The majority of cells within the basal compartment stained positive for CD59a, and CD1d+ cells fell mainly in the CD59ah area. (C) CD1d MaSCs are the most label-retaining cells within the mammary gland. Prepubescence mice were injected with BrdU (50 mg/kg body weight) for 5 d. Glands were either harvested from mice on the last day of BrdU injection to evaluate the total BrdU incorporation (week 0) or harvested after a 12 wk BrdU chase. Single-cell suspensions were stained with antibodies against CD24, CD29, and CD1d and analyzed on an LSRII Cell Analyzer (BD Bioscience). BrdU incorporation was measured in total MaSC (CD24+CD29h) and CD1d MaSC (CD24+CD29hCD1d+) populations. (D and E) Histological analysis of mammary gland CD1d MaSCs outgrowths. Cleared fat pads from prepubescent female mice were injected with (D) either total MaSCs or CD1d MaSCs and (E) 25 CD1d MaSCs cells harvested from glands pretransplanted with CD1d MaSCs. Glands were harvested 12 wk after transplantation and embedded in agarose, and endogenous GFP signal was imaged. Images display outgrowths from two distinct glands injected with CD1d MaSCs cells (D) or secondary transplanted CD1d MaSCs (E).
We next sorted CD1d+ MaSCs for RNAseq and compared their gene expression profile with those cell populations described in Fig. 2. Cluster analysis of all RNAseq libraries suggests that the CD1d MaSC expression signature is closer to the expression pattern found for H2b-GFP MaSCs than for any other cell type (Fig. S3C). These results could be suggestive that the common expression signature between CD1d+ MaSCs and H2b-GFPh MaSCs defines the stem cell state of mammary gland cells.
To ask whether CD1d+ MaSCs are slowly dividing cells, we performed BrdU label retention experiments. We injected BrdU into eight prepubescence female mice (3 wk old) over 5 consecutive d. Cells were harvested on the day of the last injection (week 0) from one-half of the mice and after 12 wk from the remaining mice (Fig. 3C). FACS analysis showed that ∼20% of the total MaSC population retained BrdU, and up to 60% of CD1d MaSCs were BrdU-retentive. This result adds confidence to the use of CD1d as a cell surface marker to represent the H2b-GFP MaSCs, because it is the most label-retaining cells and perhaps, therefore, the most enriched for stem-like cells within the mammary gland.
We then went on to repeat the mammary gland reconstruction assays, but this time, we compared CD1d+ MaSC transplantation efficiency with the transplantation efficiency displayed by the total MaSC (Lin−CD24+CD29h) population using cells from the H2b-GFP mice off DOX. Comparing donor-derived outgrowths (identifiable by GFP expression) between injection with CD1d+ MaSCs and injection with total MaSCs, we found that, despite bringing the injected cell number down to single digits, CD1d+ MaSCs effectively gave rise to GFP outgrowths in the majority of graft recipients (Fig. 3D and Dataset S3). This result gave a predicted MRU frequency of ∼1/8 CD1d MaSCs compared with the 1/44 MRU frequency from total MaSCs (Dataset S3). FACS collection of CD1d+ MaSCs from a reconstructed gland also effectively gave rise to a gland when serially transplanted into another mouse, showing that these cells also have the capacity to self-renew in addition to regenerating the gland (Fig. 3E).
MaSC-Focused shRNA Screen.
To identify genes and pathways necessary for the maintenance of MaSC reconstitution potential, we selected a set of abundantly and differentially expressed genes from RNAseq libraries of H2b-GFPh MaSCs and CD1d+ MaSCs and targeted them in shRNA-mediated knockdown experiments. We used shRNAs identified by a prediction algorithm developed in our laboratory, taking, on average, two hairpins per gene. Hairpins targeting nondifferentially expressed genes were also included as well as depletion control hairpins targeting Rpa3 and Polr2b and neutral control hairpins targeting Firefly luciferase and Renilla luciferase. All genes were targeted in a one-by-one approach in an assay lasting ∼3 wk (Fig. S4A).
shRNAs were introduced into the immortalized mammary gland cell line, Comma-Dβ (18). These cells give rise to both luminal and myoepithelial compartments in colony-forming and transplantation assays, independent of the method of MaSC enrichment (19–21). In addition, ∼50% of Comma-Dβ cells stain positive for Cd1d, placing them in our improved MaSCs isolation profile (Fig. S4B).
