Abstract
The membrane–cytosol interface is the major locus of control of actin polymerization. At this interface, phosphoinositides act as second messengers to recruit membrane-binding proteins. We show that curved membranes, but not flat ones, can use phosphatidylinositol 3-phosphate [PI(3)P] along with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] to stimulate actin polymerization. In this case, actin polymerization requires the small GTPase cell cycle division 42 (Cdc42), the nucleation-promoting factor neural Wiskott–Aldrich syndrome protein (N-WASP) and the actin nucleator the actin-related protein (Arp) 2/3 complex. In liposomes containing PI(4,5)P2 as the sole phosphoinositide, actin polymerization requires transducer of Cdc42 activation-1 (toca-1). In the presence of phosphatidylinositol 3-phosphate, polymerization is both more efficient and independent of toca-1. Under these conditions, sorting nexin 9 (Snx9) can be implicated as a specific adaptor that replaces toca-1 to mobilize neural Wiskott–Aldrich syndrome protein and the Arp2/3 complex. This switch in phosphoinositide and adaptor specificity for actin polymerization from membranes has implications for how different types of actin structures are generated at precise times and locations in the cell.
Keywords: endocytosis, BAR domain, cytoskeleton
The original discovery of actin as the scaffold in muscle upon which myosin exerts its force obscured for some time that a major role of actin in nonmuscle cells involves its intimate association with cellular membranes. Through morphological studies of the actin cytoskeleton, the close association of actin polymerization and the specification of actin polarity with respect to membranes came to the fore. As the regulatory biochemistry of actin was unraveled, a striking role for regulators with substantial, and regulated, residence in the membrane, such as phosphoinositides and small G proteins, emerged. In the past decades, the extraordinary dynamics of the cytoskeleton as well as the equally extraordinary dynamics of membrane traffic have become centerpieces of cell biological understanding. That an important structural linkage between the dynamics of the membrane and cytoskeleton exists is evident, but how actin regulators are targeted to specific membrane sites is less clear.
Phosphoinositides are chemically dynamic lipids that are important upstream regulators of actin assembly (1–3); they can exert their effects through specific protein domains, such as the FYVE (Fab1, YOTB, Vac1, EEA1) domain in EEA1 (early endosome antigen 1) that binds phosphatidylinositol 3-phosphate [PI(3)P]; the pleckstrin homology (PH) domain in Akt that binds phosphatidylinositol (3,4) bisphosphate; and the PH domain in phospholipase Cδ that binds phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2]. The actin polymerization machinery is vast and complex, consisting of nucleators, transducers, cappers, bundlers, and more, many of which are regulated by phosphoinositides (4, 5) and are good candidates to test for biochemical coordination of membrane processes. The fluidity and curvature of membranes can also have important consequences for their interaction with cytosolic proteins (6). We have recently shown that frog egg extracts combined with supported lipid bilayers generate focal, bundled actin structures that contain actin polymerizing factors not previously seen in similar studies using small unilamellar liposomes (7). The composition and structure of these bundles suggest that these structures assembled on supported bilayers resemble filopodia (7). In contrast, actin assembled with liposomes was diffuse rather than bundled; these actin structures could be recapitulated with a small set of purified components (8, 9): the FCH-Bin–amphiphysin–Rvs (F-BAR) domain protein transducer of cell cycle division 42 (Cdc42)-dependent actin assembly-1 (toca-1), neural Wiskott–Aldrich syndrome (N-WASP), and the actin-related protein (Arp) 2/3 complex. The identity of these components suggests that this reconstituted pathway mimics events in endocytosis (10, 11).
To reveal the mechanism by which actin assembly is coordinated with dynamic membrane events, such as endocytosis and filopodia formation, we compared the assembly of actin from liposomes with that from supported bilayers. The in vitro results reveal interplay among membrane presentation, phosphorylation on the 3-position of phosphatidylinositol, and the localization of regulators of actin nucleation.
Results
Differences in Actin Assembly Between Liposomes and Supported Bilayers.
When exposed to frog egg extracts, liposomes stimulate the assembly of diffuse actin structures, whereas supported lipid bilayers generate filopodial-like structures, filament bundles anchored at the glass surface by proteins like formins and vasodilator-stimulated phosphoprotein (VASP) found in filopodial tips (7). In seeking to understand the difference, we noted that actin polymerisation stimulated from liposomes and supported lipid bilayers had slightly different lipid requirements; both contained phosphatidylcholine (PC) (48%) and PI(4,5)P2 (4–10%). However, in the supported bilayer experiments, we used phosphatidylserine (PS) (48–42%) to mimic the inner leaflet of the plasma membrane, whereas in liposomes we used phosphatidylinositol(PI) (48–42%) (1). We now find the presence of PI instead of PS in liposomes leads to a twofold increase in the rate and a 1.5-fold increase in the extent of actin polymerization. The trend was reversed for the supported bilayers: the rate of assembly was 40% lower with PI than with PS (Fig. 1 A and B). Using the same batches of liposomes to make supported lipid bilayers, we could control for differences in total lipid composition. The distinction between PI and PS is unexpected because they have the same net charge at neutral pH and similar fatty acid chain compositions. The obvious difference is that PI can be modified to generate other phosphoinositides, whereas PS cannot.
