Abstract
The Na+-Ca2+ exchanger (NCX) is a ubiquitously expressed plasma membrane protein. It plays a fundamental role in Ca2+ homeostasis by moving Ca2+ out of the cell using the electrochemical gradient of Na+ as the driving force. Recent structural studies of a homologous archaebacterial exchanger, NCX_Mj, revealed its outward configuration with two potential ion permeation pathways exposed to the extracellular environment. Based on the symmetry of NCX_Mj structure, an atomic model of an inward-facing conformation was generated showing similar pathways but directed to the cytoplasm. The presence of these water-filled cavities has yet to be confirmed experimentally, and it is unknown if the mammalian exchanger adopts the same structure. In this study, we mutated multiple residues within transmembrane segments 2 and 7 of NCX1.1 (cardiac isoform) to cysteines, allowing us to investigate their sensitivity to membrane-impermeable sulfhydryl reagents as exchanger current block. By trapping NCX1.1 in the inward-facing configuration, we have mapped two differently sized cytoplasmic aqueous cavities, the access of which is modified during exchange. This data reveals movements of the protein associated with ion transport. Electrophysiological characterization shows that the conserved residues within transmembrane segments 2 and 7, coordinating Na+ and Ca2+ ions in NCX_Mj, play a fundamental role in NCX1.1. Our results suggest a similar architecture between the mammalian and archaebacterial exchangers.
Keywords: sodium–calcium exchanger, MTS reagents, ion coordination sites, ion translocation pathways
The Na+-Ca2+ exchanger (NCX) is a widely expressed electrogenic antiporter that exchanges one Ca2+ ion for three Na+ ions across the plasma membrane (1), thereby playing a fundamental role in Ca2+ homeostasis (1–3). Despite intensive investigation, how Na+ and Ca2+ access their binding sites is unknown, and there is limited knowledge about the residues involved in ion transport (4–6). The recent high-resolution crystal structure of an archaebacterial NCX homolog (NCX_Mj) and its modeled inward configuration (7) have shown two potential pathways for ions to gain access to the core of the exchanger where one identified Ca2+ binding site and three inferred Na+ binding sites reside. Of twelve residues forming these sites, four residues are in transmembrane segment (TMS) 2, four residues are in TMS7, and two residues each are in TMS3 and -8. These TMSs, referred to as α-repeats (8), are conserved within the extended exchanger family (3) and important for ion translocation in both the mammalian NCX (4–6, 9) and the Na+/Ca2+-K+ exchanger (10).
The structure of NCX_Mj represents a milestone in the study of exchangers. However, for the mammalian homolog, functional studies supporting this structure are absent, and it is unknown if it adopts a similar architecture. Moreover, there is no information on the protein conformational changes that may occur during ion transport. The goals of this work were to identify the ion translocation pathways of the eukaryotic NCX (cardiac isoform NCX1.1) (11), their potential structural rearrangements during ion transport, and the residues involved in ion binding. Such studies allow determination of whether NCX1.1 functionally parallels the structural evidence from NCX_Mj and validate it under more physiological conditions. To these ends, we applied cysteine accessibility analysis to TMS2 and -7 of NCX1.1. Our results show that several residues lining TMS2 and -7 are exposed to the cytoplasm when the ion binding sites of NCX1.1 face the intracellular side of the cell (inward-facing configuration) but are inaccessible during transport. Results indicate that TMS2 and -7 contribute to the cytoplasmic ion permeation pathway and that they undergo conformational changes during ion translocation. Comparison between our accessibility data and the structural model of the inward-facing NCX_Mj model supports the presence of two cytoplasmic-facing aqueous cavities in NCX1.1. By using thiol-reactive compounds of different bulkiness, we also determined that these aqueous openings are of different sizes. Finally, electrophysiological analyses of the cysteine mutants reveal that those residues inferred to coordinate Na+ in NCX_Mj play an important role in the transport properties of NCX1.1. This data provides strong functional evidence for the identity of the residues forming the Na+ binding sites in the exchanger family. Similarly, mutations of residues analogous to those positions found to coordinate Ca2+ in NCX_Mj drastically alter NCX1.1 activity, further underlying similarity between the two proteins. The data indicate that the α-repeats of NCX play a fundamental role in transport function and that their architecture is maintained among distant family members.