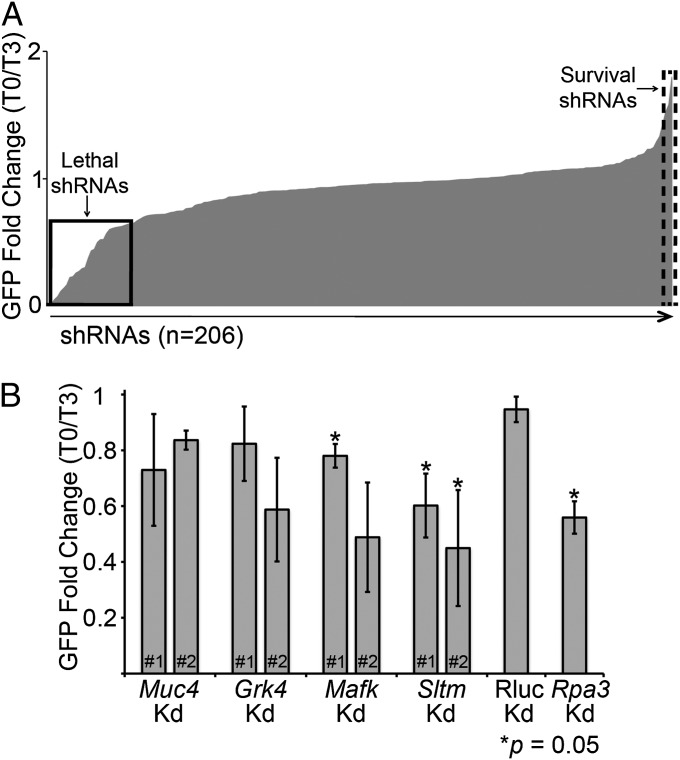
Cells were monitored for GFP expression (as a proxy for shRNA expression), and changes in the proportion of GFP-expressing cells would be indicative of a relevant gene function. The majority of screened shRNAs did not alter GFP levels during the 3-wk screening period (Fig. 4A), which could suggest that the correspondent genes were not essential for growth maintenance of Comma-Dβ cells. However, a distinct set of shRNAs altered the maintenance of GFP-expressing cells by either depleting GFP+ cells (Fig. 4A, lethal shRNAs) or promoting expansion of GFP+ cells (Fig. 4A, survival shRNAs) over time.
Fig. 4.
Mammary gland focused screen. (A) One by one screen; 206 shRNAs, covering ∼56 genes, were tested. The solid line square shows the fold change of shRNAs considered to be lethal, because GFP percent for these cells decreased overtime, whereas the dashed line square highlights data from shRNAs considered to show survival preferences in cells, because GFP percent increased overtime. (B) Screen hits validation. Two new hairpins targeting four genes selected as lethal hits from the first screen. The chart represents results of two independent experiments. *P = 0.05 by the t test.
We decided to further investigate a subset of genes that interfered with Comma-Dβ growth, because our focus was to understand the spectrum of genes that might block normal mammary gland biogenesis. Among the selected genes were mucin-like gene (Muc4), G protein-coupled receptor gene family member (Grk4), and transcription factors (MafK and Sltm). An additional set of hairpins for these genes was rescreened in Comma-Dβ cells with GFP levels followed for 10 d. No clear effect on the percentage of GFP-positive cells was observed when cells expressed the new shRNAs against Muc4 and Renilla luciferase control, whereas an shRNA-dependent response was observed, according to GFP frequency, when the genes Grk4 and Mafk were targeted (Fig. 4B). In addition, both new shRNAs against the gene Sltm consistently decreased GFP-expressing cells to levels comparable with the depletion achieved by Rpa3, the lethal control. Interestingly, Sltm encodes a transcription factor-like protein that binds both DNA (scaffold attachment factor-box DNA binding motif) and RNA (RNA binding domain) in response to estrogen levels (22). We are currently investigating the implications caused by loss of Sltm expression during normal mammary gland development and tumorigenesis.
Discussion
The ongoing interest in stem cells and more recently, cancer stem cells highlights the need for improvements in purification and analysis of this rare but important population. Our previous understanding of MaSCs has been clouded by the limited capability to obtain a pure population devoid of contaminating, more differentiated cells. Here, we took advantage of a previously used system to identify relatively quiescent cells (6) in the mammary gland. We propose that the label-retaining cells from the K5tTa-H2b-GFP mouse represent a subset of active MaSCs, displaying increased mammary gland reconstitution ability over previously published cell populations identified as MaSCs.