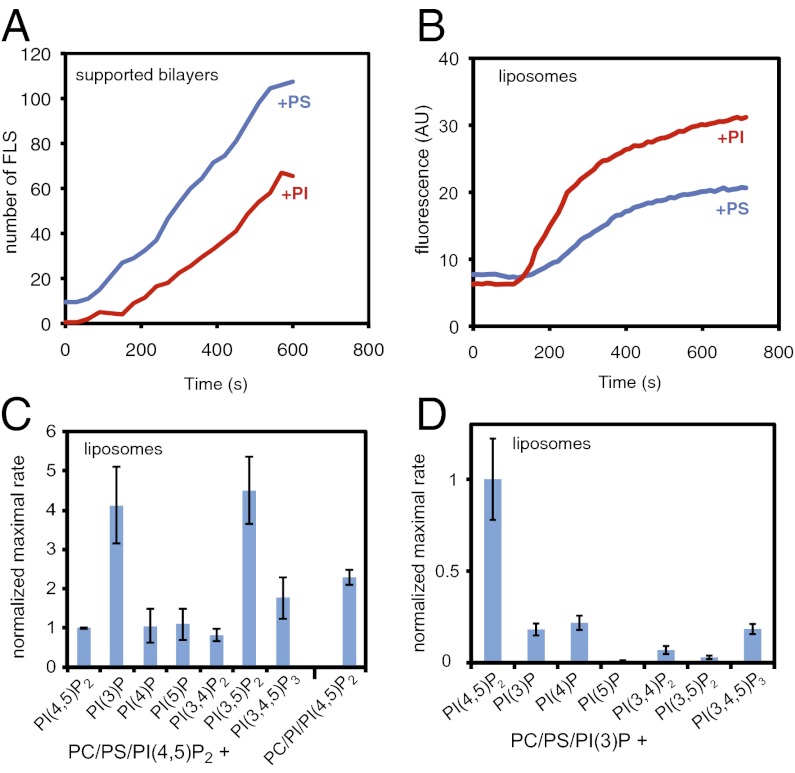
Fig. 1.
PI and PI(3)P contribute to actin polymerization from liposomes. (A) Numbers of filopodia-like structures from supported lipid bilayers showing that filopodia-like structure formation is favored by PS and not PI. The lipid compositions were 48% PC, 48% PS or PI, and 4% PI(4,5)P2. (B) Pyrene actin assay of actin polymerization from liposomes showing that it is favored by PI rather than PS, with the same liposomes used in A. (C) Bar chart showing the maximal rate of actin polymerization (slope of a pyrene actin assay) with a lipid composition of 48% PC, 47% PS, 4% PI(4,5)P2, and 1% of the test phosphoinositide, or PC/PI/PI(4,5)P2. The values are normalized against PC/PS/PI(4,5)P2 liposomes. (D) Substituting PI(4,5)P2 and keeping 1% PI(3)P constant shows that PI(4,5)P2 is essential. Data are normalized to PC/PS/PI(3)P/PI(4,5)P2 liposomes. Data in A and B is representative of four independent experiments. Data in C and D is the average of three experiments, with error bars showing the SD.
To test whether PI phosphorylation is responsible for the potency of PI to induce actin polymerization from liposomes, we substituted different phosphoinositides for PI within a standard lipid mixture of PC/PS/PI(4,5)P2 (Fig. 1C). Addition of PI(3)P or PI(3,5)P2 led to a four- to fivefold increase in the rate of polymerization from liposomes compared with PI and other phosphatidylinositol phosphates (PIPs; Fig. 1C). This augmented activity still required PI(4,5)P2 (Fig. 1D). The potency of PI(3)P to stimulate actin polymerization from liposomes was very high: half-maximal at 0.3% and saturated at 1% (Fig. S1A).
Production of PI(3)P in the Extract.
To test whether a PI3 kinase activity in the extract was responsible for the activation, we added the phosphatase domain of myotubularin to counteract all PI(3)P production; actin polymerization was inhibited 60% (Fig. S1B). Wortmannin, a PI3 kinase inhibitor with broad specificity, reduced actin polymerization by 40% (Fig. 2A), and BEZ-235, a specific class I PI3 kinase inhibitor (Fig. 2A), also inhibited. There was no inhibition by wortmannin or BEZ-235 when the liposomes were generated with PI(3)P (Fig. 2B). The amount of PI(3)P generated on the membranes was below our limit of detection. By using the rate of actin polymerization as a readout of PI(3)P production, the expected phosphorylation of the PI corresponded to 0.05% conversion, two orders of magnitude below the sensitivity of biochemical or radiochemical assays for PI(3)P.
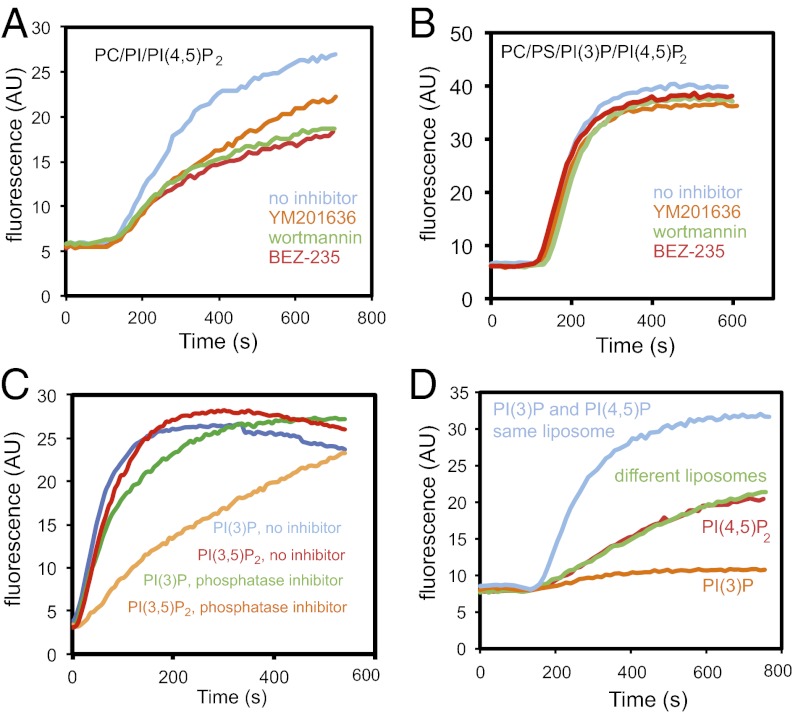
Fig. 2.