Results
Effect of Cysteine Mutations: Role of TMS2 and -7 in Ion Translocation.
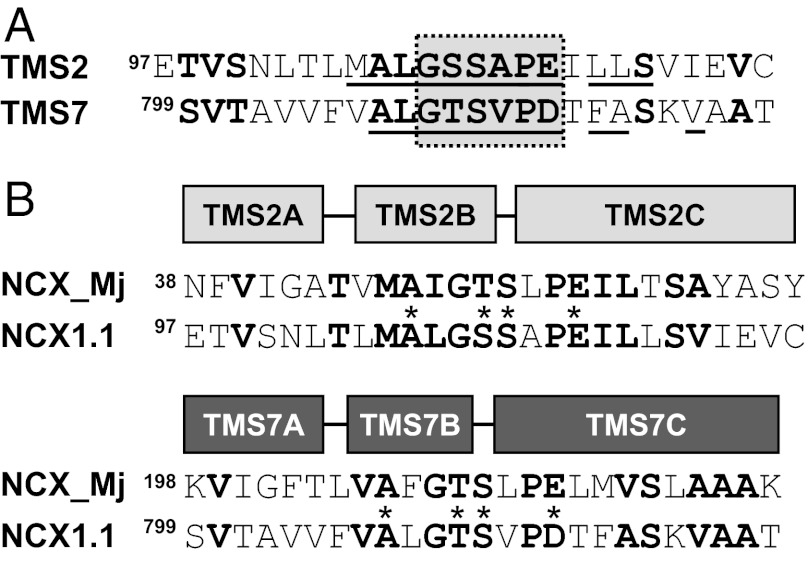
Twenty-three residues in TMS2 and -7 of NCX1.1 were individually replaced with cysteine with the final goal to investigate their reactivity with membrane-impermeable Methanethiosulfonate (MTS) reagents. First we analyzed the functional effects of these mutations, positions that are underlined in Fig. 1A. By analogy to NCX_Mj structure (7), the residues of TMS2 and -7 coordinating ion binding are Ala106, Ser109, Ser110, Glu114 and Ala807, Tyr810, Ser811, and Asp814, respectively (Fig. 1B). Some mutations produced inactive exchangers, namely E113C of TMS2 and T810C, S811C, and D814C of TMS7. Only mutant T810C could be rescued by the more conservative serine replacement. Functional data are consistent with the role of these residues in ion binding or transport. Indeed, as predicted by the crystal structure of NCX_Mj (7), Thr810 and Ser811 coordinate Na+ nearest to the external side, whereas the two negatively charged residues, E113 and D814, participate in multiple binding sites and play a central role in transport function.
Fig. 1.
Amino acid alignment of the α-repeats. (A) Intramolecular similarity between TMS2 and -7 of the α-repeats of NCX1.1. Conserved residues are shown in bold. Mutated residues used in this study are underlined. The GXXXP motif is boxed. (B) Sequence alignment between the archaebacterial NCX_Mj and the mammalian NCX1.1 exchangers TMS2 and -7. Conserved residues are in bold, and residues coordinating Na+ and Ca2+ in NCX_Mj are marked with an asterisk.
