Abstract
Dosage compensation, the equalized X chromosome gene expression between males and females in Drosophila, has also been found in triple X metafemales. Inverse dosage effects, produced by genomic imbalance, are believed to account for this modulated expression, but they have not been studied on a global level. Here, we show a global expression comparison of metafemales (XXX; AA) with normal females (XX; AA) with high-throughput RNA-sequencing. We found that the majority of the X-linked genes in metafemales exhibit dosage compensation with an expression level similar to that of normal diploid females. In parallel, most of the autosomal genes were expressed at about two-thirds the level of normal females, the ratio of inverse dosage effects produced by the extra X chromosome. Both compensation and inverse effects were further confirmed by combination of X-linked and autosomally located miniwhite reporter genes in metafemales and relative quantitative PCR of selected genes. These data provide evidence for an inverse dosage component to X chromosome compensation.
Keywords: aneuploidy, epigenetics, transcription
In Drosophila, the expression of genes on the monosomic X chromosome in males is up-regulated approximately twofold to equal the level of the two X chromosomes in females in a process referred to as dosage compensation, which has been hypothesized to be accomplished by the male specific lethal (MSL) complex (1, 2). The protein components of this complex are associated with the X chromosome in males but not in females (1, 3–5). However, there is also evidence for dosage compensation of X chromosomal genes in triple X metafemales (6–10), which have no MSL complex. When the MSL2 protein was ectopically expressed in females and metafemales, no changes of X chromosome expression were observed (10). In addition, no loss of dosage compensation occurred and many autosomal genes were up-regulated in the maleless (mle) mutant males in which the complex is dissociated (11–14). From these data, it was suggested that the MSL complex did not act as the direct mediator of dosage compensation. In contrast, inverse dosage effects produced by genomic imbalance are believed to play an important role in modulation of X expression and some other genotypes with compensation (8–10, 15–17).
The inverse dosage effect refers to the negative relationship between gene expression and chromosomal dosage. The effect has been modeled to result from the impact of altered subunit stoichiometry on the assembly and function of multisubunit molecular regulatory complexes (15–18). This response has been demonstrated by gene expression analyses in dosage series of chromosome arms in maize (15, 16, 19) to involve several modulations across the genome, both in the varied and nonvaried segments, and similarly in Drosophila (10, 20–22). The pervasiveness of this effect is illustrated by a compilation of 38 segmentally autosomal trisomic effects on only eight random gene products (23–28) (Table 1). Over 80% of the segments affected the expression of at least one gene (Table 1). Although the detected modifications may be either positive or negative correlations with the chromosome dosage, negatively acting inverse dosage effects are most prevalent (15, 16, 19) (Table 1). When chromosomal segments of substantial size are reduced from two doses to one in a species in which it is possible, dosage compensation will result for many of the genes on the varied segment via an inverse dosage effect increasing the expression of genes on the single chromosome (15, 16, 19).
Table 1.
Selected literature compilation of inverse dosage effects in segmental trisomics