Phosphorylation of PI by class I PI3 kinase is important, and PI(3)P and PI(4,5)P2 work together to stimulate actin polymerization. (A) Pyrene actin assay on addition of PC/PI/PI(4,5)P2 into extracts preincubated with 10 μM wortmannin, BEZ-235, or YM201636. (B) Pyrene actin assay on addition of 48% PC, 47% PI, 1% PI(3)P, and 4% PI(4,5)P2 liposomes in extracts that were preincubated with the drugs similarly to A showing that the 3-position stimulates actin polymerization. (C) Pyrene actin assay on addition of 48% PC, 47% PS, 1% PI(3)P or 1% PI(3,5)P2, and 4% PI(4,5)P2 in extract preincubated with a 1/100 dilution of a phosphatase inhibitor mixture. (D) Addition of liposomes where PI(3)P and PI(4,5)P2 are present in the same or distinct liposomes. Data is representative of two or three independent experiments.
Because PI(3)P and PI(3,5)P2 are each capable of stimulating actin polymerization, whereas PI(4,5)P2 and PI(3,4,5)P3 are not (Fig. 1C), it is possible that (i) PI(3)P could be a precursor for the active lipid PI(3,5)P2; (ii) PI(3)P might be the active lipid generated from PI(3,5)P2; or (iii) both PI(3)P or PI(3,5)P2 might stimulate actin polymerization. Specificity might be mediated by a protein that binds the 3-position, but whose binding is blocked by phosphorylation at the 4- but not the 5-position. To establish whether PI(3)P functioned only as a precursor for PI(3,5)P2, we asked whether a PI(3)P 5-kinase inhibitor, YM201636, inhibits actin polymerization (Fig. 2A); it reduced polymerization but less effectively than the class I PI 3-kinase inhibitor, BEZ-235 (Fig. 2A), suggesting that PI(3,5)P2 can stimulate actin polymerization, but that PI(3)P is also effective. To evaluate whether YM201636 might be simply interfering, we added YM201636 when PI(3)P was present in the liposomes; it had no effect (Fig. 2B). We asked whether a phosphatase in the extract could account for the activity of PI(3,5)P2. In the absence of specific lipid phosphatase inhibitors, we tested a nonspecific phosphatase inhibitor mixture (Fig. 2C). Phosphatase inhibitors suppressed the actin polymerization resulting from PI(3,5)P2 liposomes greater than for PI(3)P liposomes, consistent with the fact that PI(3,5)P2 is dephosphorylated by the extract and that PI(3)P is the active lipid.
PI(3)P and PI(4,5)P2 Act in cis.
Phosphoinositides are thought to be enriched in distinct membranes [e.g., PI(3)P at the endosome, PI(4,5)P2 at the plasma membrane], so it is possible in our assays that PI(3)P and PI(4,5)P2 act at disparate sites and communicate by soluble factors. Alternatively, both phosphoinositides may have to be present in cis. To address the cis vs. trans action of PI(3)P and PI(4,5)P2, we generated separate liposomes with PI(3)P or PI(4,5)P2. As expected from earlier results, liposomes containing just PC/PS/PI(3)P are incapable of stimulating actin polymerization (Fig. 1D), and liposomes containing PC/PS/PI(4,5)P2 stimulate actin polymerization at a much reduced level (Fig. 1C). On mixing, the rate and extent of actin polymerization is not elevated compared with the PC/PS/PI(4,5)P2 liposomes alone. Thus, the two lipids have no observable effect in trans (Fig. 2D).
Role of Membrane Curvature in Actin Polymerization.
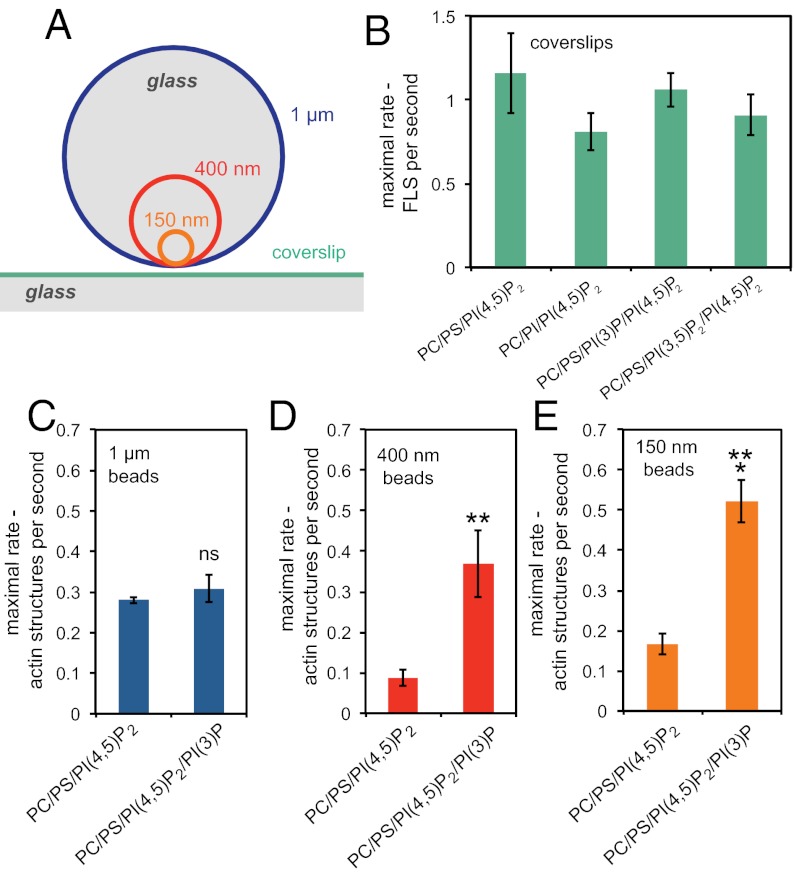
Major changes in membrane curvature accompany the formation of membrane trafficking vesicles; these dramatic modifications are thought to involve actin, suggesting that physical features of the membrane might in themselves provide cues for the recruitment of actin regulatory proteins. For example, deformability of the membrane substrate is important for the activation of N-WASP, and membrane curvature is also known to influence the activity of PI3 kinase (9, 12). The use of small glass beads as the bilayer support (Fig. 3A) allowed us to ask whether the curvature is the key factor responsible for the ability of the actin machinery to respond to the presence of PI(3)P. As expected from the relative reduction in actin polymerization by substituting PS for PI in studies from supported bilayers (Fig. 1A), we found no stimulation in the rate of generating filopodia-like structures on flat supported bilayers with PI or by adding PI(3)P or PI(3,5)P2 (Fig. 3B). To test the influence of different membrane curvatures, we coated silica microspheres of 150 nm, 400 nm, and 1 μm in diameter with PC/PS/PI(4,5)P2 or the same composition containing PI(3)P (13). At higher curvatures there is an increase in the number of actin structures polymerized from the surface of the beads when the membranes contain PI(3)P, whereas with 1-μm (essentially flat) beads, the presence of PI(3)P makes no difference (Fig. 3 C–E; Fig. S2). Hence, machinery downstream of PI(3)P seems to favor a curved surface for activation.
