Abstract
Chaperonins are cage-like complexes in which nonnative polypeptides prone to aggregation are thought to reach their native state optimally. However, they also may use ATP to unfold stably bound misfolded polypeptides and mediate the out-of-cage native refolding of large proteins. Here, we show that even without ATP and GroES, both GroEL and the eukaryotic chaperonin containing t-complex polypeptide 1 (CCT/TRiC) can unfold stable misfolded polypeptide conformers and readily release them from the access ways to the cage. Reconciling earlier disparate experimental observations to ours, we present a comprehensive model whereby following unfolding on the upper cavity, in-cage confinement is not needed for the released intermediates to slowly reach their native state in solution. As over-sticky intermediates occasionally stall the catalytic unfoldase sites, GroES mobile loops and ATP are necessary to dissociate the inhibitory species and regenerate the unfolding activity. Thus, chaperonin rings are not obligate confining antiaggregation cages. They are polypeptide unfoldases that can iteratively convert stable off-pathway conformers into functional proteins.
Keywords: molecular chaperones, protein aggregation, protein misfolding, protein unfolding
Newly synthesized polypeptides, or stress-labile proteins destabilized by mutations, may fail to properly reach their native state and instead form misfolded species that may coalesce further into stable toxic aggregates (1–3). Molecular chaperones, such as the cage-like eukaryotic chaperonin containing t-complex polypeptide 1 (CCT/TRiC) and GroEL/GroES chaperonins and the heat shock protein (Hsp)70/Hsp40s (4, 5), form a first line of cellular defenses against early misfolded species on the cytotoxic protein aggregation pathway, leading to degenerative conformational diseases and aging (6). In vitro, chaperonins may prevent the aggregation of artificially unfolded proteins and use ATP hydrolysis to promote their native refolding but fail to act upon misfolded species already entangled into large aggregates (7, 8).
Eukaryotic cytosolic CCTs, bacterial GroEL, and mitochondrial Hsp60 are double-ring complexes with two central cavities. Their key role in cellular proteostasis is generally thought to be the ATP-driven transient confinement of aggregation-prone polypeptides within the central cavities, allowing “in-cage” protein refolding to the native state. The mechanistic steps first involve spontaneous binding of an unfolded or misfolded polypeptide to exposed hydrophobic residues on the apical domains facing the upper passageway of the chaperonin cavity (9). Next, the ATP-regulated movements of the apical domains cause the dissociation of the tightly bound polypeptide, either within the confined space of the cavity or directly into the external solution, especially in cases of large polypeptides (10). The transient capping of the cavity by a helical lid domain in CCT, or a GroES7 cover in the case of GroEL14 (5, 11), is generally thought to be an obligatory step to confine the released polypeptide within the cage, where it presumably needs to reach its native state while being secluded from other aggregating polypeptides. An allosteric signal from the empty trans-ring then prompts the uncapping of the substrate-containing cis-cavity (12, 13), causing the release of the natively refolded protein. Thus, ATP is thought to drive the reaction cycle of GroEL and CCT by timely alternating between an in-cage sequestration phase, to promote by confinement the spontaneous refolding of an unfolded polypeptide, and a dissociation phase, to release the natively refolded protein from the cage into the solution. However, exceptions also have been reported of large proteins being released directly from the GroEL cage without encapsulation, upon GroES binding to the opposite ring, possibly leading to out-of-cage refolding (14, 15).
Here, we revisit the obligate link between the mechanism by which chaperonins can convert a stable misfolded polypeptide into a native protein and the mechanism of encaging to avoid aggregation of the substrate during the various steps to native refolding. To this aim, we used as a unique type of stringent chaperone substrates in the form of stable misfolded inactive polypeptides that without chaperonins tended neither to aggregate nor to refold spontaneously to the native state. We found that both apoGroEL and apoCCT acted as efficient polypeptide unfolding molecular machines that could rapidly convert an excess of misfolded polypeptide substrates into unfolded intermediates that were released from the chaperonin to slowly reach the native state in solution. However, following several full turnovers of binding, unfolding, release, and out-of-cage refolding, both GroEL and CCT activities gradually became stalled by over-sticky intermediates, whose dissociation required the action of an ancillary regeneration mechanism that depended on ATP hydrolysis and, in the case of GroEL, also on GroES mobile loops (16).
Results
Freeze–Thaw Denatured Rhodanese Is a Stringent Substrate That Is Not Aggregation Prone.
Unstable guanidinium HCl- or urea-unfolded rhodanese has been used in classic in vitro chaperonin assays by virtue of its high propensity to aggregate readily upon removal of the denaturant and form resistant species, unless readily assisted by a large molar excess of GroEL, GroES, and ATP (17). In contrast, here we used freeze–thaw (FT) denatured rhodanese (FTrho) as a unique type of substrate generated beforehand in the absence of GroEL, by iterative FT cycles. Similar to FT-denatured luciferase (FTluc) (18), inactive FTrho was found to be composed of stable, soluble, and mostly monomeric species (Fig. S1A) (19). Indicative of the presence of stable misfolded β-structures, FTrho bound 3.5 times more thioflavin T (Th-T) than native rhodanese (Nrho) (Fig. S1B). Without chaperones, less than 3% spontaneously converted into Nrho in 60 min at 25 °C. Remarkably, no FTrho converted into light-scattering aggregates in 18 h (Fig. S1C). The FTrho species were more resistant than Nrho to urea unfolding (Fig. S2), and FTIR spectroscopy showed the presence of intramolecular misfolded β-sheets different from Nrho (19). Therefore, FTrho was a stringent candidate substrate with a potential to differentiate between the chaperonin’s ability to mediate protein refolding per se and its ability to prevent aggregation.
ApoGroEL Can Catalytically Unfold FTrho Without ATP.