| Trehalase |
||||||||||
| Region | αGPDH | αGPO | SDH | Ddc | Dnase-1 | Fumarase | Kf | Abdomen | Soluble thorax | Insoluble thorax |
| 21A 25A | − | |||||||||
| 25A 27E | 1.45 | 0.69 | ∼1.23 | 0.75 | ||||||
| 27E 30F | 1.2 | |||||||||
| 30F 35BC | 0.78 | |||||||||
| 35BC 38C | 0.76 | 0.77 | ||||||||
| 36EF 37D | 1.51 | |||||||||
| 37D 38C | 0.83 | |||||||||
| 38C 41 | 0.44 | |||||||||
| 41 43C | − | |||||||||
| 40 43C | 0.8 | 0.83 | 0.84 | |||||||
| 43C 45F | 0.65 | 0.85 | 0.81 | 0.74 | ∼0.78 | 0.77 | ||||
| 45F 47E | 0.68 | 0.82 | ||||||||
| 47E 50C | ||||||||||
| 50C 52E | 1.46 | − | ||||||||
| 52E 54F | 0.73 | |||||||||
| 54F 57B | ||||||||||
| 56D 57B | 0.84 | |||||||||
| 57B 59B | 0.73 | |||||||||
| 59B 60F | ||||||||||
| 61A 64E | − | 0.73 | ||||||||
| 64E 67C | 0.79 | − | ||||||||
| 67C 70C | 0.82 | + | 0.77 | |||||||
| 70C 74A | 0.71 | − | ∼0.68 | 0.81 | ||||||
| 72D 76B | ∼0.83 | |||||||||
| 74A 76F | ||||||||||
| 76E 79D | − | |||||||||
| 74A 79D | 0.52 | 0.84 | 0.71 | |||||||
| 79D 83CD | 0.77 | 0.8 | 0.85 | 0.81 | ||||||
| 83EF 86B | 0.85 | + | 0.85 | |||||||
| 86B 88C | 0.65 | 0.77 | 0.85 | |||||||
| 88C 91B | 0.76 | 0.64 | ||||||||
| 90E 91B | − | |||||||||
| 91B 93F | 0.62 | 0.78 | ∼1.25 | |||||||
| 93F 96A | 0.73 | − | 0.8 | ∼0.83 | 0.75 | |||||
| 96A 97F | − | |||||||||
| 96A 97F female | 0.8 | |||||||||
| 97F 100F | 0.73 | 0.7 | 0.61 | |||||||
| 101A 102F | ||||||||||
Enzyme activity ratio of segmental trisomics to euploids is shown. Underlining or “+” indicates increased expression indicative of a dosage effect and location of the gene, and “−” indicates inverse dosage effect (values <0.85). Data compiled from specific references were as follows: α-glycerophosphate dehydrogenase (αGPDH) (23), α-glycerophosphate oxidase-1 (αGPO) (23), succinate dehydrogenase (SDH) (23), dopa decarboxylase (Ddc) (24), DNase (Dnase-1) (25), fumarase (26), kynurenine formamidase (Kf) (27), and Trehalase (28).
Results
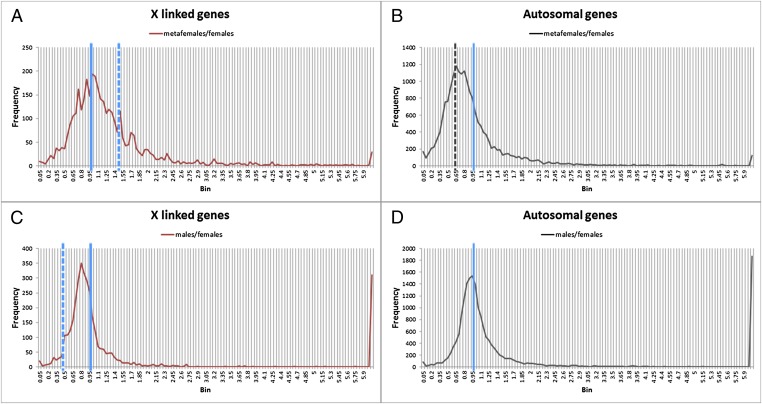
To address whether dosage compensation and inverse effects are present on a global genomic scale in triple X females, we performed high-throughput RNA-sequencing experiments using total mRNA from third instar larvae of metafemales (XXX), females (XX), and males (XY) from the same cultures (Fig. S1A). Gene reads from three biological replicates were averaged, and a ratios distribution analysis for all expressed isoforms was conducted by comparing the metafemales (XXX) with normal females (XX) (Fig. 1), which were plotted in bins of 0.05 increments. This type of analysis provides strong power to determine trends of gene expression because hundreds of data points (transcript isoforms) are present in the major bins. From the frequency distribution of X-linked genes (Fig. 1A), we found that the majority of the genes on the triple X chromosomes have similar total expression levels as normal diploid females, with the highest peak at a ratio of 1.0 (no change). Furthermore, the whole distribution is roughly centered around 1.0, which indicates that dosage compensation of the X chromosomes occurs in metafemales; otherwise, the distribution would be centered around 1.5, the value indicative of gene dosage effects (3/2). Indeed, some gene dosage effects were detected as a minor peak at a ratio of 1.5, representing a set of genes that do not compensate. Another minor peak is present near the ratio 0.75, which is indicative of overcompensation and is likely to represent X-linked genes doubly inversely affected (see below). Similar cases of dosage compensation have been reported in other aneuploids (15, 16, 20, 22).
Fig. 1.