Fig. 3.
Membrane curvature contributes to actin polymerization from PI(3)P/PI(4,5)P2 membranes. (A) Relative sizes of glass beads (to scale) compared with a coverslip. (B) Bar chart showing that the rate of appearance of filopodia-like structures is unaltered by the presence of PI(3)P or PI(3,5)P2. The lipid compositions were 48% PC, 47% PS, 1% PI(3)P or PI(3,5)P2, and 4% PI(4,5)P2. (C) Rate of appearance of actin structures growing on bilayers composed of 48% PC, 48% PS, 4% PI(4,5)P2 and 48% PC, 47% PS, 4% PI(4,5)P2, and 1% PI(3)P supported on 1-μm-diameter glass beads showing no significance difference between the compositions. (D) Rate of appearance of actin structures growing on bilayers supported on 400-nm-diameter glass beads showing a very significant increase in the presence of PI(3)P (**P = 0.008). (E) Number of actin structures growing on bilayers supported on 150-nm-diameter glass beads showing a significant increase in the presence of PI(3)P (***P = 0.0001). Data are the mean of six experiments; error bars show SEM.
Target of PI(3)P Mobilization of Actin Assembly.
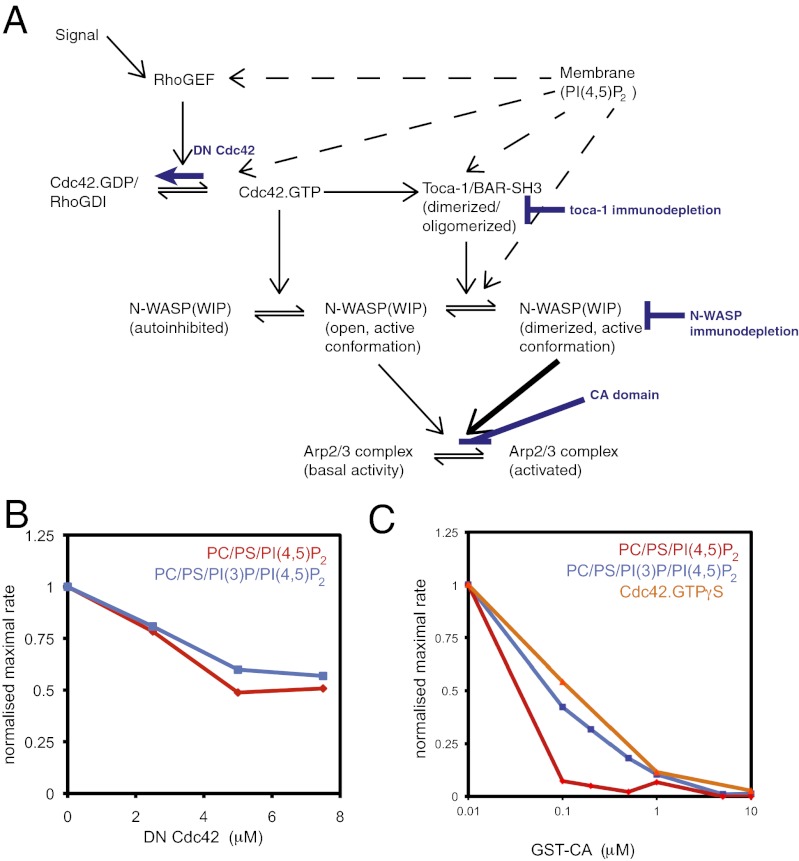
PI(4,5)P2 is known to stimulate actin assembly in frog egg extracts via a site on N-WASP, via recruitment of the F-BAR domain protein toca-1 to membranes, and by activating Cdc42 (Fig. 4A). To test whether this same pathway is involved in PI(3)P stimulation of actin assembly, we applied inhibitors at different points in the pathway and compared the relative responses of PI(3)P liposomes to liposomes where the PI(3)P is replaced by PS (Fig. 4A). To assay whether Cdc42 is the target of PI(3)P, we tested whether dominant-negative Cdc42 inhibits the activation by PI(3)P. Cdc42 is similarly required in both environments (Fig. 4B). We also asked whether PI(3)P stimulates actin polymerization through the Arp2/3 complex, by testing the sensitivity of polymerization to the conjugated central acidic (GST-CA) domain, a fragment of N-WASP that inhibits the Arp2/3 complex activity. Actin polymerization can be totally inhibited by addition of GST-CA showing that the PI(3)P pathway is mediated by the Arp2/3 complex (Fig. 4C). Significantly, it takes 5- to 10-fold higher concentration of GST-CA to inhibit actin assembly when PI(3)P is present, suggesting that the function of PI(3)P is to stimulate Arp2/3 complex activation (Fig. 4C). This process is similar to the effect of stimulating actin polymerization by adding Cdc42.GTPγS to extracts, which also operates via the Arp2/3 complex (Fig. 4C).
Fig. 4.