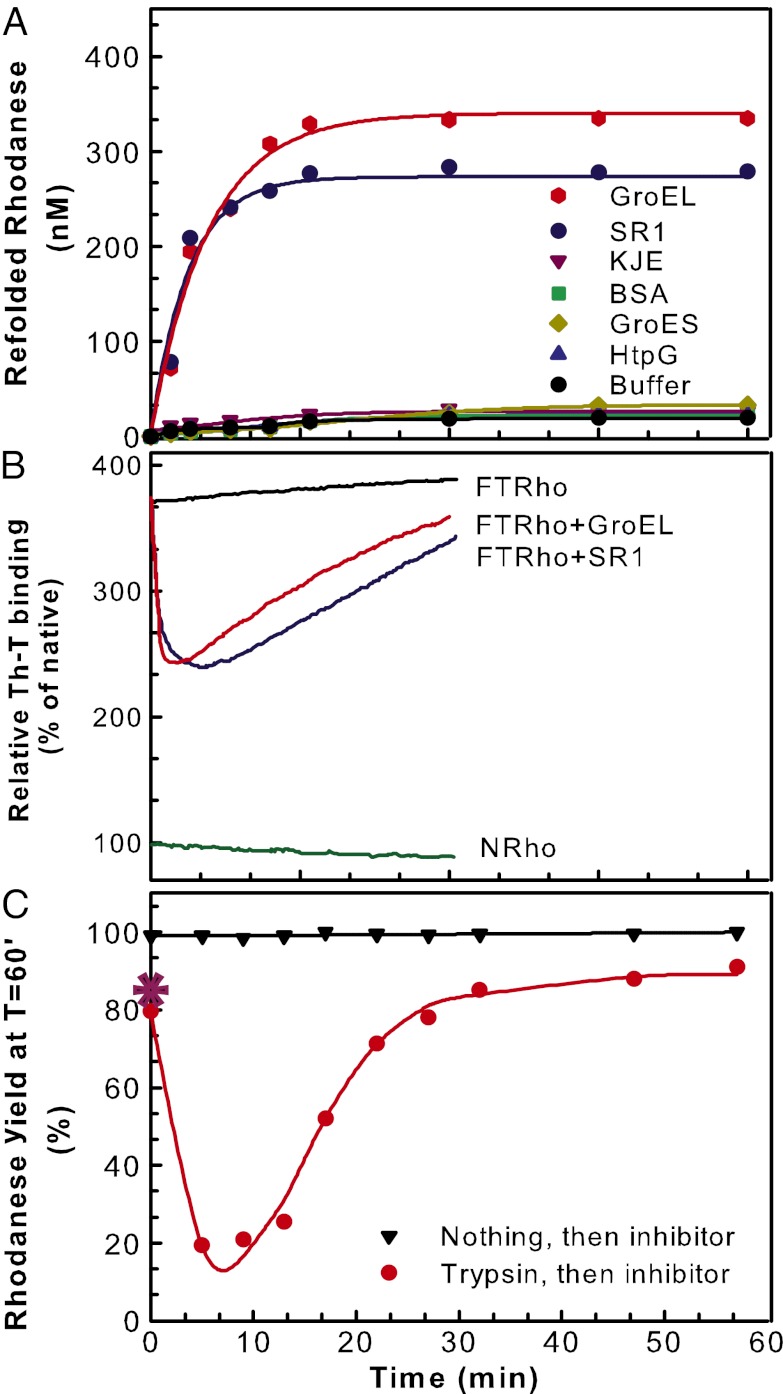
Whereas during 18 h, up to 13% of the FTrho was converted spontaneously to Nrho, addition of equimolar apoGroEL (1 µM protomers, i.e., 71.4 nM of GroEL14) produced the same amount of Nrho in 3 min, corresponding to a 360-fold acceleration of the native refolding reaction. Whereas without ATP other molecular chaperones and proteins in equimolar amounts [DnaK, DnaJ, Caseinolytic peptidase B protein (ClpB), HtpG, GroES, BSA] remained ineffective, up to a third of the substrate (330 nM) was refolded rapidly by apoGroEL in 15 min (T50 = 4 min 30 s; Fig. 1A) and similar high refolding yields were observed with equimolar single-ring GroEL7-mutant SR1 mutants (20). On average, each GroEL7 ring converted half the FTrho substrate per minute, generating 2.3 native products in 30 min. FTrho thus allowed chaperonins to work through multiple turnovers, overcoming the single-turnover limitation observed in previous in vitro refolding assays with aggregation-prone substrates (7, 17). Enzyme activity, kinetics of deuterium exchange, NMR, and FRET spectroscopy previously showed that mere binding to apoGroEL may cause the unfolding of metastable native or misfolding proteins, such as pre–β-lactamase (21), heat-denatured cyclophilin (22), barnase (23), or urea-denatured Ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) (24). However, here we show that a single apoGroEL ring may carry several complete, consecutive turnovers including substrate binding, unfolding, release, and native refolding.
Fig. 1.
GroEL is an ATP-independent catalytic polypeptide-unfolding enzyme. (A) Time course of stringent chaperonin-mediated refolding (reactivation) of FTrho (1 µM) at 22 °C without or with equimolar (expressed in protomers) GroEL14, single GroEL7 ring SR1, GroES7, DnaK + DnaJ + GrpE (KJE), HtpG2, or BSA. (B) Time-dependent net relative changes in FTrho Th-T fluorescence in the presence of equimolar GroEL14, SR17, KJE, or only buffer as in A. Nrho is 100%. (C) Time-lapse trypsin sensitivity of FTrho. FTrho (1 µM) was incubated with equimolar GroEL (1 µM protomers) as in A. Three minutes before the indicated time points, 0.02 mg/mL trypsin was added, and at the indicated time points, trypsin activity was stopped by adding 100 µM trypsin inhibitor (TLCK). Refolding was allowed to continue until 60 min after GroEL addition, and the refolding relative yields (from A, 330 nM = 100%) were plotted against the time at which the (3-min) trypsin treatments were ended by TLCK addition. TLCK was added without (black ▼) or following (red ●) a prior 3-min trypsin treatment. Trypsin and inhibitor pretreatment of GroEL before FTrho addition at T = 0 min did not affect refolding yields (*). Lines in A and C are simple guides for the eyes.