Unlike previous methods, where cell selection is based on the presence of constitutive fluorescence in cells (3, 23), the use of a cell state-dependent GFP system allows for a more biological relevant fluorescence reliability. The extended time between halting GFP expression and analysis and also, selection of only the brightest cells decrease the possibility that the GFP protein might be detected in a cell cycling at a normal rate, despite the fact that the GFP expression is switched off. This system allowed for the possibility of a much more stringent selection process; however, it is acknowledged that there are limitations with using this mouse model on a routine basis for enriching for MaSCs. The need to use a transgenic mouse and however reduced, the level of heterogeneity within cells selected—evident by <100% regrowth efficiency—illustrate the need for the cell surface marker, CD1d, identified in our study.
Cd1d is known to be expressed as a cell surface marker on a variety of antigen-presenting cells belonging to a cluster of glycoproteins involved in T-cell antigen presentation (13). Because we physically remove all hematopoietic cells using magnetic beads before FACS, we are confident that this differentially expressed marker does not simply reflect contaminating cells. This statement is supported by the presence of CD1d+ cells within the normal-like mammary gland cell line, Comma-1Dβ. In fact, 50% of these cells, isolated during midpregnancy, were positive for CD1d when stained with two distinct antibodies (Fig. S4B).
We, therefore, propose that CD1d is a genuine marker for MaSCs and when used combined with the cell surface markers CD24 and CD29, greatly enhances the purification of reconstituting cells above and beyond those cells selected based on label retention alone and those selected based on previously published markers. We perhaps did not exhaust all of the possibilities presented by our RNAseq data for the description of novel MaSC markers, but our findings do support CD1d as being a valuable component for purifying true MaSCs.
We found the proportion of CD1d+ cells (1%) within the basal compartment to be greater than the proportion of H2b-GFPh cells (0.2%) in this same compartment. This observation draws to light another drawback of relying solely on this particular label-retaining mouse model and in the same context, relying on GFP expression of a gene reporter mouse to identify MaSCs. The cytokeratin K5 (Krt5), for example, although shown to be expressed by basal-type cells, may not be expressed by all cells in this compartment. In addition, GFP expression may also be disrupted in some cells, perhaps by suppression of the transgene promoter; alternatively, some cells could fail to shut down GFP expression on DOX treatment. Had we only selected cells based on GFP expression from the K5 promoter, we would not be selecting all—or solely—those cells capable of self-renewal. This exclusion of MaSCs has been illustrated previously, where cells negative for a reporter GFP were able to still proliferate and regenerate into a new gland (3), something that we also see to a small degree with the K5tTa-H2b-GFP mouse model.
The identification of CD1d as a unique marker for this MaSC population and the distinct transcriptome of the Cd1d MaSCs suggest that these cells perform a function distinct from progenitors and more differentiated cells. Despite their gene expression profile clustering more closely with myoepithelial cells, they are still able to produce a new gland. It is unclear, however, if all of the CD1d MaSCs are multipotent stem cells or if they represent a combination of the recently described luminal and myoepithelial unipotent MaSCs (23). Because we are not using lineage tracing here, we cannot say for certain if all of the injected CD1d MaSCs would give rise to both compartments when allowed to repopulate the gland and if they are themselves the precursors to the cells that are largely responsible for tissue maintenance.
Identifying the genes involved in maintaining a stem cell and their self-renewing capabilities is vital to furthering our understanding of how these genes might be involved in abnormal gland development and tumorigenesis. Our current knowledge on this hypothesis, however, is, at best, limited; until now, it has been difficult to segregate myoepithelial cells and MaSCs, because they share common cell surface markers and very similar gene expression profiles (1, 24). The large number of shared genes expressed among cells identified by standard markers would mask any true differential patterns expressed by those cells with self-renewing properties. CD1d MaSCs cells only become divergent when the expression patterns of a relatively small number of genes are considered, a fact that would be overlooked if not using a more refined selection process. In addition to improving gene profiling as a whole for this minority population, the use of CD1d to isolate single cells for profiling could provide clues to gene expression changes between hypothesized MaSC states. For example, the complete loss of label-retaining cells after pregnancy suggests that these cells have undergone a more extensive process of cell division than in a virgin gland. However, CD1d+ cells are still present, unaltered to some extent, and using this marker, it would be possible to monitor gene expression changes during pregnancy and involution.