Actin polymerization stimulated by PI(3)P and PI(4,5)P2 uses Cdc42 and is enhanced by GTPγS. (A) Schematic diagram of signaling from membranes through Cdc42, N-WASP, toca-1, and Arp2/3 complex and highlighting the steps that were inhibited. (B) Dose–response of actin polymerization in a pyrene actin assay in the presence of increasing concentrations of dominant negative Cdc42 with PI(4,5)P2 and PI(3)P/PI(4,5)P2 stimuli (composition used in Fig. 3). (C) Dose–response of actin polymerization in response to Cdc42.GTPγS, PI(4,5)P2, PI(3)P/PI(4,5)P2 stimuli with increasing concentrations of Arp2/3 complex-activation inhibitor, GST-CA domain. PI(3)P/PI(4,5)P2 and Cdc42.GTPγS are more resistant to the inhibitor.
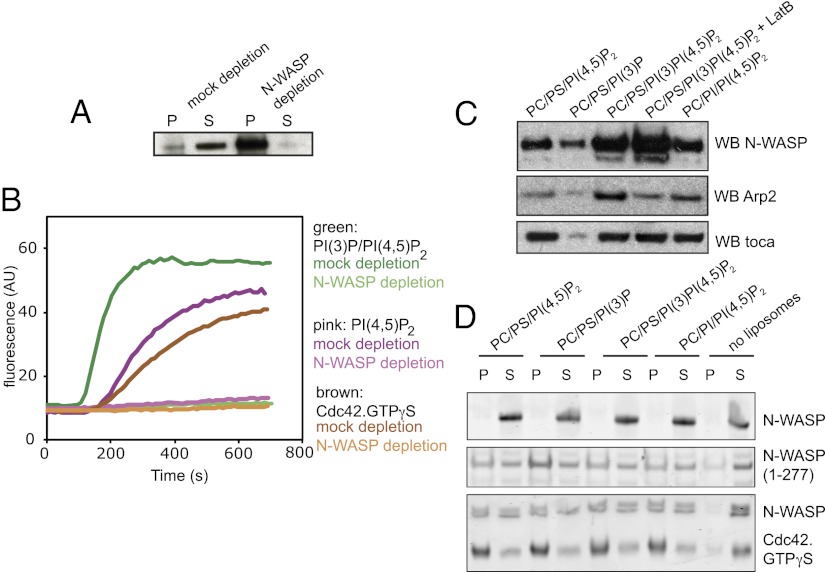
The Arp2/3 complex can be stimulated by several nucleation promoting factors, including Wiskott–Aldrich syndrome protein verprolin homologous (WAVE) and Wiskott–Aldrich syndrome protein and SCAR homologue (WASH) (14). To determine whether N-WASP is a relevant factor for PI(3)P-stimulated actin polymerization, we immunodepleted N-WASP from the extract and observed that actin polymerization was completely blocked (Fig. 5 A and B). Actin polymerization stimulated by PI(3)P/PI(4,5)P2 was also inhibited by wiskostatin (Fig. S3).
Fig. 5.
Actin polymerization stimulated by PI(3)P and PI(4,5)P2 uses N-WASP; however, the specificity for PI(3)P/PI(4,5)P2 binding is not within N-WASP itself. (A) Western blot of an immunodepletion showing the bead pellet and extract in the supernatant with control IgG and anti–N-WASP antibodies. (B) Pyrene actin assays on mock and N-WASP–depleted extract shows that N-WASP is critical for actin polymerization with Cdc42.GTPγS, PI(3)P, and PI(3)P/PI(4,5)P2 liposomes. (C) Liposome sedimentation assay from addition of liposomes into extract showing that although Arp2/3 complex and N-WASP binding to the liposomes is increased by the presence of PI(3)P, toca-1 binding is not. Use of latrunculin shows that N-WASP sedimentation is not due to bulk actin polymerization. Arp2 recruitment is driven by both membrane and polymerized actin. (D) Liposome sedimentation assays with the indicated lipid compositions with purified N-WASP, active N-WASP fragment (1–277), or N-WASP and activated Cdc42. There is no specificity in binding for PI(3)P/PI(4,5)P2-containing liposomes, suggesting an external recruitment factor. P, pellet; S, supernatant.
Native N-WASP is in an autoinhibited conformation, where the active C terminus (which can activate the Arp2/3 complex) is occluded by folding back and binding to the protein in a way that prevents its binding to the Arp2/3 complex. A two-stage activation process is envisaged, where (i) binding to Cdc42 and PI(4,5)P2 on the membrane leads to an initial deinhibition of N-WASP, and then (ii) oligomeric BAR–SRC homology 3 (SH3) domain proteins bind and cluster multiple active N-WASP molecules on the membrane, which display pairs of verprolin central acidic (VCA) domains to recruit and activate the Arp2/3 complex (15). The activation of N-WASP and Arp2/3 complex to stimulate actin polymerization takes place at the membrane surface, and this is where new actin monomers are added (7, 16, 17). To determine how different lipid compositions affect the recruitment of the toca-1/N-WASP/Arp2/3 complex pathway in the extracts, we performed a sedimentation assay with liposomes of different composition and blotted for toca-1, N-WASP, and Arp2/3 complex. The combined presence of PI or PI(3)P plus PI(4,5)P2 caused increased recruitment of N-WASP and Arp2/3 complex, but did not cause an increase in the amount of toca-1 on the surface of the liposomes compared with liposomes containing PS/PI(4,5)P2 (Fig. 5C). Recruitment of N-WASP to PI(3)P/PI(4,5)P2 liposomes was not dependent on the presence of polymerized actin, as shown by adding latrunculin B; Arp2 also still sedimented, although at reduced levels, as expected from the assembly of the Arp2/3 complex at junctions between mother and daughter actin filaments (Fig. 5C).
PI(3)P enhancement of actin assembly on vesicles could be due to increased recruitment of N-WASP, rather than via toca-1 (Fig. 4A); this could occur directly through N-WASP, which is known to contain a basic region that binds membranes, so it might contain a cryptic PI(3)P binding site. Using a completely purified system, we assayed whether N-WASP alone, a truncated mutant of N-WASP missing the autoinhibitory region, and N-WASP plus activated Cdc42 bound better to liposomes containing PI(3)P (Fig. 5D). We found no enhanced reactivity with liposomes containing PI(3)P, showing that PI(3)P stimulation is probably not through increased binding of N-WASP itself directly to membranes, or by an increased ability of N-WASP to interact more productively with Cdc42 (Fig. 5D).