Fluorescence spectroscopy showed that apoGroEL caused a rapid net loss of the high FTrho Th-T signal (T50 = 1 min), indicating that upon GroEL binding, FTrho lost a significant fraction of its intramolecular misfolded β-structures (18, 19), a result compatible with some degree of unfolding. Within 2 min, the apparent unfolding signal leveled and was followed by a slower net regain of Th-T signal, corresponding to the formation of some native β-structures (Fig. 1B), as suggested by a parallel regain of rhodanese activity (Fig. 1A). Time-lapse transient trypsin digestions at various time points of the reaction confirmed that binding of apoGroEL caused an initial decompaction and partial unfolding of the bound FTrho substrate. FTrho thus was incubated before and after GroEL addition with 0.02 mg/mL trypsin and, 3 min later, with an excess of the trypsin inhibitor N-α-tosyl-L-lysine chloromethyl ketone hydrochloride (TLCK). A prior short trypsin treatment of GroEL alone had no effect on its subsequent unfolding/refolding activity, and a prior short trypsin treatment of FTrho or Nrho alone followed by GroEL addition produced 82% and 91% Nrho, respectively, in 60 min (compared with the apoGroEL-mediated yield without trypsin pretreatment set to 100%; Fig. 1C). The similar elevated levels of trypsin resistance of FTrho and Nrho imply that they were comparably compact. Remarkably, when transient trypsin digestion of FTrho was applied 2 min after GroEL addition, only 18% of the refolding yield (mediated by apoGroEL in 60 min) was observed. A marked transient increase of sensitivity of FTrho to trypsin, despite a possible partial recruitment of trypsin by the added GroEL and a possible partial sequestration of the substrate in the cavity, is strong evidence that upon binding, GroEL caused a decompaction and, therefore, the partial unfolding of the compact misfolded FTrho species. When the trypsin digestions were applied later in the GroEL reaction, they progressively became less effective, and the GroEL-mediated refolding yields recovered up to a maximal 87%, with a T50 = 17 min (Fig. 1C). Thus, following an initial rapid drop of the compactness of the substrate, its subsequent slow regain of trypsin resistance is evidence of its slow conversion into new compact native species, as attested by its regained enzymatic activity.
In this reaction, each GroEL heptameric ring thus acted as an ATP-independent enzyme, which first rapidly unfolded the misfolded polypeptide substrate upon binding, then rapidly released it as a species highly sensitive to the trypsin molecules of the external solution, where it slowly refolded to the more compact protease-resistant native state. Other misfolded substrates in excess, by virtue of their low propensity to aggregate, could patiently wait their turn to be unfolded and released by the chaperonin in several successive turnovers. The same results were found with the single-ring SR1 mutants, indicating that the release step did not depend on an allosteric signal from the trans-ring.
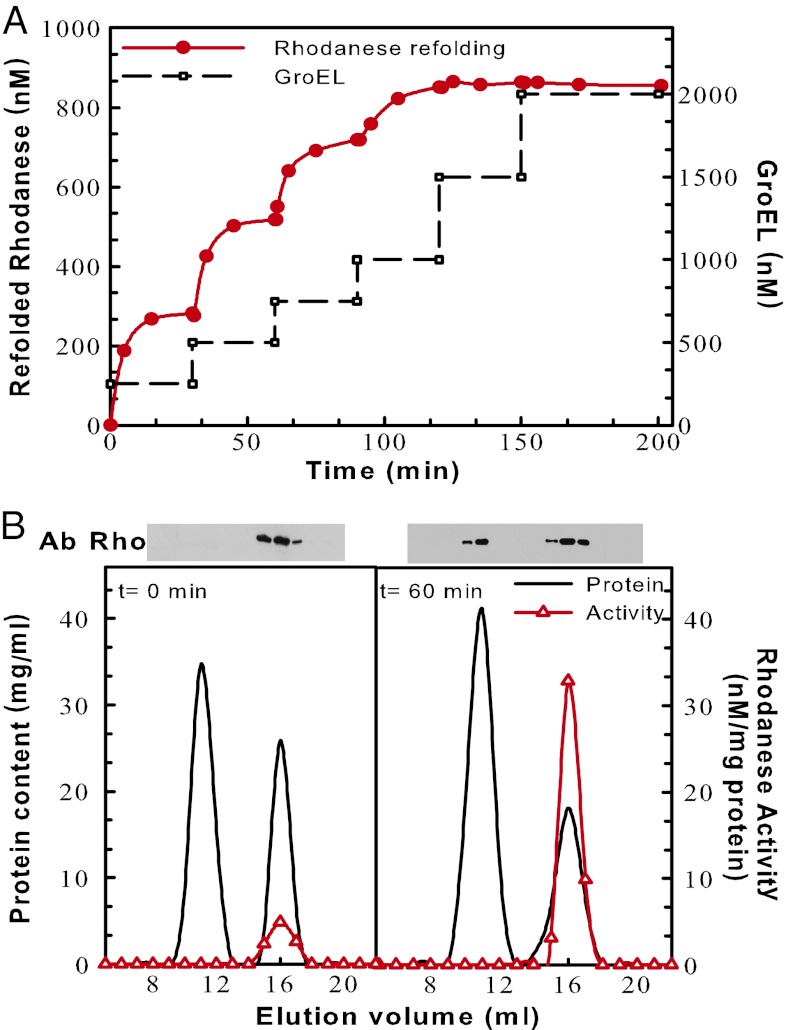
Although exceptionally effective, the catalytic unfolding action of apoGroEL gradually halted after processing two to three misfolded polypeptides per ring. To address the reasons for arrest, we titrated the unfolding/refolding activity of increasing substoichiometric amounts of apoGroEL sequentially added to the reaction every 30 min, to a constant excess FTrho (2 µM). As little as 250 nM GroEL (i.e., 36 nM of GroEL7 active sites) was found to refold up to 280 nM rhodanese in 25 min, corresponding to seven to eight turnovers per GroEL7 ring; then, it stopped (Fig. 2A). The subsequent addition of fresh GroEL (36 nM GroEL7 sites) produced nearly the same amount of Nrho, indicating that the first refolding reaction was not arrested because all the substrate had been consumed or turned into GroEL-resistant species, but rather that the initial GroEL molecules became inhibited. Further additions of substoichiometric amounts of fresh GroEL expectedly processed less and less FTrho, until equimolar GroEL (286 nM of GroEL7 catalytic sites) was found saturating, yielding up to 46% of stable Nrho (Fig. 2A). Gel filtration chromatography, activity, and Western blots of FTrho preincubated with apoGroEL for 0 min or 60 min (Fig. 2B) confirmed that the catalytic unfoldase action of apoGroEL became stalled by inactive over-sticky rhodanese species acting as inhibitors of the unfoldase catalysis.