Global endogenous gene expression ratio distributions of metafemales (XXX; AA) compared with normal females (XX; AA). Metafemales and normal females were obtained from crosses of C(1)DX, y w f/Y females with y+ w+ f+/Y males. The global expression pattern of these genotypes was obtained by mRNA-sequencing and analyzed by generating the ratio distributions. Ratio distributions of endogenous genes from metafemales were compared with those of normal females, separated into X-linked genes (A) and autosomal genes (B). Ratio distributions of expressed endogenous genes from males were compared with those of normal females, separated into X-linked genes (C) and autosomal genes (D). The solid blue line represents the ratio “1.0” (no change), the dashed blue line represents the ratios “1.5” (A) and “0.5” (C) (the ratio of gene dosage effects), and the dashed black line (B) shows the ratio “0.67” [the ratio of inverse dosage effects (2/3)]. Three biological replicates were performed for each genotype. A K-S test of the X chromosome distribution revealed a significant deviation from a normal distribution for a dosage effect surrounding a value of 1.50 (P < 1.0 × 10−17). A K-S test for autosomal genes revealed a significant deviation from a normal distribution for no effect surrounding the value of 1.00 (P < 1.0 × 10−17).
As illustrated previously (10, 15, 16, 18, 19, 29), these inverse dosage effects modulate genes not only in the varied segments but elsewhere in the genome. Our analysis of triple X females illustrates this relationship. We analyzed the ratios distribution in the various comparisons for autosomal genes (Fig. 1B). The highest frequency of the bin distribution for metafemale/female ratios is centered around 0.70, which is almost identical to the ratio of an inverse dosage relationship (2/3) caused by the triple X chromosomes compared with diploid females (XX), suggesting the expression of many autosomal genes is shifted downward. A shoulder of the distribution is present at 1.0, which represents no change. A minor peak surrounds the ratio of 0.44, which is a doubled inverse effect that has also been observed in segmental trisomic studies (23) (Table 1) and suggests independent and additive effects. As noted above, the minor peak at about 0.75 for X-linked genes likely represents the same phenomenon. Selected gene expressions from RNA-sequencing with different locations on the X chromosome and autosomes were validated using relative quantitative PCR (Table 2) with an exogenous control, from which similar ratios were found.
Table 2.
Validation of RNA-sequencing using relative quantitative PCR
| Gene name | Location | RNA-sequencing, metafemale/female | Relative quantitative PCR | Max | Min |
| Ssp3 | 2L | 0.67 | 0.602 | 0.755 | 0.48 |
| Jet | 2L | 1 | 1.179 | 1.325 | 1.048 |
| Ssrp | 2R | 0.67 | 0.524 | 0.576 | 0.477 |
| Magu | 2R | 1 | 1.132 | 1.273 | 1.007 |
| Lysp | 3L | 0.167 | 0.114 | 0.136 | 0.097 |
| CG16758 | 3L | 0.512 | 0.259 | 0.304 | 0.221 |
| Oxt | 3L | 1 | 1.043 | 1.311 | 0.829 |
| Kap-alpha1 | 3L | 1 | 0.942 | 1.028 | 0.863 |
| Ac76E | 3L | 1.98 | 1.34 | 1.607 | 1.117 |
| Sp7 | 3R | 0.37 | 0.214 | 0.252 | 0.181 |
| Gish | 3R | 0.67 | 0.595 | 0.655 | 0.541 |
| Nup358 | 3R | 0.67 | 0.693 | 0.791 | 0.607 |
| Rpl3 | 3R | 1.18 | 1.046 | 1.23 | 0.89 |
| Sw | X | 1.5 | 1.341 | 1.435 | 1.253 |
| Out | X | 1.5 | 1.313 | 1.443 | 1.196 |
| Ag5r2 | X | 1.5 | 1.571 | 1.675 | 1.473 |
| TLK | X | 1 | 0.932 | 1.01 | 0.86 |
| Ing3 | X | 1 | 0.954 | 1.204 | 0.756 |
| CG9577 | X | 1 | 1.028 | 1.192 | 0.886 |
| Myb | X | 1 | 0.908 | 1.08 | 0.763 |
| Eo-ry | X | 1.4 | 1.475 | 1.719 | 1.265 |
| fog | X | 0.96 | 0.928 | 1.126 | 0.764 |
| Karl | X | 0.829 | 0.574 | 0.671 | 0.492 |
| CG15771 | X | 1.76 | 1.529 | 1.924 | 1.215 |
Ratios of selected gene expression when metafemales (XXX; AA) were compared with normal females (XX; AA) are shown. Three biological and technical replicates were applied to each genotype in RT-PCR. The relative quantification (RQ) for each pair of primers was measured based on the ΔCt analysis according to the instructions from the manufacturer (7300 Real-time PCR system, sequence detection software version 1.3.1; Applied Biosystems). The data were analyzed with calculation of the 95% confidence interval. The average RQ of each gene with its maximum RQ (Max) and minimal RQ (Min) is shown.