Sorting nexin 9 and Not Toca-1 Is Important for PI(3)P-Activated Actin Polymerization.
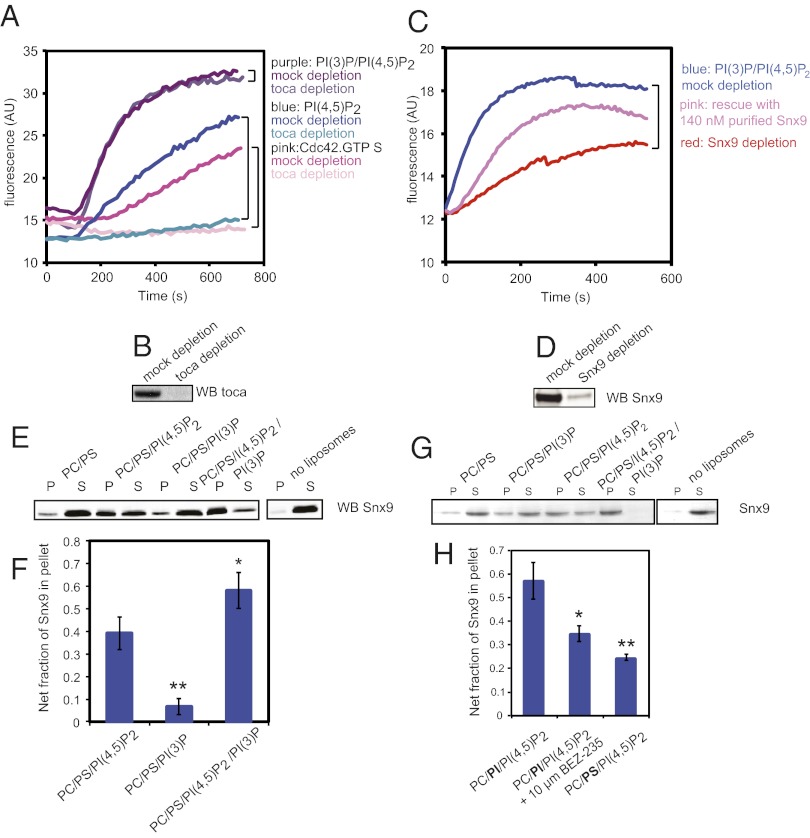
In frog egg extracts, toca-1 is essential for N-WASP–mediated actin assembly on vesicles containing PI(4,5)P2, and is involved in filopodia-like structure formation on supported bilayers (7, 8). We verified that with the PS/PI(4,5)P2 liposomes, and also with Cdc42-GTPγS, toca-1 is essential (Fig. 6A, blue and pink traces). However, is toca-1 also essential for liposomes containing PI(3)P? When we depleted toca-1 we found that in stark contrast to PI(4,5)P2 and Cdc42-GTPγS stimulation, PI(3)P stimulation of actin polymerization is independent of toca-1 (Fig. 6A, purple traces, and Fig. 6B).
Fig. 6.
PI(3)P/PI(4,5)P2-containing liposomes stimulate actin polymerization by recruiting additional N-WASP and Arp2/3 complex to the liposome surface via Snx9. (A) Pyrene actin assay in response to GTPγS.Cdc42, PI(4,5)P2, or PI(3)P/PI(4,5)P2 from mock-depleted or toca-1–depleted extracts showing that PI(3)P/PI(4,5)P2 stimulates actin polymerization independently of toca-1. (B) Western blot of toca-1 in mock- and toca-1–depleted extract used showing effective immunodepletion. (C) Pyrene actin assay in response to PI(3)P/PI(4,5)P2 from mock-depleted extract, Snx9-depleted extract, and Snx9-depleted extract supplemented with 140 nM purified Snx9. (D) Western blot of Snx9 from mock- and Snx9-depleted extract showing effective immunodepletion. (E) Sedimentation of Snx9 from extracts with indicated liposome compositions showing that it binds preferentially to liposomes containing both PI(3)P and PI(4,5)P2. (F) Quantification of Snx9 that is pelleted in the liposome compositions in E, adjusted for binding to control PC/PS liposomes alone, determined by Western blotting. *P = 0.037 and **P = 0.0022 are significant differences by t test. Data are representative of three independent experiments; error bars are SD. (G) Sedimentation of purified Snx9 with indicated liposome compositions showing that it binds preferentially to liposomes containing both PI(3)P and PI(4,5)P2. (H) Liposome sedimentation assay showing the quantification of Snx9 in extract pelleted in PC/PI/PI(4,5)P2 in the absence or presence of 10 μM BEZ-235, and in control PC/PS/PI(4,5)P2 liposomes. *P = 0.037 and **P = 0.0061 indicate a significant and very significant difference in binding relative to PC/PI/PI(4,5)P2 liposomes alone by t test. Data are representative of four experiments; error bars show SEM.
In considering what might replace toca-1 in PI(3)P-induced polymerization, we appreciated that toca-1 is part of a large group of proteins that contain BAR domains and SH3 domains. Many of these proteins probably act redundantly; few have been assayed for their role in actin nucleation. Sorting nexin 9 (Snx9) is of particular interest, because it has previously been shown to both stimulate N-WASP activation of Arp2/3 complex and to have a Phox homology domain that was implicated in binding to PI(3)P, although this domain seems to act somewhat promiscuously (18). To test whether Snx9 could be involved in transducing the signal from PI(3)P to N-WASP and potentially activate the Arp2/3 complex in frog egg extracts we depleted Snx9 from the extract (Fig. 6 C and D); this led to a fivefold decrease in the initial rate of actin polymerization, with the remaining signal consistent with the level of the toca-1/PI(4,5)P2-dependent assembly. Readdition of 140 nM of purified Snx9 to the Snx9-depleted extract substantially increased the rate of actin polymerization (Fig. 6C, pink trace). The decrease in rate relative to mock-depleted extract is likely due to coimmunodepletion of N-WASP along with Snx9.
