Online communities are using the power of play to solve complex research problems.
The Foldit Void Crushers Group sounds more like a noisy art-rock band than a research team. However, there it is, listed as coauthor of an article in the October 2011 issue of Nature Structural & Molecular Biology.
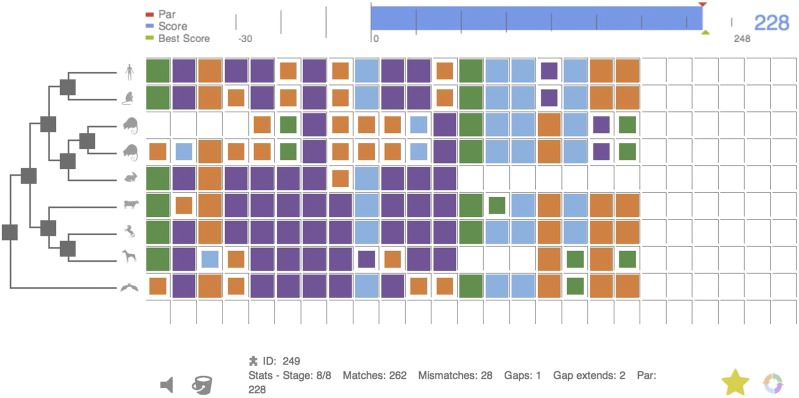
Players of Phylo compare the genome sequences of many animals to find genetic segments that are similar. Copyright © 2012 McGill University. All rights reserved.
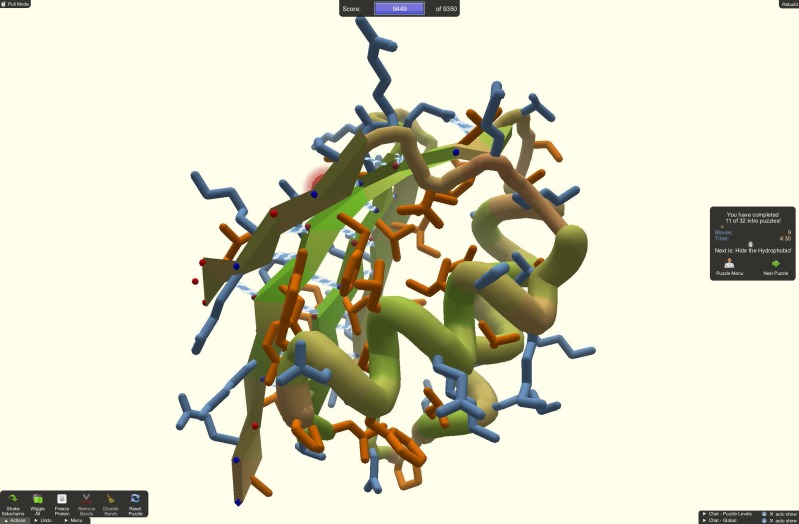
Foldit asks players to find the most stable configurations of folded proteins, a task at which humans can beat the best computer algorithms. Image courtesy of the University of Washington, Seattle.
The group is made up of online game players who had a critical role in the paper’s complex achievement—solving a protein structure that had vexed biologists for a decade. Similar appearances in the pages of respected journals mark the growing importance of such “citizen scientists,” whose collective brain power is tackling some of science’s most difficult challenges.
More than a passing fad, researchers say that blending human and computer strengths through games—gamification—may be the best way to process some otherwise daunting data sets, particularly those where innate human capabilities can fill the gaps left by imperfect algorithms.
Humans around the world spend an estimated 3 billion hours playing online games every week, which amounts to a staggering waste of neurological computing cycles. However, crowdsourcing and citizen science efforts in the form of games offer the enticing possibility of shifting some of that energy toward useful work—helping to cure cancer, for instance, instead of “ridding a game world of zombies” as one team put it in a recent paper (1).
The first computer game to make a significant impact in biology, called Foldit, launched in 2008. Foldit players can twist, stretch, and rearrange protein backbones and side chains to find the most stable configurations. Those players who perform the best are allowed to graduate to tougher research challenges. “We wanted to see if, by letting people do some puzzle fitting and spatial reasoning about how the pieces of the protein fit together, we could combine humans and computers to solve problems they weren’t able to solve alone,” says lead Foldit designer Seth Cooper, at the University of Washington in Seattle.