It has been suggested that stem cells within the mammary gland contribute in some way to the proposed notion of a cancer stem cell. In mouse mammary tumor virus (MMTV)-Wnt1 and p53−/− mice, for example, a preneoplastic mammary gland was seen to have an increased number of functional MaSCs, and ectopic expression of wnt-1 enhanced the self-renewing capabilities of cells, leading to cancers (24). CD1d itself has even been linked to breast cancer. In one study, antibodies against CD1d, combined with anti death receptor 5 (anti-DR5), a TNF-related apoptosis inducing ligand (TRAIL) receptor, led to rejection of tumor growth after injection of 4T1 tumor cells, a mouse breast cancer cell line (25), into a syngeneic mouse fat pad (26). Whether this observation was a result of the proposed interaction with natural killer cells or a disruption of the ability of the cancer to self-renew remains to be seen. The latter could be possible, because we show that the 4T1 mouse breast cancer cell line (Fig. S4C) and primary mouse breast cells (27) (Fig. S4D) display a population of cells that is positive for Cd1d. In humans as well, CD1d plays some unknown role. Down-regulation of CD1d expression has been shown to correlate with increasing metastasis in a mouse breast cancer model (28) and disease progression in multiple myeloma (29). Our own studies have shown that CD1d is expressed as a cell surface marker in some but not all of the human breast cancer cell lines tested (Fig. S4E). Those cell lines that showed CD1d+ cells were from basal-like breast cancers; luminal-like cancer cell lines, however, showed no CD1d+ cells.
With the ability to now purify a more homogeneous self-renewing population of MaSCs, it is possible to delve more deeply into the biology of these cells. We have not only appointed CD1d as a marker of MaSCs but also used this information to draw out gene targets for disrupting mammary gland development and possibly, malignancies. It is also unknown yet if these specific cell markers are a cause or effect of the ability of the cell to retain stemness; interrupting their expression and studying the effect on gland development and cancer are critical topics of future study.
Materials and Methods
Mice.
K5tTa-H2b-GFP heterozygote mice (6) were bred, and 20-d-old pups were checked for GFP expression using the IVIS100 in vivo imaging system (Caliper). CD-1 female mice were purchased from Charles River. Basal-like mouse mammary gland tumors were obtained from the transgenic mouse mammary tumor model C3-tag (27) (a gift from Mikala Egeblad, Cold Spring Harbor Laboratory, New York). All experiments were performed in agreement with and approved by the Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee.
Two-Photon Microscopy.
Mammary glands were harvested and defatted by three rounds of acetone treatment (20 min each). Defatted mammary glands were embedded and imaged according to previously published methods (30). In short, experiments were performed on a high-speed multiphoton microscope with integrated vibratome sectioning (TissueCyte 1000; TissueVision, Inc.). 3D scanning of 5-mm Z-volume stacks was achieved with a microscope objective piezo (PI E-665 LVPZT amplifier and P-725 PIFOC long-travel objective scanner), which translated the microscope objective with respect to the sample. Each optical section was imaged as a mosaic of individual fields of view equal to 0.83 × 0.83 mm and reconstructed posthoc using Fiji and custom-written Matlab software.
Antibodies.
Antibodies for flow cytometry were purchased from eBioscience unless otherwise specified, and they include anti-CD24 eFluor@ 450, biotinylated and PE-conjugated anti-CD45, biotinylated and phycoerythrin (PE)-conjugated anti-CD31, biotinylated and PE-conjugated anti-Ter119, PE-Cy7–conjugated anti-CD29, FITC- and PE-conjugated anti-CD61, antigen-presenting cell-conjugated anti-CD133, PerCP-CY5.5– and PE-conjugated anti-Cd1d (clones 1B1 and K253, respectively; BioLegend), PE-conjugated anti-CD22, monoclonal CD59a (Hycult Biotech), PE-conjugated human anti-Cd1d, 7-AAD viability staining solution (BioLegend), FITC-conjugated mouse IgG, and PE-conjugated rabbit IgG. Antibodies for immunostaining were chicken anti-GFP (Invitrogen), mouse monoclonal Cytokeratin 18 (SCTB), anti-chicken–IgG-Alexa Fluor 647 (Invitrogen), and anti-mouse–IgG Alexa Fluor 568 (Invitrogen).