We tested the recruitment of Snx9 to liposomes containing PI(3)P and PI(4,5)P2, using a sedimentation assay (Fig. 6 E and F). Binding to liposomes is significantly increased by the combination of PI(3)P and PI(4,5)P2 compared with conditions using PI(4,5)P2 alone and PI(3)P alone. As a test of whether the dual PI(3)P/PI(4,5)P2 specificity is recapitulated by purified Snx9, we performed a liposome sedimentation assay; indeed, purified Snx9 binds preferentially to the PI(3)P/PI(4,5)P2 mixture (Fig. 6G).
Because Snx9 binds more strongly to PI(4,5)P2/PI(3)P-containing liposomes than liposomes containing PI(3)P, we also assayed its ability to bind liposomes composed of PC/PI/PI(4,5)P2, in the absence and presence of the class I PI (3) kinase inhibitor BEZ-235 (Fig. 6H), to verify that PI(3)P production from PI is indeed the mechanism stimulating actin polymerization from liposomes. Snx9 binding to PC/PI/PI(4,5)P2 liposomes is significantly increased compared with control PC/PS/PI(4,5)P2 liposomes. In addition, preincubation of the frog egg extract with 10 μM BEZ-235 leads to a significant decrease of Snx9 binding. This data is consistent with PI(3)P generated from PI by PI3 kinase present in the extract.
Discussion
To a large degree, the questions about what controls the placement, structure, and behavior of the actin cytoskeleton can be attributed to the recruitment of specific proteins and lipids to the membrane and their posttranslational modification. In vitro approaches to understanding mechanism have been especially important because manipulating the membrane composition inside cells in a temporally and spatially controlled manner is virtually impossible. Specific mechanistic questions are particularly elusive, because recruitment of components can be tied to structural differences of the membrane, such as curvature and fluidity, as well as chemical properties.
By observing that actin assembly in complex extracts responds differently to liposomes than to flat supported lipid bilayers, we were able to distinguish an important role for PI(3)P and its precursor PI in organizing actin polymerization. PI(4,5)P2 is indispensible in both contexts. Furthermore, by using supported bilayers on small glass beads, we could show that a curved context is important for the response of actin polymerization to PI(3)P present in the membrane, suggesting that BAR domain proteins might be involved in the pathway. Focusing on how actin nucleation is promoted by PI(3)P, we were able to identify Arp2/3 complex as the relevant actin nucleator, N-WASP as the nucleation promoting factor, and Cdc42 as the small GTPase involved in PI(3)P-dependent assembly in frog egg extracts. We narrowed the possible mechanisms of actin control by PI(3)P to an assembly process on membranes, i.e., the recruitment and activation of N-WASP and the Arp2/3 complex, and demonstrated that Snx9 is the key BAR domain adaptor protein involved. Where this process differs from the previously described pathway to actin polymerization in frog egg extracts is in the use of BAR domain protein toca-1. Toca-1 is essential for the mobilization of activated N-WASP on curved liposomal vesicles when PI(3)P is not present. However, when PI(3)P is present, Snx9 becomes essential for the increased actin polymerization, and toca-1 is not needed. The toca-1–dependent pathway is still expected to be active because PI(4,5)P2 is still present in the membranes. Snx9 and toca-1 are similar in that they both contain BAR domains and SH3 domains, but they differ in Snx9 containing a PX domain and toca-1 the homology region 1 domain, which interacts with Cdc42.GTP.
An important question is what physiological event in the cell is recapitulated by this process of PI(3)P-dependent assembly, and what cellular processes require actin polymerization activated by the coincidence of PI(3)P, PI(4,5)P2, and membrane curvature? Snx9 has been implicated in endocytosis, macropinocytosis, phagocytosis (particularly of apoptotic cells), and mitosis (18–21). The enrichment of PI(4,5)P2 at the plasma membrane clearly highlights the importance of that location for the Snx9 pathway, and PI(3)P is known to be produced on macropinosomes, phagosomes, and endosomes (22–24). However, the plasma membrane is likely not to be the only location of the pathway, because phosphoinositide localizations are increasingly recognized to be enrichments rather than compartments, and Snx9 activity has been implicated in endocytosis-independent processes also (21). Because the Snx9/N-WASP/Arp2/3 pathway has multifarious cellular roles, the simplified in vitro system described here provides a tractable way of dissecting the mechanism of action of the pathway, in a complex protein environment (of the extracts) without ambiguity caused by analyzing cell phenotypes that originate from a combination of distinct cell biological processes. Together with previous observations that Snx9 binding to membranes stimulated actin polymerization through N-WASP (25), our results suggest that coincident signals from membrane curvature, PI(3)P and PI(4,5)P2, are transduced through Snx9 to induce actin polymerization, and it seems likely that this plays a critical role in endocytosis.
Materials and Methods
Liposomes.
Large unilamellar liposomes filtered to 100 nm were made by standard methods (26). All lipids were natural brain or liver lipids purchased from Avanti Polar Lipids, except for the non-PI(4,5)P2 phosphoinositide phosphates, which were synthetic with C-16 chains and purchased from Echelon Biosciences. To observe the difference between PI and PS, it was necessary to use fresh (presumably unoxidized) PI. Liposomes were made to a 2-mM total lipid concentration by calculation from the average molecular weight of the lipids. The percentages are percent by moles. For filtering, liposomes were sequentially filtered through the respective size filter using an extruder and polycarbonate filters from Avanti Polar Lipids.
Pyrene Actin Assays and Supported Bilayers.