By the nature of sequencing reactions, it is necessary to adjust the read count in each lane to the total because of the high variability from one sample to another. When a majority of mRNA is modulated in the same direction in the experimental sample relative to the control, the potential exists that the magnitude of such effects would be diminished (30). In many analyses of sex chromosome dosage compensation, sex chromosome expression is specifically normalized to autosomal expression on the assumption that there is no change of the latter. Beyond the requisite sequencing normalization, we have treated the X and autosomal expression values independently because the nature of the inverse effect will partially cancel itself. This approach is justified in two ways. First, minor peaks of gene dosage effect for the X or no change for the autosomes conform to the expected values. Second, the effects were phenotypically confirmed on an absolute level as described below.
Male/female ratios were also examined for the X and the autosomes. The X-linked genes formed a major peak slightly below the value of 1.0, which would represent full compensation. A clear shoulder is present at about 0.50, which represents no compensation. Autosomal ratios surround a value of 1.0, which represents no change. A major spike to the far right represents genes with large quantitative differences between the sexes.
In the few experimentally produced aneuploids in which monosomics and the corresponding trisomics of the same chromosomal region can be produced (15, 16, 19), or in the case of single genes that produce an inverse dosage effect (18, 29), there is often but not universally the opposite effect in the monosomics vs. trisomics. To determine if this relationship exists for the X chromosome, transcript isoforms in the metafemale/female ratio distribution in the compensated range of 0.9–1.1 were matched from the male/female comparison. The distribution of this subset of isoforms, which are closely centered around the value of compensation in metafemales, is similar to the full distribution of male/female ratios. There is a small peak around the ratio of 0.50 indicating a small group of genes that are not compensated in males but are compensated in metafemales (Fig. S2). A Kolmogorov–Smirnov (K-S) test of the subset distribution compared with the parental distribution indicated that the subset distribution was significantly different (P < 5 × 10−5).
Reciprocally, the transcript isoforms that exhibit a dosage effect in metafemales in the range of 1.40–1.60 were selected from the male/female comparisons, and their distribution was plotted. The distribution is similar to the overall male/female pattern but with a peak near the range of no compensation. This analysis indicates that the genes showing a dosage effect in metafemales do not necessarily fail to show compensation in males compared with females, but there is overlap (Fig. S3). A K-S test indicated that the two distributions are not significantly different (P = 0.10).
These comparisons of compensation and dosage effects between metafemales and males should be tempered with several considerations. First, selection for sex-biased expression based on optimization for sex-specific functions might be independent to some degree of the process of dosage compensation, producing a different response in males compared with metafemales. Second, selection has been operative on male X chromosomal expression levels since the degeneration of the homolog to produce the heteromorphic sex chromosome condition, whereas there has been no natural selection operating to produce compensation in metafemales, again providing the potential for divergent responses. Third, there is evidence for a lack of gene dosage effect, or buffering, in deficiency heterozygotes (31–33) that is potentially independent of the inverse dosage effect and could account for compensation in males but not metafemales.