Mammary Gland Preparation.
Mammary glands were harvested from young female mice (6–10 wk) and dissociated according to previously published protocol (1). After dissociation, cells were resuspended in 1 mL MACS Buffer (Myltenyi Biotech) and incubated with biotinylated anti-CD45, anti-Ter119, and anti-CD31 antibodies for 20 min. Cells were washed with 10 volumes MACS Buffer and further incubated with antibiotin magnetic microbeads (Myltenyi Biotech). Labeled cells were loaded into a magnetic column attached to a magnetic field (Myltenyi Biotech), and lineage-depleted flow-through cells were collected and further stained.
Flow Cytometry.
Cells were stained for 30 min at 4 °C with antibody mix in PBS supplemented with 1% (vol/vol) FBS followed by wash with 10× volume PBS. Cells were resuspended in PBS plus 1% (vol/vol) FBS and further stained with 7-AAD immediately before sorting or analysis. Cells were sorted using a FACS ARIAII SORP (BD Bioscience). For cell analysis, LRSII (BD Bioscience) cell analyzer or MACSQuant (Myltenyi Biotech) were used. Data analysis was performed using either FloJo (Tree Star) or Diva (BD Bioscience).
Matrigel Colony Assay.
Cells were sorted into 96-well plates containing 100 μL chilled 50% (vol/vol) Matrigel Matrix (BD Bioscience), further transferred to 100% Matrigel Precoated Chamber Slides (Lab-Tek), and incubated at 37 °C for 5 min. Complete Growth Media (1) was added to the chamber and renewed every other day for 10 d. Colonies were counted using Nikon Eclipse T1 microscope (Nikon).
Mammary Gland Transplant.
Cells were sorted into 96-well plates containing 30 μL 50% (vol/vol) Growth Factor Reduced Matrigel (BD Bioscience) and 0.01% (vol/vol) Tripan Blue (Sigma). Cells were injected into inguinal glands of 3-wk-old females that had been cleared of endogenous epithelium. Recipient glands were removed for evaluation 8–12 wk after cell injection.
Mammary Transplant Analysis.
Frozen sections and/or agarose-embedded sections were fixed with 4% paraformaldehyde (Sigma) for 20 min followed by tissue permeabilization and blocking using 10% (vol/vol) goat serum (Sigma). Paraffin-embedded sections were dewaxed and subjected to antigen retrieval for 15 min in Trilogy buffer (Cell Marque) before blocking as described above. Primary antibody staining was performed overnight at 4 °C with constant agitation followed by three washes with 0.1% (vol/vol) Tween 20. Secondary antibody staining was carried out for 45 min at room temperature with constant agitation followed by three washes with 0.1% (vol/vol) Tween 20. Slides were mounted with ProLong Gold supplemented with DAPI (Invitrogen). For immunohistochemistry detection of GFP-positive outgrowths, the kit Ace IHC Detection Kit (Epitomics) was used according to the manufacturer’s instructions. Tissue sections were analyzed using either the Nikon Eclipse T1 microscope (Nikon) or Zeiss LSM 710 confocal microscope. For whole-mount images, glands were harvested, spread atop a glass slide, defatted, and stained with Carmine Aluminum solution prior image analysis. MRU frequency was estimated using the ELDA algorithm (10). Mammary gland reconstitution was considered successful if, by the time of analysis, at least one-third of the fat pad was repopulated with GFP+ structures.
RNAseq Library Preparation.
Cells were sorted into Eppendorf tubes filled with TRIzol LS (Invitrogen), and RNA purification was performed according to the manufacturer’s instructions. DNase-free RNA samples were used for the preparation of double-strand cDNA libraries using the Version 1 Ovation RNAseq System (Nugen). cDNA libraries were phosphorylated, adenylated, and ligated to Illumina adapters followed by PCR enrichment. Single-ended sequencing was performed for 36 cycles in Illumina GAII instruments (Illumina).
RNAseq Mapping and Analysis.