Fig. 2.
The chaperonin unfoldase activity becomes stalled by over-sticky FTrho species. (A) An excess of FTrho substrate (2 µM) was supplemented stepwise every 30 min, first with substoichiometric amounts, then up to equimolar GroEL protomers (black lines), and rhodanese activity was measured as in Fig. 1A at the indicated time points. (B) GroEL complexes become stalled by over-sticky inactive rhodanese species. FTrho incubated for 0 (Left) or 60 min (Right) with equimolar GroEL as in Fig. 1A was separated by gel filtration chromatography. The eluted fractions were assayed for total protein concentration (black line) and rhodanese activity (red △) and further separated by SDS-gel electrophoresis to detect the presence of rhodanese by Western blots. Lines are simple guides for the eyes.
These observations were tested and confirmed by a mathematical model based on simple thermodynamic and kinetic considerations (SI Text, section S1 and Fig. S3) capturing the basic experimental observations (SI Text, section S1 and Fig. S4), which provided quantitative estimates of the parameters governing the different steps of the catalytic unfoldase cycle mediated by an apoGroEL heptameric ring: binding, unfolding, release, and refolding (Table S1). The model provided insight into the evolution of the various intermediate species produced in the reaction (SI Text, section S1 and Fig. S5). It showed best fits with the data when assuming rapid unfolding on the upper rim of the cavity (T50 < 1 min), followed by a slow out-of-cage refolding (T50 = 6 min 18 s) of the released unfolded FTrho species in solution (Fig. S4). The model could account for the slower refolding rates we observed in the presence of excess (7 µM) compared with limiting (0.5–1 µM) apoGroEL (Figs. S4 and S5), implying that the species released from the cage were in a nonnative, on-pathway state that, at variance with the lowest-affinity Nrho state, could be recaptured transiently by the excess of apoGroEL. Modifying the model to compel the protein to undergo obligate in-cage refolding, with the released product being native and averse to GroEL rebinding, produced much worse fits than when assuming out-of-cage refolding. Moreover, it predicted that an excess of chaperonin would accelerate the reaction, whereas the experiments showed a slowing down. The optimal out-of-cage refolding model thus predicted that binding, unfolding, and release of a first substrate should have been rapid enough (T50 < 1 min) to allow the binding of the next substrate, whereas the first released product continues its slow (T50 = 6 min 18 s) native refolding in solution. This might best account for the ability of each apoGroEL ring to undergo up to eight consecutive turnovers in several minutes (as in Fig. 2A, step one) while accommodating in their cavity, at most, a single 30-kDa substrate at a time.
Role of ATP and GroES.
Because cells contain several millimolars of ATP and mitochondria contain equimolar GroEL and GroES protomers (Table S2), chaperonins will unlikely stay long without binding ATP and GroES. Therefore, next we addressed the effect of ATP and GroES on stalled chaperonins. First, we incubated apoGroEL, apoSR1, or apoCCT with FTrho for 30 min until complete unfolding/refolding arrest was achieved; then, we added ATP, GroES, or both (Figs. 3B and 4).
Fig. 3.
The sequential additive effects of ATP + GroES on GroEL-mediated unfoldase/refoldase activity. (A) The effects of ATP and GroES or mobile loop preincubation on the GroEL unfoldase/refoldase activity. One-micromolar protomers of GroEL (red ■) or SR1 (blue ▿) first were preincubated with 5 mM ATP and 1 µM GroES7 or 5 µM mobile loops (ML) (ETKSAGGIVLTGS; green ○) and subsequently supplemented at T = 0 min with 1 µM FTrho, and the time-dependent refolding rhodanese was measured. For comparison, the refolding of FTrho alone (black ●) or with GroEL (without ATP from Fig 1A; red ◇, dashed line) also is shown with illustrative schemes of the possible GroEL14GroES7, GroEL14(GroES7)2 (34–36), SR17GroES7, or GroEL14(ML7)2 complexes that may form under the various conditions. (B) Net chaperonin-assisted rhodanese refolding. FTrho (1 µM) first was incubated for 10 min in buffer at 22 °C then supplemented with equimolar GroEL protomers as in Fig 2A; then at T = 40 min it was supplemented with only ATP (5 mM), with only equimolar GroES, or with both as indicated. Lines are simple guides for the eyes.