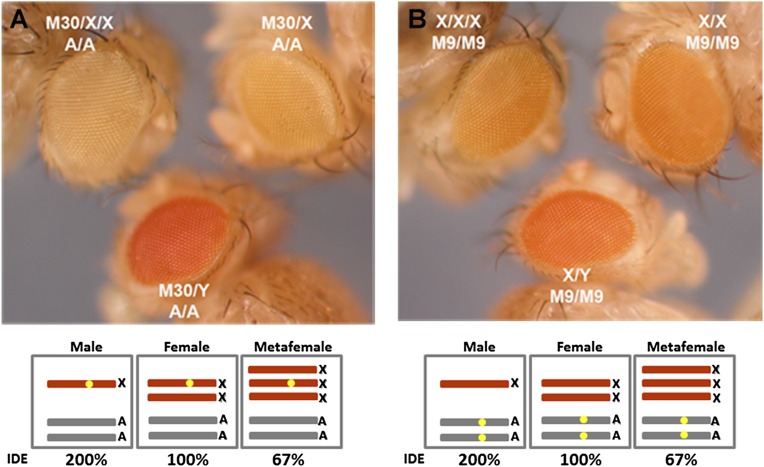
With the availability of miniwhite reporter genes on the X chromosome (M30) and the autosomes (M9) (34), we made crosses (Fig. S1 B and C) to obtain metafemales with these reporter genes (Fig. 2) to test phenotypically whether inverse dosage effects and dosage compensation occurred in metafemales. The endogenous white gene resides on the X chromosome, but it is mutant in this material, allowing white transgenes in different genomic locations to be assayed phenotypically. Metafemales rarely survive to the adult stage but were, in fact, used originally to establish that dosage compensation occurs in this genotype (6). This test provides an absolute phenotypic validation of the RNA analyses. When one copy of the X chromosome M30 reporter was present (Fig. S1B) on one X chromosome in males, females, and metafemales, the highest expression level was detected in males, an intermediate level was detected in females, and the lowest expression was detected in metafemales (Fig. 2A). If the X chromosomes are dosage-compensated with different doses (6–10), total expression (set to 200% total in diploids) will be divided by the number of chromosomes. In other words, a single X chromosome has a 200% (200%/1) expression level in males (XY), 100% (200%/2) expression level in females (XX), and 67% (200%/3) expression level in metafemales (XXX). Thus, the results with the single M30 reporter showing a gradient of eye color are consistent with an inverse dosage effect operating on white in each X chromosomal genotype. These three genotypes of flies were also produced with the autosomal M9 reporter (Fig. S1C). The varied number of X chromosomes shows an influence on the expression of this reporter with a similar gradient expression trend (Fig. 2B), which further confirmed the autosomal expression near 67% found in metafemales.
Fig. 2.
Expression comparisons of reporter genes in males (XY; AA), females (XX; AA), and metafemales (XXX; AA). Metafemales with the reporter genes were obtained from the described crosses (Fig. S1). (A) Eye color comparisons of an X-linked reporter, M30. One copy of the miniwhite reporter was present in males (M30/Y; A/A) (Lower), females (M30/X; A/A) (Upper Right), and metafemales (M30/X/X; A/A) (Upper Left). (B) Eye color comparisons of an autosomal reporter, M9. The homozygous miniwhite reporter gene was examined in males (X/Y; M9/M9) (Lower), females (X/X; M9/M9) (Upper Right), and metafemales (X/X/X; M9/M9) (Upper Left). Each genotype with the reporter gene is shown in the box, and the reporter gene is designated by the yellow dot. IDE, Inverse Dosage Effect.
Discussion
Given the generality of the inverse effect across phylogenetic distances, it seems likely that other cases of sex chromosomes might use this effect to mediate compensation or, alternatively, must mute the effect to prevent detrimental effects of the change of sex chromosomal dosage. The inverse effect is prevalent in data from maize and Drosophila, which are obviously widely separated phylogenetically. This fact suggests that the inverse effect is the reflection of generalized mechanisms of gene expression in eukaryotes. Studies of gene expression in yeasts and mammals in disomics or trisomics have reported differing results as to compensation and trans-effects (e.g., refs. 35–38) vs. claims of primarily dosage effects of the varied genes (e.g., refs. 39, 40). Here, a substantial fraction of the Drosophila genome (∼20%) was varied and a mosaic pattern of effects was observed, but with a majority of genes being inversely affected. Thus, the impact might be considered to be greater than varying individual chromosomes in other species that constitute much smaller fractions of the genome. Single chromosomes or segments might therefore be expected to exhibit a spectrum of effects. Indeed, the inverse effect can be reduced to the action of single genes whose function is integral to gene expression (e.g., ref. 18). Because of the heterogeneous nature of the genes that produce an inverse dosage effect, it has been hypothesized that the effect is a reflection of the kinetics of macromolecular complex function rather than a particular type of activity (17). Thus, it seems unlikely that the kinetics responsible would be unique to different taxa unless some other process counteracts these effects. As noted above, the inverse effect, by its nature, will be obscured if expression values of the varied chromosome are normalized to the remainder of the genome and produce falsely generated dosage effects for the varied chromosome. The fact that the inverse effect can be visualized phenotypically and reduced to the action of single genes (refs. 8, 9, 18, 22, 29, 41 and this work) illustrates it is not itself an artifact of normalization.