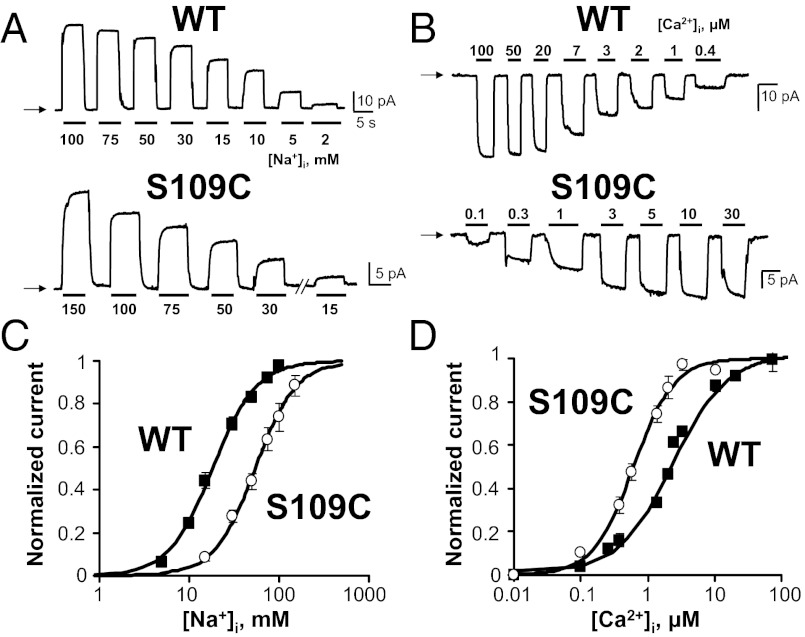
Mutagenesis at the remaining ion coordinating sites generated active exchangers, although with somewhat reduced ionic currents as compared to WT. Exchangers A106C, S109C, and S110C of TMS2 and A807C and T810S of TMS7 were analyzed for their response to application of cytoplasmic Na+ and Ca2+. Fig. 2 displays an example showing the altered transport properties of the S109C mutant, namely a decreased affinity for cytoplasmic Na+ (K1/2, millimolar, values are S109C: 57 ± 5, n = 4; WT: 18 ± 1, n = 9) and increased affinity for Ca2+ (K1/2, millimolar, values are S109C: 0.64 ± 0.08, n = 4; WT: 2.1 ± 0.1, n = 10). The corresponding residue in NCX_Mj, Thr50, was inferred to coordinate Na+ at a site nearest to the cytoplasm, and our functional data support this role for Ser109.
Fig. 2.
Mutation of Ser109 within TMS2 of the mammalian NCX1.1 alters transport properties. Representative outward (A) and inward (B) currents from the WT NCX1.1 and S109C mutant. Outward exchange currents (A) were measured in the presence of 8 mM Ca2+ in the pipette (extracellular) and varying cytoplasmic Na+ concentrations, whereas inward currents (B) were obtained with 100 mM Na+ in the pipette and varying cytoplasmic Ca2+ concentrations. C and D show corresponding Na+ (C) and Ca2+ (D) dependency curves. Measurements were obtained after removal of Ca2+ and Na+ secondary regulation with chymotrypsin. Currents were normalized to their maxima and fitted to a Hill function I = 1/(1 + (K1/2/[M]N)), where [M] is the concentration of Na+ or Ca2+, K1/2 is the concentration of Na+ or Ca2+ needed to give half-maximal activation, and N is the Hill coefficient. Squares and circles indicate WT and mutant exchangers, respectively. Experiments were conducted at VH = 0 mV at 35 °C. Each point is the average between four and nine experiments. Values are in Fig. 3.
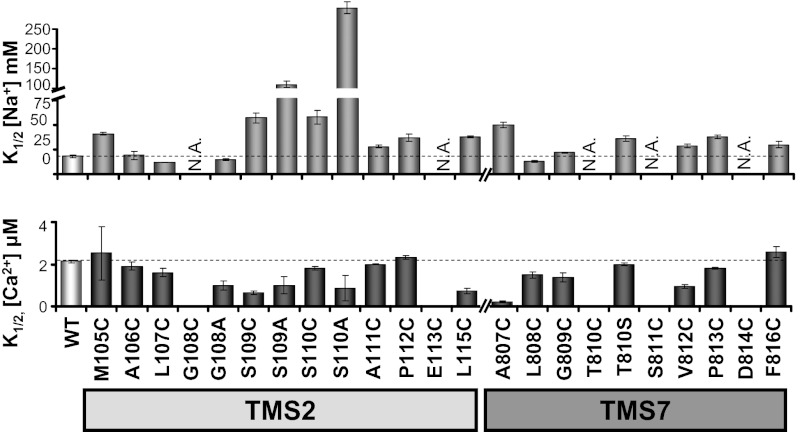
The remaining mutants were examined, and Fig. 3 summarizes the concentrations of Na+ and Ca2+ needed to activate 50% of exchanger current (Hill coefficient values are reported in Fig. S1). Exchangers S109C, S110C, and A807C showed the most significant decrease in apparent affinity to intracellular Na+. Side chain size and polarity were also investigated by replacing Ser109 and Ser110 with the apolar residue alanine (S109A and S110A). Such replacement further decreased Na+ binding and reciprocally enhanced Ca2+ binding affinity. The result is consistent with Na+ and Ca2+ antagonizing each other when bound to their respective binding sites within the core of NCX1.1 (7, 12).