Fig. 4.
CCT can mediate the ATP-independent refolding of both FTrho and FTluc. Time course of stringent chaperonin-mediated refolding of FTrho (1 µM) (A) or FTluc (B) at 22 °C in the presence of buffer (□) or 1 µM bovine CCT hetero-oligomers (protomers) without (blue ○) or with 5 mM ATP (red ●). Lines are simple guides for the eyes.
Remarkably, apoCCT, but not the other chaperones tested, showed the same ability to drive the catalytic refolding of FTrho, with a similar number of turnovers and yields as apoGroEL (Fig. 4A), suggesting that catalytic unfolding/refolding is a particular functional feature of both classes of chaperonins. The presence of ATP from the start (Fig. 4A) or the subsequent addition of ATP to FTrho-stalled–apoCCT complexes caused the release and refolding of about 102 nM Nrho (Fig. S6A). This confirmed that ATP suffices to decrease the affinity for over-sticky polypeptides that may stall the catalytic unfoldase sites of CCT chaperonins. Addition of ATP alone to FTrho-stalled–apoGroEL complexes was less effective at releasing refoldable inhibitory species from GroEL or SR1, suggesting that the ATP-fueled upward and sidewise twisting of the GroEL apical domains (11) remained predominantly frustrated by the bound over-sticky species. GroES addition to FTrho-stalled apoGroEL released a more important fraction of stalling species. This showed that GroES can bind GroEL even without ATP and stabilize the low-affinity state for over-sticky substrates, as previously shown to be the case in GroEL prebound with heat-denatured malate dehydrogenase (MDH) (25). As expected, addition of both ATP and GroES caused an effective net release and native refolding of about 210 nM Nrho (Fig. 3B), demonstrating synergism between the two at dissociating high-affinity species (25). In the case of SR1 single rings, addition of ATP and GroES resulted only in a minor FTrho release and refolding (40 nM Nrho) (Fig. S6B). These species were previously shown to be in-cage refolded under the sealed GroES cap of SR1 (20). Remarkably, GroES and ATP-mediated refolding of apoGroEL-stalled species was six times more effective because of the ability of GroEL trans-ring to mediate the timely dissociation of GroES. Thus, although possible, obligate in-cage refolding appears to be considerably less effective than out-of-cage refolding, as independently suggested from model-based simulations of the data (Figs. S3–S5).
Whereas apoSR1 was as effective as apoGroEL in refolding FTrho, preincubation of SR1 with GroES and ATP before FTrho addition completely inhibited refolding (Fig. 3A). Thus, the known irreversible binding of GroES to SR1, not only prevents the timely early exit of refolding species to complete optimal out-of-cage refolding, but also prevented new misfolded substrates from accessing the binding sites. This suggests that helices H and I, which are involved in substrate binding on the GroEL apical domains (11), also serve as the catalytic sites for unfolding.
To address the role of GroES binding in GroEL regeneration further, we used a minimal fragment of the GroES mobile loop, NH2-ETKSAGGIVLTGS-COOH, that binds GroEL with high specificity but with a lower affinity than whole GroES7 complexes (26). Remarkably, a fivefold molar excess of mobile loop peptides recovered GroEL-stalled FTrho nearly as effectively as equimolar GroES (in protomers; Fig. 3 and Fig. S6D), especially with ATP. Thus, mere binding of mobile loops to apoGroEL sufficed to drive the productive dissociation of sticky inhibitory species from the catalytic unfoldase sites of GroEL14, leading to native refolding, an effect that was however poorly efficient with the SR1 mutants (Fig. S3C). Because individual mobile loops bind far from the cavity’s entry (11, 26), encapsulation and in-cage refolding under sealed GroES7 caps are not obligate steps of the basic catalytic unfolding mechanism by which apoGroEL may convert stable misfolded polypeptides into natively refoldable proteins.
In the case of CCT, ATP alone sufficed to dissociate and refold 106 nM of over-sticky intermediates in 60 min (T50 = 8 min) (Fig. S6A). This difference from GroEL was not unexpected, given that CCTs encode for small apical loops with putative GroES functions (27). Thus, GroEL and CCT strongly resemble each other in terms of ATP-independent catalytic unfoldase/refoldase activity. They both converted unbound misfolded substrates into free out-of-cage native proteins, and when they became stalled by sticky intermediates, they activated an ancillary regeneration mechanism with ATP (and mobile loops) to evict chaperonin-bound intermediates and convert them into free, natively refolded proteins.
FTrho Is Not an Unique Substrate.
Because our results were obtained with a new type of substrate, FTrho, we next questioned whether other chaperonin substrates might be generated with similar characteristics. Early observations suggested the presence of FTrho-like chaperonin-amenable species in native protein stocks, as addition of GroEL + GroES + ATP to presumably “all-native” stocks of rhodanese and pre–β-lactamase recovered 165% and 200% of active enzymes, respectively (21, 28). Here, we further tested whether such inactive species in native stocks might be processed by apoGroEL alone. Addition of apoGroEL + GroES to native stocks of Nrho or MDH produced 5% and 7% more native species, respectively (Fig. S7 A and B). Thus, native protein stocks in general may contain significant amounts of stable inactive misfolded species that can be refolded by apoGroEL. Following dilution of urea-unfolded rhodanese and completion of spontaneous refolding, which reached 30% Nrho (Fig. S7C), addition of apoGroEL also produced a net 4.5% increase of Nrho, and adding apoGroEL + GroES without ATP produced some additional Nrho (Fig. S7C), suggesting that a subpopulation of sticky urea-denatured species binds apoGroEL with an affinity similar to that of the GroEL-stalling FTrho species. Similarly, mild heat denaturation also generated some stable misfolded rhodanese species amenable to apoGroEL (Fig. S7D). Importantly, whereas apoGroEL remained ineffective at refolding FTluc (18), apoCCT was found to efficiently form up to 15%, and in the presence of ATP up to 22%, native luciferase (Fig. 4B). Thus, stable FTrho monomers are not exceptional substrates. Other in vitro denaturing conditions and different proteins may accumulate as apoGroEL-amenable stable inactive species. It is tempting to speculate that in stressed cells too, early misfolded species similar to FTrho, FTluc, and MDH may form and become as readily unfolded by GroEL or CCT and thus rehabilitated into nontoxic functional proteins at a very low ATP cost (6).