Bend It, Shape It
Protein folding is fabulously difficult to predict because each protein can potentially adopt an astronomical number of different shapes. Computers can theoretically run through every one, but that could take decades. So researchers design algorithms that cut corners, such as using known folding patterns for similar proteins as starting points. This delivers useful results but also leaves room for improvement, not least because computers cannot dream up creative ways to shape a protein. “People are intrinsically better than computers at designing things,” says the University of Washington’s David Baker, a biologist and Foldit cocreator.
About 300,000 people now apply those innate design skills playing Foldit. Besides competing, they also collaborate and discuss various problems and solutions from their gameplay, and the Foldit team actively encourages this community atmosphere because it leads to scientific success.
One example involved the Mason–Pfizer monkey virus retroviral protease, an enzyme that plays a key role in the monkey equivalent of AIDS. Researchers are working with this class of proteases in hopes of developing new AIDS treatment strategies. However, researchers had not been able to figure out the enzyme’s structure, in part because a critical component folds in an unusual way. However, when researchers set the problem as a Foldit challenge, players accomplished the task in a few weeks (2). The Foldit Void Crushers Group was one of two Foldit player teams that played a key role, hence their author listing.
Players working together virtually have also developed protein-folding algorithms that sometimes beat those created by the experts (3). And more recently, the Foldit research team began giving players the chance to not just manipulate existing proteins, but to take on more open-ended challenges. They’ve already redesigned a model enzyme called Diels-Alderase, vastly improving its chemical activity. “There’s no way a computer could have done that,” says Baker. The work won the entire Foldit users group authorship on the resulting paper (4).
Pattern Recognition
In the midst of Foldit’s successes, other games were emerging to address similarly challenging biological problems. Some of Baker’s student collaborators on the Foldit project went on to develop another popular game, called EteRNA, that involves folding RNA molecules. And researchers at the Massachusetts Institute of Technology in Cambridge have about 50,000 people from 100 countries busily mapping the complex connections between retinal neurons using a game called Eyewire.
These games all give players access to a stripped-down version of the underlying scientific data. However, Jérôme Waldispühl, a bioinformatician at McGill University in Montreal, and his colleagues took a decidedly different approach when they set out in 2010 to gamify the daunting process of multiple sequence alignment (MSA). This involves comparing the genome sequences of many animals to find genetic segments that are similar, if not identical. Identifying these common genes allows researchers to home in on those that are the most functionally important across various species.
The game is called Phylo, and, apart from an initial explanation of the science, it involves nothing more than moving colored blocks around. Each represents a DNA nucleotide, but players do not need to understand that to participate—they just look for the closest color pattern matches.
Within hours of launching the game in November 2010, thousands of people had signed up to play. Servers crashed. “We were very surprised by the impact that this work has had on the public,” says Waldispühl. After 6 months, the team found that some 70% of the DNA regions analyzed by players were better aligned than they had been using just computer algorithms (5).
Audience Participation
Others are explicitly targeting scientists to play their games, rather than seeking help from the untrained masses. Andrew Su, a bioinformatician at the Scripps Research Institute in La Jolla, CA, and his colleagues developed Dizeez, which taps the knowledge of genomics researchers.
Dizeez asks players to link genes with associated diseases and awards points according to how the answer matches existing database information. Such play can help to confirm or discount gene roles already suspected, but Su hopes to eventually create a version that seeks and compiles new associations not yet recorded in databases, which inevitably lag behind the latest research findings.
The Dizeez prototype, released in 2012, has attracted hundreds of players already. Su suspects that it would be much more difficult to persuade so many scientists to share what they know through more conventional channels, such as surveys. “Some people have spent hours on our games because they love that dopamine hit when they get points,” he says.
However, if dopamine hits were the only draw for citizen scientists, then it’s unlikely that science-based games could ever win out over other offerings, says Chris Lintott, an astronomer and citizen science proponent at the University of Oxford, United Kingdom. “I don’t think anybody sits down and says, ‘I’ve had a hard day at work. I want to play a game. Should I play World of Warcraft or Foldit?’” he jokes. “We’re filling a different void in people’s lives.”