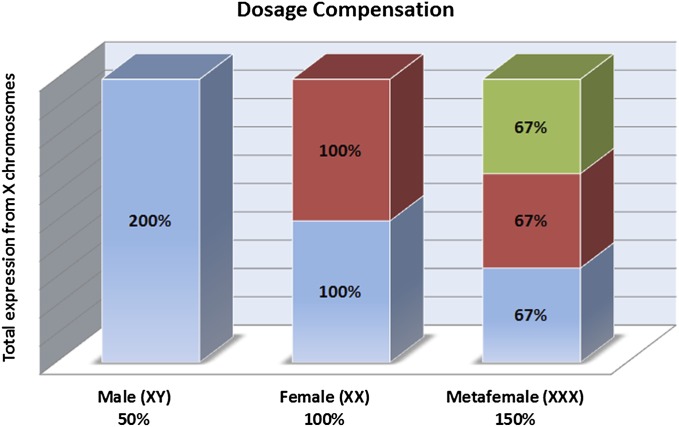
The combination of a gene dosage effect and an inverse effect will result in dosage compensation. When a gene dosage effect of structural genes is set to 100% in normal diploid females, the effect will be 50% in single X males and 150% in triple X metafemales. However, when the inverse dosage effect on each X chromosome in normal diploid females is set to 100%, the effect produced by the varied X chromosome will be 200% in males and 67% in metafemales (Fig. 3). The potential reduction of structural gene expression to one-half of the diploid level in males would be brought back to the normal female diploid level by the twofold increase. Reciprocally, the increase of gene expression via one extra X chromosome in metafemales would be cancelled to achieve compensation by the two-thirds reduced expression operating on each copy.
Fig. 3.
Dosage compensation in males (XY), females (XX), and metafemales (XXX). If gene dosage effects in the various genotypes are set to the gene copy relationship (horizontal axis percentage) and inverse dosage effects on each X chromosome (designated percentages in boxes) are set to 100% in normal diploid females (XX), the total expression of X chromosomes will be 200%. Once one X chromosome is lost over evolutionary time in males, the gene dosage effects will be at 50%, whereas the inverse dosage effects produced by this monosomic X chromosome will be 200%, by which dosage compensation is achieved. On the other hand, the increased gene dosage effects (150%) in metafemales are cancelled by the inverse dosage effects (67%) to equalize their expressions in normal females.
Dosage compensation also occurs in triploid flies that have one or two copies of the X chromosome compared with the three copies present in triploid females (42, 43). However, the magnitude of modulation is different from males and metafemales. The magnitude of change to account for compensation is such that the inverse effect can also account for the differential modulation of the X chromosome in metamales (X; AAA) and triploid intersexes (XX; AAA) that exhibit compensation relative to triploid females (XXX; AAA).
A popular hypothesis to explain X chromosome dosage compensation in Drosophila involves the MSL complex (1). This complex is composed of at least five proteins and two noncoding RNAs, and it is highly enriched on the male X chromosome. One of the components is the product of the males absent on the first (mof) gene, which is a histone acetyl transferase that catalyzes elevated levels of H4 Lys16Ac. Because acetylation fosters open chromatin and is characteristic of high gene expression, this hypothesis is straightforward.
However, the MSL hypothesis does not account for dosage compensation in metafemales, which is documented on the global level in this study. Compensation in males involves a twofold up-regulation, whereas compensation in metafemales involves a two-thirds down-regulation. There is no MSL complex in metafemales (10), and, indeed, its presumed action would not predict an involvement. Furthermore, as noted above, there is evidence for compensation in triploid metamales with one X chromosome and triploid intersexes with two X chromosomes compared with triploid females with three sets of all chromosomes. The different magnitudes of modulation of the X chromosome in these cases, which are also different from those in diploid males, are not addressed by the MSL hypothesis.