Fig. 3.
TMS2 and -7 mutations alter the mammalian NCX1.1 transport properties. Summary of NCX1.1 mutant apparent affinities for cytoplasmic Na+ and Ca2+. Error bars indicate the SEMs. Mutants with no measurable activity are indicated as N.A. Hill coefficient values are reported in Fig. S1.
The amino acids within TMS2 and -7 found to coordinate Na+ and Ca2+ ions in NCX_Mj contribute to two highly conserved stretches of residues consisting of 108GSSAPE113 and 809GTSVPD814 in TMS2 and -7, respectively (Fig. 1A). Within these sequences, the glycines and prolines do not directly participate in ion coordination (7). Nevertheless, these residues are highly conserved, suggesting an important role in NCX1.1 function. Although mutation of Gly108 to cysteine (G108C) abolished transport activity, insertion of an alanine (G108A) yielded an active exchanger with an unaffected apparent affinity for cytoplasmic Na+ (K1/2, millimolar, values are G108A: 15 ± 1, n = 5; WT: 18 ± 1, n = 9) but a significantly increased Ca2+ sensitivity (K1/2, micromolar, values are G108A: 0.9 ± 0.2, n = 9; WT: 2.1 ± 0.1, n = 10). Similarly, the equivalent mutation in TMS7, G809C, increased the cytoplasmic Ca2+ sensitivity (K1/2, micromolar, values are G809C: 1.39 ± 0.21, n = 6; WT: 2.1 ± 0.1, n = 10). These results indicate that a larger side chain at these two sites alters transport properties, presumably by altering the architecture of the adjoining ion binding sites. Introduction of cysteines at prolines 112 and 813 mainly decreased the sensitivity of NCX1.1 to Na+ (Fig. 3) without, however, preventing its ability to transport, indicating that these prolines are not essential for function.
Cytoplasmic Permeation Pathways of NCX1.1.
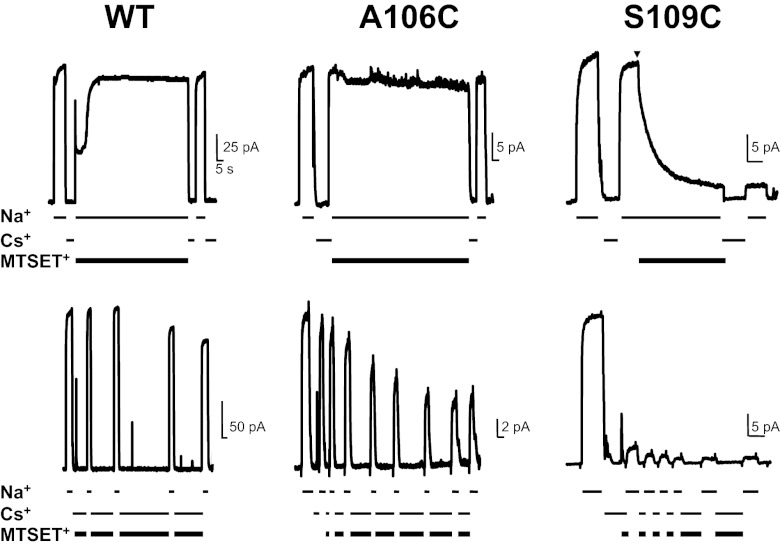
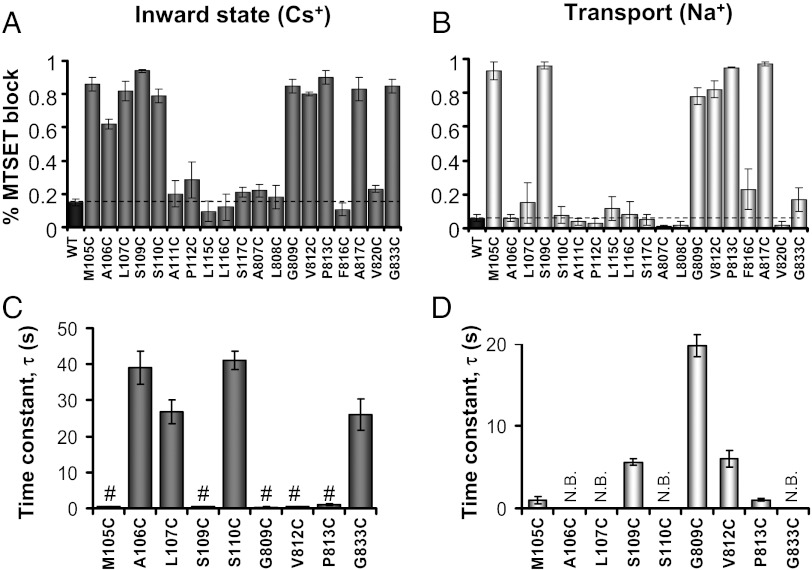
Based on structural information from NCX_Mj (7), the mammalian NCX1.1 is expected to have two aqueous cavities allowing cytoplasmic Na+ and Ca2+ to reach their binding sites. The presence of these water-filled crevices has not been confirmed experimentally. We applied cytoplasmic membrane impermeant 2-(Trimethylammonium)ethyl Methanethiosulfonate Bromide (MTSET+) to define these aqueous pathways within NCX1.1. Blockage of exchanger current after reagent washout is used as an indication of residue reactivity and thus, its exposure to an aqueous environment. MTSET+ was applied either in the presence or absence of cytoplasmic Na+ while maintaining extracellular Ca2+ (pipette) constant. In the absence of Na+ (replaced by Cs+), no transport occurs, and the exchanger ion binding sites are trapped facing the cytoplasm (inward-facing state) (12, 13). Indeed, using these ionic conditions, only a half-reaction cycle of the exchanger can occur; external Ca2+ is transported inward and released. In the absence of Na+, the inward-facing ion binding site remains empty, and the reaction cycle cannot