Discussion
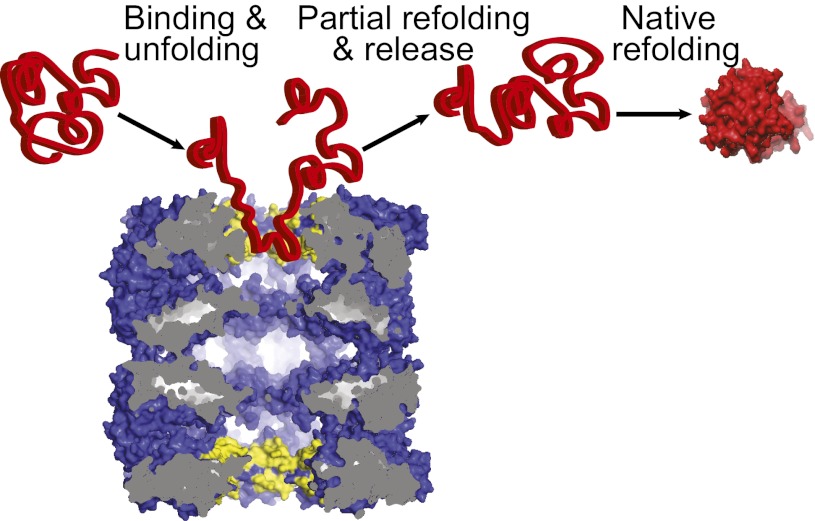
We found that both classes of double-ring cage-like chaperonins act as polypeptide-unfolding enzymes, converting stable high-affinity misfolded polypeptide substrates into unstable high-affinity unfolded intermediates that subsequently refold into low-affinity native products. The unfoldase mechanism did not necessitate ATP or the in-cage confinement of the substrate. This was evidenced by the kinetic model that best fit the data and by comparing the volume of Nrho to that of the cavity in apoGroEL (Fig. 5). FTrho was a compact, structurally damaged inactive species that needed the ATP-dependent action of a large molar excess of DnaK chaperones to become unfolded enough to thereafter undergo spontaneous refolding to the native state (19); yet, we found here that it could become as sufficiently unfolded by equimolar apoGroEL or apoCCT (protomers) without ATP. Our data with FTrho also showed that without nucleotides, GroES7 could cause the release and native refolding of GroEL-bound inactive species, confirming initial observations with GroEL–MDH complexes (25). Although this observation is important for the understanding of the chaperonin mechanism, it is likely irrelevant in cells that contain millimolars of ATP.
Fig. 5.
Scheme of apoGroEL acting as an unfolding catalyst. ApoGroEL (Blue) mediates iterative cycles of binding to the upper cavity (yellow), unfolding, release, and out-of-cage refolding, thereby converting high-affinity misfolded polypeptide substrates (Left) into partially unfolded intermediates (Center) that fold spontaneously in solution into low-affinity native products (Right). Here, native rhodanese is more voluminous than the collapsed upper cavity of apoGroEL.
When FTrho was compelled to confinement in SR1 under a sealed GroES7 cap, this was counterproductive compared with wild-type GroEL14, indicating that for particular substrates, unrestricted out-of-the-cage refolding may be more effective than restricted in-cage refolding. The unfoldase activity was also found to be prone to gradual inhibition by over-sticky intermediates, and the regeneration of the catalytic activity necessitated ATP and the binding of GroES mobile loops. This further indicates that following binding and unfolding, tight confinement under apical loops in CCT, or a whole GroES7 cap in GroEL, is not mandatory for the release of the intermediate. Moreover, our data suggest that when free in solution, folding intermediates may be more at liberty to sample various partially extended conformations to reach the native state than when detrimentally confined deep inside chaperonin cages. Thus, in general, chaperonins do not need to use their cage-like structures to carry their main activity as catalytic polypeptide unfoldases and to avert the formation of early off-pathway misfolded species. This does not exclude that in particular cases the cage-like structures may also act to prevent the aggregation of unfoldase-resistant misfolded polypeptides into potentially more toxic species, but such antiaggregation activity would expectedly inhibit the catalytic unfolding activity.
Relevance of Chaperonins Acting as Unfoldases in the Cell.
A particular class of substrates has been identified by immune pull-down on the basis of their selective ability to remain tightly associated either to GroES-less GroEL particles or exclusively to GroES–GroEL–ADP complexes (29, 30), suggesting that similar to our in vitro data, some polypeptide substrates in cells also may require assistance from GroEL without GroES. Noticeably, we showed here that once unfolded, yet another class of FTrho-like substrates might readily dissociate from GroEL and consequently fail to be identified by immune pull-downs as GroEL substrates.