We used the Refseq transcriptome (mm9 mouse assembly) downloaded through the University of California, Santa Cruz (USCS) Table Browser (31). Reads were mapped in two stages: first, they were mapped to sequences constructed using all annotated Refseq exons with overlapping exons collapsed, and second, they were mapped to all possible junctions formed from all pairs of exons for the same gene. Mapping was done with RMAP (32) and allowed up to three mismatches in 36 bases. Reads mapping ambiguously (including mapping to an exon and a junction) were discarded. For each Refseq transcript, we counted the number of reads with mapping location that was inside the transcript's exons (allowing a given read to be counted for two distinct transcripts as long as the location is unique) or through one of the transcript's junctions. Reads per kilobase per million (RPKM) calculations discarded duplicate reads and corrected gene size for the portion of the gene that cannot be uniquely mapped. Differential expression between two RNAseq experiments was computed using a 2 × 2 contingency table and either a χ2 statistic or Fisher exact test to obtain a P value for differential expression. Briefly, the contingency tables contained, for each gene, the counts of reads mapping into the gene and the counts of reads mapping outside the gene for both experiments. The P values were corrected for multiple testing using the Bonferroni correction. The genes that remained were called as differentially expressed (corrected P > 0.01), and rankings for differentially expressed genes were based on ratios of RPKM values.
Quantitative PCR.
Cells were sorted into 96-well plates containing 30 μL Cell-To-Ct lysis buffer (Ambion). cDNA synthesis was performed according to the manufacturer’s instruction. Real-time PCR was performed using specific Taqman probes (Applied Biosystems) for each gene and Gapdh mRNA as an endogenous control. Samples were run on a 7900 Real-Time PCR System (Applied Biosystems).
BrdU Experiment.
BrdU label-retaining experiments were performed using the BrdU-APC Flow Kit (BD Bioscience); 3-wk-old female mice were injected with BrdU (one time per day for 5 consecutive d, 50 mg/kg body weight), and mammary glands were harvest at specified time points. Mammary gland cells were prepared according to the BrdU manufacturer’s recommendations. Cells were analyzed with an LSRII Cell Analyzer (BD Bioscience), and 1 million cells were recorded per sample. For each experiment (n = 2), three glands were analyzed at week 0 (last day of BrdU injection), and three glands were analyzed at week 12 after BrdU injection.
Knockdown Experiment.
shRNAs against 56 selected genes were pulled and transferred from pGIPz (LMN vector) lentiviral backbone (Open Biosystems) to MSCV-miR30-PGK-NEO-IRES-GFP retroviral backbone (a gift from Christopher R. Vakoc, Cold Spring Harbor Laboratory, New York). Plasmid was transfected into Plat-E cells (33) using Lipofectamine 2000 and vesicular stomatitis virus g-protein (VSVG), and virus was collected 24 and 36 h posttransfection. Cells were infected by spin infection and allowed 2 d for recovery. GFP levels were measured using MACSQuant Cell analyzer (Miltenyi Biotech) from 10,000 cells. Hairpins used on validation experiments were ordered as oligonucleotides from Integrated DNA Technologies (IDT) and used as the template for PCR reactions using KOD hot-start polymerase (EMD Milipore) and the primers 5MIR (5′-CAGAAGGCTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3′) and 3MIR (5′-CTAAAGTAGCCCCTTGAATTCCGAGGCAGTAGGCA-3′). PCR products were column-purified (Qiagen), digested with EcoRI and XhoI enzymes, and cloned into predigested LMN vectors using T4 rapid ligase (Promega).
Supplementary Material
Acknowledgments
We thank Elaine Fuchs, Mikala Egeblad, and Christopher R. Vakoc for valuable mouse stains and reagents. This work was supported by National Institutes of Health Grand Opportunity Award #1 RC2 CA148507 and P01 Award 2P01CA013106.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited at the National Center for Biotechnology Information Short Read Archive (NCBI SRA) [submission study SRP018968 regarding the following samples: Myo differentiated SRX246984 (SRR768419 and SRR768420); Myo progenitor SRX246985 (SRR768421 and SRR768422); luminal alveolar SRX246986 (SRR768423 and SRR768424); luminal progenitor SRX246987 (SRR768425 and SRR768426); luminal ductal SRX246988 (SRR768427 and SRR768428); H2b GFP MaSC SRX246989 (SRR768429, SRR768430, and SRR768431); CD59 MaSC SRX246990 (SRR768432); and CD1d MaSC SRX246991 (SRR768433)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303919110/-/DCSupplemental.