That added incentive is an altruistic desire among many to make a contribution to some larger good, instead of dedicating their time to games and phone apps with no return.
Good Citizens
Lintott became a pioneer of citizen science when he and his colleague created Galaxy Zoo in 2007, which helps to reveal how galaxies form and evolve. They were overwhelmed with participants willing to look at hundreds of images of galaxies to categorize their shapes. However, they also got plenty of inquiries from other scientists asking for help setting up their own games. Lintott’s team launched a citizen science Web site called Zooniverse in 2009, in part to formalize the process for scientists requesting his team’s help. Last year they got about 100 good proposals for new projects in various fields.
The diversity of projects shows that there is not a single solution for applying human computing power to biological or other problems. Most of what Zooniverse hosts are not even games in a traditional sense, but instead are designed to tap altruistic motivation and scientific fascination.
Working with the London-based research charity Cancer Research UK (CRUK), the Zooniverse team developed an application called Cell Slider. Users do not score points— they simply help cancer researchers sift through a mountain of drug trial data by identifying cancerous cells in slides. Amy Carton, who runs the citizen science program at CRUK, says that the first data set released on Cell Slider would have taken researchers 18 months to process; instead, the Cell Slider citizen scientists got it done in 3 months.
“We know what motivates our users,” says Lintott. “They want to make a contribution to research, and they are motivated by doing good, so we build projects to capitalize on that and keep people in that mode.”
Lintott is a great fan of more competitive games like Foldit but says that model would not fit everywhere. One of the reasons Foldit works well is that there is a straightforward way to offer a meaningful score, based on the energy state of the protein—lower energy means a more stable configuration, the goal for folding. However, with galaxies or cell slides, for instance, the significance of a person’s actions on screen may not be known for quite some time.
Lintott’s team’s experience has been that people quit playing straightforward point-scoring games sooner and don’t come back. “You could criticize that by saying they were badly designed games,” he says. “But that just highlights that game design is hard.”
Click Here to Cure Cancer
CRUK is also working on a project that lends itself to a more competitive game experience, and the outcome could well become the highest profile biology effort yet. Tentatively titled GeneRun, players will help to process cancer patients’ anonymized genetic data so that researchers can learn more about gene–cancer connections. “We have a lot of data to get through,” says Carton. “To get through it quicker, we need people to come and help us.”
She has enlisted help from groups at Google, Amazon, and Facebook to help build the game. At a “hackathon” in March, held at a Google campus in the United Kingdom, more than 50 code writers broke into teams and created about a dozen prototype games. Carton says her team will soon pick one game to develop further and aims to launch it this summer. “I think it’s a great way to engage with the public,” says cancer researcher Carlos Caldas at CRUK, whose group is providing data for Cell Slider. “I do think it will have an impact in science.”
One obvious concern is whether new games can continue to build significant player communities as more are released, but Cooper believes that “there’s definitely more to be done.” Although 300,000 Foldit players may seem like a lot, the game Angry Birds has hundreds of millions of active users and Facebook about a billion. Nobody expects science games to climb that high, but the numbers show there’s a massive pool of potential players to draw from.
Lintott agrees. “I think [gamification] will become a standard way of dealing with large data sets,” he says. “I believe it’s that important.”
References
- 1.Good BM, Su AI. Games with a scientific purpose. Genome Biol. 2011;12(12):135. doi: 10.1186/gb-2011-12-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khatib F, et al. Foldit Contenders Group Foldit Void Crushers Group Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nat Struct Mol Biol. 2011;18(10):1175–1177. doi: 10.1038/nsmb.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatib F, et al. Algorithm discovery by protein folding game players. Proc Natl Acad Sci USA. 2011;108(47):18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eiben CB, et al. Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat Biotechnol. 2012;30(2):190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawrykow A, et al. Phylo players Phylo: A citizen science approach for improving multiple sequence alignment. PLoS ONE. 2012;7(3):e31362. doi: 10.1371/journal.pone.0031362. [DOI] [PMC free article] [PubMed] [Google Scholar]