Given that chaperonins hydrolyze ATP very slowly (0.1–0.5 min−1), this raises the possibility that despite the presence in human cells of millimolars of ATP and equimolar protomers of GroES (Table S2), the misfolded polypeptides sporadically forming during synthesis and under stress may become readily unfolded and released in solution before ATP is significantly hydrolyzed by the chaperonins (Fig. 5). ATP hydrolysis would thus strictly be necessary to fuel structural changes in the chaperonins to increase, against a gradient of free energy, the time they stay in the low-affinity releasing state and thus to recover over-sticky intermediates as native proteins. Together with FTluc and MDH, FTrho might serve as an attractive paradigm for very early-misfolded species on the proteotoxic aggregation pathway, to study the role of chaperones in preventing and curing protein conformational diseases.
Our observation that stable misfolded FTrho-like polypeptides, similar to the 82-kDa aconitase (14), need not fully enter the chamber and stay under a sealed GroES lid suggests that large polypeptides with several misfolded domains might be catalytically unfolded domain by domain, as shown to be the case with chimerical rhodanese fused to GFP or dihydrofolate reductase (31). CCT also was suggested to assist multidomain protein refolding in a domain-by-domain manner, thus mimicking optimal cotranslational folding (32).
Chaperonins are 2% (wt/vol) of the total mass of intracellular proteins in human (HeLa) cells (Table S2) (33). Further experiments beyond the scope of this work are needed to assess the relative importance of iterative catalytic unfolding/refolding and prevention of aggregation by sequestration as complementary mechanisms to delay the onset of protein misfolding diseases and aging (34).
Methods
Proteins.
GroEL and GroES were purified according to standard laboratory procedure (35). His-tagged luciferase was purified as described previously (18) and stored in 15% (vol/vol) glycerol at −80 °C. Bovine rhodanese and pig heart mitochondrial MDH (mtMDH) were purchased from Sigma–Aldrich. Mobile loop peptides were from GenScript. All protein concentrations were estimated by the Bradford Assay and mentioned as protomer concentrations unless otherwise mentioned.
Protein Activity Measurements.
Rhodanese activity was measured by a colorimetric method (monitored at 460 nm) based on the formation of the complex between ferric ions and one of the reaction products, thiocyanate (17). Luciferase activity was measured using a Victor Light 1420 Luminescence Counter (Perkin–Elmer) as described previously (18). The activity of MDH was measured by following the time-dependent oxidation of NADH by mtMDH at 340 nm (25).
Chaperone Refolding Assays.
Refolding assays were performed in refolding buffer [50 mM Tris acetate (pH 7.8), 150 mM KCl, 20 mM MgCl2] in the presence or in the absence of ATP (5 mM) with different chaperone concentrations, as mentioned in the figure legends. For rhodanese, the refolding buffer also included 50 mM Na2S2O4, 10% (vol/vol) glycerol, 2 µM BSA, and 20 mM DTT. Rates of rhodanese, luciferase, and MDH refolding were derived from the linear phase of the time curves of recovered enzymatic activity.
Trypsin Sensitivity of Rhodanese.
For trypsin digestion, 1 µM FTrho was incubated with trypsin (0.02 mg/mL) for 3 min and the digestion was stopped by adding 100 µM of the trypsin inhibitor TLCK (Sigma–Aldrich). The trypsin digestion during GroEL-assisted unfolding/refolding of FTrho at different time points was followed by measuring yields 60 min after GroEL addition, regardless of when the transient trypsin treatment was applied. Time 0: the 3′ trypsin treatment was applied to FTrho and stopped by TLCK addition before GroEL addition at T = 0 min.
Th-T fluorescence and light-scattering measurements were as described earlier (19).
Denaturation of Rhodanese.
The misfolded monomeric luciferase was prepared by freeze–thawing as described in ref. 18. The misfolded monomeric rhodanese was prepared in general according to Natalello et al. (19). In short, 2 µM Nrho in 20 mM Na phosphate (pH 7.5), 12.5 mM Na thiosulfate, and 20 mM DTT was denatured by five to eight consecutive cycles of rapid freezing at −160 °C and slow thawing at 18 °C. When inactivated more than 90%, aggregates were removed by 5′ centrifugation at 20,000 × g and the supernatant was concentrated to a final concentration of 2–3 µM of mostly inactive FTrho species. For heat denaturation, rhodanese (5 µM) was incubated for 10 min at 62 °C in 50 mM Tris⋅HCl, 150 mM KCl, and 20 mM MgCl2, pH 7.5. The residual activity after heat exposure was <2% of the initial. For urea-unfolded rhodanese, rhodanese was incubated in different concentrations of urea up to a maximum of 8 M for 1 h at 25 °C.
Size-Exclusion Chromatography.
To isolate GroEL stalled with misfolded inhibitory rhodanese, mixtures of GroEL and rhodanese at time 0 and 60 min were passed through size-exclusion chromatography in buffer (50 mM Tris⋅HCl, 150 mM potassium chloride, and 20 mM magnesium chloride, pH 7.5) using a Superose 6 HR10/30 gel filtration column (Amersham Pharmacia Biotech, GE Healthcare) at a flow rate of 0.5 mL/min. The absorbance was monitored at 280 nm. Apparent molecular weights were estimated by gel filtration of standard proteins (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Elena Bochkareva for the gift of GroEL and GroES, José Maria Valpuesta for the gift of CCT, Matthias Mayer for HtpG, Avital Parnas for unshown preliminary experiments with acid-denatured MDH, Dan S. Tawfik for discussions, and Alberto Merli and Anika Braune for discussions and technical assistance. We also thank the Herbette Foundation of the University of Lausanne for partial financing of P.G.’s research in the A.A. laboratory. This project was financed by Swiss National Science Foundation Grants 125502/1 and 140512/1 (to P.G.) and Israel Science Foundation Grant 452/09 (to A.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219867110/-/DCSupplemental.