References
- 1.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 3.Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24(17):1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu Z, et al. Embryonic and hematopoietic stem cells express a novel SH2-containing inositol 5′-phosphatase isoform that partners with the Grb2 adapter protein. Blood. 2001;98(7):2028–2038. doi: 10.1182/blood.v98.7.2028. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE, Lindeman GJ. Mammary stem cells and mammopoiesis. Cancer Res. 2006;66(20):9798–9801. doi: 10.1158/0008-5472.CAN-06-2254. [DOI] [PubMed] [Google Scholar]
- 6.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikaelian I, et al. Expression of terminal differentiation proteins defines stages of mouse mammary gland development. Vet Pathol. 2006;43(1):36–49. doi: 10.1354/vp.43-1-36. [DOI] [PubMed] [Google Scholar]
- 8.Smalley M, Ashworth A. Stem cells and breast cancer: A field in transit. Nat Rev Cancer. 2003;3(11):832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 9.Neville MC. Introduction: Transplantation of the normal mammary gland: Early evidence for a mammary stem cell. J Mammary Gland Biol Neoplasia. 2009;14(3):353–354. doi: 10.1007/s10911-009-9152-6. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Asselin-Labat ML, et al. Delineating the epithelial hierarchy in the mouse mammary gland. Cold Spring Harb Symp Quant Biol. 2008;73:469–478. doi: 10.1101/sqb.2008.73.020. [DOI] [PubMed] [Google Scholar]
- 12.Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16(6):700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Adams EJ, López-Sagaseta J. The immutable recognition of CD1d. Immunity. 2011;34(3):281–283. doi: 10.1016/j.immuni.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Qin X, et al. Genomic structure, functional comparison, and tissue distribution of mouse Cd59a and Cd59b. Mamm Genome. 2001;12(8):582–589. doi: 10.1007/s00335-001-2060-8. [DOI] [PubMed] [Google Scholar]
- 15.Haas KM, et al. CD22 ligand binding regulates normal and malignant B lymphocyte survival in vivo. J Immunol. 2006;177(5):3063–3073. doi: 10.4049/jimmunol.177.5.3063. [DOI] [PubMed] [Google Scholar]
- 16.Greenlee MC, Sullivan SA, Bohlson SS. CD93 and related family members: Their role in innate immunity. Curr Drug Targets. 2008;9(2):130–138. doi: 10.2174/138945008783502421. [DOI] [PubMed] [Google Scholar]
- 17.Becker-Herman S, Arie G, Medvedovsky H, Kerem A, Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol Biol Cell. 2005;16(11):5061–5069. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina D, Oborn CJ, Kittrell FS, Ullrich RL. Properties of mouse mammary epithelial cell lines characterized by in vivo transplantation and in vitro immunocytochemical methods. J Natl Cancer Inst. 1986;76(6):1143–1156. [PubMed] [Google Scholar]
- 19.Danielson KG, et al. Clonal populations of the mouse mammary cell line, COMMA-D, which retain capability of morphogenesis in vivo. In Vitro Cell Dev Biol. 1989;25(6):535–543. doi: 10.1007/BF02623566. [DOI] [PubMed] [Google Scholar]
- 20.Deugnier MA, et al. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev Biol. 2006;293(2):414–425. doi: 10.1016/j.ydbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21(24):3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CW, et al. A novel member of the SAF (scaffold attachment factor)-box protein family inhibits gene expression and induces apoptosis. Biochem J. 2007;407(3):355–362. doi: 10.1042/BJ20070170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 24.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12(2):R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dexter DL, et al. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38(10):3174–3181. [PubMed] [Google Scholar]
- 26.Teng MW, et al. Combined natural killer T-cell based immunotherapy eradicates established tumors in mice. Cancer Res. 2007;67(15):7495–7504. doi: 10.1158/0008-5472.CAN-07-0941. [DOI] [PubMed] [Google Scholar]
- 27.Green JE, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: Ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19(8):1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 28.Hix LM, et al. CD1d-expressing breast cancer cells modulate NKT cell-mediated antitumor immunity in a murine model of breast cancer metastasis. PLoS One. 2011;6(6):e20702. doi: 10.1371/journal.pone.0020702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spanoudakis E, et al. Regulation of multiple myeloma survival and progression by CD1d. Blood. 2009;113(11):2498–2507. doi: 10.1182/blood-2008-06-161281. [DOI] [PubMed] [Google Scholar]
- 30.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9(3):255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AD, et al. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25(21):2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.