In a recent study, different types of targeting of the MSL complex to reporters or to the X chromosomes failed to provide any evidence that association with the MSL complex would cause a twofold up-regulation compared with the progenitor state without the complex (34). In addition, dissociation of the MSL complex does not cause a loss of compensation, but there is evidence that autosomal gene expression is increased (11–14). Other cases of compensation in the absence of the complex have been reported for the male germline (44) and partially in early embryos before the MSL complex is sequestered to the X chromosome (45).
These considerations lead to the idea that a modulation inversely related to the dosage of the X chromosome relative to the autosomes provides a mechanistic solution to explain compensation in the different genotypes of the X chromosome that compensate with or without the MSL complex (34, 46). The sequestration of the MSL complex to the X chromosome might have evolved to mute the potential inverse effect that the monosomic X chromosome would otherwise produce on the autosomes (12, 14, 34). With such sequestration to high levels, the greatly increased amount of acetylation would tend to overcompensate the X chromosome. Thus, an additional activity is hypothesized to have evolved in conjunction with the complex to override the impact of acetylation on gene expression. Such an activity would explain why targeting of the complex has no effect on associated genes (34). Taken together, the data suggest that X chromosome compensation has capitalized on processes that typically result from genomic imbalance (47) to achieve equalized expression and that the MSL complex modifies these effects.
Materials and Methods
Samples were obtained using genetic crosses and collected based on phenotypes of balancer chromosomes and marker genes. Adult metafemales were separated by the phenotype of y+ f+, and the miniwhite reporter expressions were compared. Total RNA was isolated using TRIZOL Reagent (Invitrogen), and the mRNAs were purified and high-throughput sequencing services were performed at the University of Missouri DNA Core Facility. The global expression patterns of the samples were analyzed and compared. Statistical analysis used the MATLAB R2012a program.
Supplementary Material
Acknowledgments
High-throughput sequencing services were performed at the University of Missouri DNA Core Facility. Research was supported by National Institutes of Health Grant R01GM068042.
Footnotes
The authors declare no conflict of interest.
Data deposition: Alignment and processed files have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE41679), and raw RNA-sequencing output data have been deposited in the Sequence Read Archive (SRA accession no. SRP016518).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305638110/-/DCSupplemental.
References
- 1.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: Epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13(2):123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 2.Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66(5):935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 3.Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69(2):375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 4.Bone JR, et al. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994;8(1):96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 5.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16(8):2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern C. Dosage compensation-development of a concept and new facts. Can J Genet Cytol. 1960;2(2):105–118. [Google Scholar]
- 7.Lucchesi JC, Rawls JM, Jr, Maroni G. Gene dosage compensation in metafemales (3X;2A) of Drosophila. Nature. 1974;248(449):564–567. doi: 10.1038/248564a0. [DOI] [PubMed] [Google Scholar]
- 8.Birchler JA, Hiebert JC, Krietzman M. Gene expression in adult metafemales of Drosophila melanogaster. Genetics. 1989;122(4):869–879. doi: 10.1093/genetics/122.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birchler JA. Expression of cis-regulatory mutations of the white locus in metafemales of Drosophila melanogaster. Genet Res. 1992;59(1):11–18. doi: 10.1017/s0016672300030123. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Birchler JA. Interaction study of the male specific lethal (MSL) complex and trans-acting dosage effects in metafemales of Drosophila melanogaster. Cytogenet Genome Res. 2009;124(3-4):298–311. doi: 10.1159/000218134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiebert JC, Birchler JA. Effects of the maleless mutation on X and autosomal gene expression in Drosophila melanogaster. Genetics. 1994;136(3):913–926. doi: 10.1093/genetics/136.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhadra U, Pal-Bhadra M, Birchler JA. Role of the male specific lethal (msl) genes in modifying the effects of sex chromosomal dosage in Drosophila. Genetics. 1999;152(1):249–268. doi: 10.1093/genetics/152.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhadra U, Pal-Bhadra M, Birchler JA. Histone acetylation and gene expression analysis of sex lethal mutants in Drosophila. Genetics. 2000;155(2):753–763. doi: 10.1093/genetics/155.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhadra MP, Bhadra U, Kundu J, Birchler JA. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics. 2005;169(4):2061–2074. doi: 10.1534/genetics.104.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birchler JA. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics. 1979;92(4):1211–1229. doi: 10.1093/genetics/92.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics. 1981;99(2):247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24(8):390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Xie W, Birchler JA. Identification of Inverse regulator-a (Inr-a) as synonymous with Pre-mRNA cleavage complex II protein (Pcf11) in Drosophila. G3: Genes, genomes. Genetics. 2012;2:701–706. doi: 10.1534/g3.112.002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266(5193):1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- 20.Devlin RH, Holm DG, Grigliatti TA. Autosomal dosage compensation Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc Natl Acad Sci USA. 1982;79(4):1200–1204. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin RH, Holm DG, Grigliatti TA. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics. 1988;118(1):87–101. doi: 10.1093/genetics/118.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birchler JA, Hiebert JC, Paigen K. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics. 1990;124(3):679–686. [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien SJ, Gethmann RC. Segmental aneuploidy as a probe for structural genes in Drosophila: Mitochondrial membrane enzymes. Genetics. 1973;75(1):155–167. doi: 10.1093/genetics/75.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgetts RB. The response of dopa decarboxylase activity to variations in gene dosage in Drosophila: A possible location of the structural gene. Genetics. 1975;79(1):45–54. doi: 10.1093/genetics/79.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Detwiler C, MacIntyre R. A genetic and developmental analysis of an acid deoxyribonuclease in Drosophila melanogaster. Biochem Genet. 1978;16(11-12):1113–1134. doi: 10.1007/BF00484532. [DOI] [PubMed] [Google Scholar]
- 26.Pipkin SB, Chakrabartty PK, Bremner TA. Location and regulation of Drosophila fumarase. J Hered. 1977;68:245–252. [Google Scholar]
- 27.Moore GP, Sullivan DT. Biochemical and genetic characterization of kynurenine formamidase from Drosophila melanogaster. Biochem Genet. 1978;16(7-8):619–634. doi: 10.1007/BF00484718. [DOI] [PubMed] [Google Scholar]
- 28.Oliver MJ, Huber RE, Williamson JH. Genetic and biochemical aspects of trehalase from Drosophila melanogaster. Biochem Genet. 1978;16(9-10):927–940. doi: 10.1007/BF00483744. [DOI] [PubMed] [Google Scholar]
- 29.Rabinow L, Nguyen-Huynh AT, Birchler JA. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila. Genetics. 1991;129(2):463–480. doi: 10.1093/genetics/129.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovén J, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8(2):e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone JH, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13(4):r28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenberg P, et al. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 2009;5(5):e1000465. doi: 10.1371/journal.pgen.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, et al. Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc Natl Acad Sci USA. 2013;110(9):E808–E817. doi: 10.1073/pnas.1222542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aït Yahya-Graison E, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: Impact on disease phenotypes. Am J Hum Genet. 2007;81(3):475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altug-Teber O, et al. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119(3-4):171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- 37.Nawata H, et al. Dysregulation of gene expression in the artificial human trisomy cells of chromosome 8 associated with transformed cell phenotypes. PLoS ONE. 2011;6(9):e25319. doi: 10.1371/journal.pone.0025319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kravets A, et al. Widespread occurrence of dosage compensation in Candida albicans. PLoS ONE. 2010;5(6):e10856. doi: 10.1371/journal.pone.0010856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 40.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322(5902):703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabl JF, Birchler JA. Dosage dependent modifiers of white alleles in Drosophila melanogaster. Genet Res. 1993;62(1):15–22. doi: 10.1017/s0016672300031517. [DOI] [PubMed] [Google Scholar]
- 42.Lucchesi JC, Rawls RM., Jr Regulation of gene function: A comparison of X-linked enzyme activity levels in normal and intersexual triploids of Drosophila melanogaster. Genetics. 1973;73(3):459–464. doi: 10.1093/genetics/73.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucchesi JC, Belote JM, Maroni G. X-linked gene activity in metamales (XY; 3A) of Drosophila. Chromosoma. 1977;65:1–7. [Google Scholar]
- 44.Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299(5607):697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lott SE, et al. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9(2):e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birchler JA. X chromosome dosage compensation in Drosophila. Science. 1996;272(5265):1190–1191. [PubMed] [Google Scholar]
- 47.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.