Pyrene actin and filopodia-like structure assays were performed as previously described (7, 26). For the pyrene assays, typically extracts were used at 5 mg/mL, diluted in XB, and pyrene actin supplemented into the extract to a concentration of 0.12 mg/mL; further energy mix was then added, plus DTT to 5 mM. Assays were started by adding in typically 1.5 μL of liposomes made at 2 mM total lipid concentration (calculated using the average molecular weights). For Figs. 1, 2 A–C and E, 4, 5, and 6A, a Varian Cary Eclipse fluorometer was used with 5-nm slits measuring at an excitation wavelength of 365 nm and emission wavelength of 406 nm. For Figs. 2D and 6C, a BMG Labtech Fluostar Optima microplate reader equipped with a 360-nm excitation and a 410-nm emission filter was used. All graphs were plotted using Excel. Silica microspheres were obtained from Bangs Labs. A total of 250 μg of 1-μm beads or 50 μg of 400-nm and 150-nm beads were sonicated in the presence of excess liposomes labeled with Rhodamine PE. The pellets were resuspended in 100 μL XB by sonication and then rediluted 20-fold for observation. The 10-μL beads suspension was deposited on the coverslip and left to sediment. The reaction mixture was then added on top of the beads, and images were acquired every 30 s for 15 min. Confocal images were acquired using an inverted Nikon TiE with a CFI Plan Apo total internal reflection fluorescence 100× 1.49 N.A. objective, equipped with an X-light Nipkow spinning disk and Lumencor Spectra X LED illumination. Images were collected using a Photometrics Evolve Delta EM-CCD camera and Metamorph software (Molecular Devices). Rhodamine PE was visualized using a green LED with a 560/25 excitation filter and 585/50 emission filter, and Alexa 647 using a red LED with a 628/40 excitation filter and 700/75 emission filter. The images were thresholded, and the number of actin structures was determined using the Analyze Particle tool in ImageJ.
Inhibitor Experiments.
The extract mix was preincubated with the relevant inhibitor for ∼10 min before commencing the assay. Assays were started by addition of liposomes into the assay mixture. GST-CA was made by standard glutathione Sepharose affinity methods after expression in BL21 E. coli. Wiskostatin was purchased from Enzo Life Sciences. YM201636 was from Chemdea, wortmannin from Sigma-Aldrich, and BEZ-235 from LC Laboratories. Phosphatase inhibitor mixture 2 was purchased from Sigma-Aldrich. Myotubularin phosphatase domain was purified using a standard Ni-NTA agarose (Qiagen) purification using the His-tag after expression in bacteria (27). Full-length His-tagged Xenopus laevis Snx9 was expressed using the FreeStyle expression system (Invitrogen) and purified using Ni-NTA agarose.
Immunodepletions.
Immunodepletions were performed as previously described using protein G- or protein A-coated magnetic beads (Dynabeads), control IgG, and antibodies against full-length N-WASP and toca-1 that have been previously described (8, 26, 28). The Snx9 antibody was raised against the GST-tagged SH3 domain (Cambridge Research Biochemicals).
Liposome Sedimentation Assays.
Prenylated Cdc42 and N-WASP fragments were purified as previously described (26, 28), and 5 μg was used per assay with 30 μL of 1-mM liposomes of the indicated concentrations. For sedimentations from extracts, 20 μL of 1-mM liposomes were added to 120-μL assay mix containing 5 mg/mL egg extract. Samples were run on SDS/PAGE, and Arp2, N-WASP, toca-1, and Snx9 were detected by Western blotting using an antibody from Santa Cruz (Arp2, sc-15389), those previously described for N-WASP and toca-1 (8, 28) and rabbit anti-serum against the SH3 domain of Snx9. Quantification of Western blots were performed using LiCor Odyssey scanning and intensity measurements.
Supplementary Material
Acknowledgments
We thank Jack Dixon for the kind gift of the myotubularin plasmid; Florian Hollfelder and Albert Kwok for access to the microplate reader; Julia Mason for technical assistance; and Nick Brown for a critical reading of the manuscript. This work was funded by National Institutes of Health Grants R01 GM26875 (to M.W.K.) and R01 GM41890 (to L.C.C.); a European Molecular Biology Organization Long-Term Fellowship; European Research Council Grant 281971; and Wellcome Trust Research Career Development Fellowship WT095829AIA (to J.L.G.). J.L.G. acknowledges the core funding provided by the Wellcome Trust (092096) and CRUK (C6946/A14492).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305286110/-/DCSupplemental.
References
- 1.Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140(5):1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17(2):181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 3.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36(3):512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90(1):259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J. 2010;98(10):2327–2336. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329(5997):1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho HY, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118(2):203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27(21):2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, et al. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat Cell Biol. 2010;12(9):902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9(3):e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubner S, et al. 1998. Enhancement of phosphoinositide 3-kinase (PI 3-kinase) activity by membrane curvature and inositol-phospholipid-binding peptides. Eur J Biochem 258(2):846–853.
- 13.Bayerl TM, Bloom M. Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys J. 1990;58(2):357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilney LG, Mooseker MS. Actin filament-membrane attachment: Are membrane particles involved? J Cell Biol. 1976;71(2):402–416. doi: 10.1083/jcb.71.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J Cell Biol. 1985;101(2):597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarar D, Waterman-Storer CM, Schmid SL. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell. 2007;13(1):43–56. doi: 10.1016/j.devcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Almendinger J, et al. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS ONE. 2011;6(4):e18325. doi: 10.1371/journal.pone.0018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu N, Shen Q, Mahoney TR, Liu X, Zhou Z. Three sorting nexins drive the degradation of apoptotic cells in response to PtdIns(3)P signaling. Mol Biol Cell. 2011;22(3):354–374. doi: 10.1091/mbc.e10-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma MP, Chircop M. SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J Cell Sci. 2012;125(Pt 18):4372–4382. doi: 10.1242/jcs.105981. [DOI] [PubMed] [Google Scholar]
- 22.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394(6692):494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 23.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoncu R, et al. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136(6):1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9(1):133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 26.Lebensohn AM, Ma L, Ho HY, Kirschner MW. Cdc42 and PI(4,5)P2-induced actin assembly in Xenopus egg extracts. Methods Enzymol. 2006;406:156–173. doi: 10.1016/S0076-6879(06)06013-7. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97(16):8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.