References
- 1.Sharma SK, De Los Rios P, Goloubinoff P. Probing the different chaperone activities of the bacterial HSP70-HSP40 system using a thermolabile luciferase substrate. Proteins. 2011;79(6):1991–1998. doi: 10.1002/prot.23024. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 3.Jaenicke R. Protein stability and protein folding. Ciba Found Symp. 1991;161:206–216. discussion 217–221. [PubMed] [Google Scholar]
- 4.Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: An unfolding story. Curr Protein Pept Sci. 2009;10(5):432–446. doi: 10.2174/138920309789351930. [DOI] [PubMed] [Google Scholar]
- 5.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 6.Hinault MP, Ben-Zvi A, Goloubinoff P. Chaperones and proteases: Cellular fold-controlling factors of proteins in neurodegenerative diseases and aging. J Mol Neurosci. 2006;30(3):249–265. doi: 10.1385/JMN:30:3:249. [DOI] [PubMed] [Google Scholar]
- 7.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 8.Buchner J, et al. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991;30(6):1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- 9.Fenton WA, Kashi Y, Furtak K, Horwich AL. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371(6498):614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 10.Motojima F, Yoshida M. Polypeptide in the chaperonin cage partly protrudes out and then folds inside or escapes outside. EMBO J. 2010;29(23):4008–4019. doi: 10.1038/emboj.2010.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clare DK, et al. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell. 2012;149(1):113–123. doi: 10.1016/j.cell.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horovitz A, Willison KR. Allosteric regulation of chaperonins. Curr Opin Struct Biol. 2005;15(6):646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Horovitz A, Fridmann Y, Kafri G, Yifrach O. Review: Allostery in chaperonins. J Struct Biol. 2001;135(2):104–114. doi: 10.1006/jsbi.2001.4377. [DOI] [PubMed] [Google Scholar]
- 14.Farr GW, et al. Folding with and without encapsulation by cis- and trans-only GroEL-GroES complexes. EMBO J. 2003;22(13):3220–3230. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inbar E, Horovitz A. GroES promotes the T to R transition of the GroEL ring distal to GroES in the GroEL-GroES complex. Biochemistry. 1997;36(40):12276–12281. doi: 10.1021/bi9714870. [DOI] [PubMed] [Google Scholar]
- 16.Landry SJ, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Gierasch LM. Characterization of a functionally important mobile domain of GroES. Nature. 1993;364(6434):255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza JA, Rogers E, Lorimer GH, Horowitz PM. Chaperonins facilitate the in vitro folding of monomeric mitochondrial rhodanese. J Biol Chem. 1991;266(20):13044–13049. [PubMed] [Google Scholar]
- 18.Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. 2010;6(12):914–920. doi: 10.1038/nchembio.455. [DOI] [PubMed] [Google Scholar]
- 19.Natalello A, et al. Biophysical characterization of two different stable misfolded monomeric polypeptides that are chaperone-amenable substrates. J Mol Biol. 2013;425(7):1158–1171. doi: 10.1016/j.jmb.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 20.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84(3):481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 21.Laminet AA, Ziegelhoffer T, Georgopoulos C, Plückthun A. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the beta-lactamase precursor. EMBO J. 1990;9(7):2315–2319. doi: 10.1002/j.1460-2075.1990.tb07403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahn R, Spitzfaden C, Ottiger M, Wüthrich K, Plückthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994;368(6468):261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]
- 23.Zahn R, Perrett S, Fersht AR. Conformational states bound by the molecular chaperones GroEL and secB: A hidden unfolding (annealing) activity. J Mol Biol. 1996;261(1):43–61. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z, Madan D, Rye HS. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol. 2008;15(3):303–311. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Zvi AP, Chatellier J, Fersht AR, Goloubinoff P. Minimal and optimal mechanisms for GroE-mediated protein folding. Proc Natl Acad Sci USA. 1998;95(26):15275–15280. doi: 10.1073/pnas.95.26.15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LL, Sigler PB. The crystal structure of a GroEL/peptide complex: Plasticity as a basis for substrate diversity. Cell. 1999;99(7):757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- 27.Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. Chaperonins: Two rings for folding. Trends Biochem Sci. 2011;36(8):424–432. doi: 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Smith KE, Fisher MT. Interactions between the GroE chaperonins and rhodanese. Multiple intermediates and release and rebinding. J Biol Chem. 1995;270(37):21517–21523. doi: 10.1074/jbc.270.37.21517. [DOI] [PubMed] [Google Scholar]
- 29.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402(6758):147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 30.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122(2):209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Kipnis Y, Papo N, Haran G, Horovitz A. Concerted ATP-induced allosteric transitions in GroEL facilitate release of protein substrate domains in an all-or-none manner. Proc Natl Acad Sci USA. 2007;104(9):3119–3124. doi: 10.1073/pnas.0700070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol. 2005;12(3):233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 33.Finka A, Goloubinoff P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 2013 doi: 10.1007/s12192-013-0413-3. 10.1007/s12192-013-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinault MP, et al. Stable alpha-synuclein oligomers strongly inhibit chaperone activity of the Hsp70 system by weak interactions with J-domain co-chaperones. J Biol Chem. 2010;285(49):38173–38182. doi: 10.1074/jbc.M110.127753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd MJ, Viitanen PV, Lorimer GH. Hydrolysis of adenosine 5′-triphosphate by Escherichia coli GroEL: Effects of GroES and potassium ion. Biochemistry. 1993;32(33):8560–8567. doi: 10.1021/bi00084a024. [DOI] [PubMed] [Google Scholar]
- 36.Azem A, Diamant S, Kessel M, Weiss C, Goloubinoff P. The protein-folding activity of chaperonins correlates with the symmetric GroEL14(GroES7)2 heterooligomer. Proc Natl Acad Sci USA. 1995;92(26):12021–12025. doi: 10.1073/pnas.92.26.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt M, et al. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265(5172):656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.