be completed. Addition of Na+ allows for the full transport cycle (cycling), possibly altering the conformation of NCX1.1 and thereby, the accessibility of cysteine residues. An example is shown in Fig. 4, which shows the sensitivity to cytoplasmic MTSET+ of outward currents recorded from WT, A106C, and S109C exchangers. Cytoplasmic application of MTSET+ had minimal effect on WT currents both while cycling (Na+ present) (14) or in the inward-facing state (Cs+ present). In contrast, mutants A106C and S109C were strongly inhibited by MTSET+ when held in the inward-facing state, showing their exposure to the cytoplasmic aqueous environment. However, although S109C current was also inhibited by MTSET+ during cycling, A106C was not, indicating the state-dependent accessibility of this position. Similarly, we investigated the accessibility of nearby residues within TMS2 (M105C, L107C, and S110C) to MTSET+ under cycling and inward-facing state conditions as summarized in Fig. 5 A and B. The mutant M105C was blocked independently of NCX1.1 state, whereas L107C and S110C currents were inhibited only in the inward-facing state. Positions 105 and 109, which face the same surface of TMS2, seem to have greater exposure to the cytoplasm than residues 106, 107, and 110, allowing MTSET+ to also react with their side chains during cycling. Consistent with this hypothesis is the high rate of reactivity of M105C and S109C exchangers to MTSET+ compared to mutants A106C, L107C, and S110C. These results are summarized in Fig. 5 C and D, which compare the time course of the MTSET+ block of several NCX1.1 mutants when cycling and held in the inward-facing state. As shown, reactivity of cysteines at positions 106 and 110 is ∼10-fold slower than the reactivity of M105C and S109C, suggesting limited accessibility of these residues.
Fig. 4.
State-dependent accessibility of mutants A106C and S109C. Representative outward currents from WT, A106C, and S109C exchangers. Intracellular MTSET+ (5 mM) was applied either in the presence (Upper) or absence (Lower) of Na+. Ionic currents recorded from A106C were only blocked by MTSET+ when perfused in the inward-facing state (Cs+ present). In contrast, S109C was inhibited both while cycling (Na+ present) and in the inward state (Cs+ present). WT traces are shown for comparison.
Fig. 5.
Reactivity of strategically introduced cysteines to MTSET+. Summary data for the block of MTSET+ in the absence (A) and presence (B) of cytoplasmic Na+ with Ca2+ present in the pipette (extracellular). In the absence of Na+ (replaced with Cs+), the ion binding sites are exposed to the cytoplasm (inward state configuration). MTSET+ (5 mM) was applied from the intracellular side of the membrane, and the percent of block was measured after reagent washout following up to 1 min continuous exposure, such as in the case of unresponsive mutants. A large number of residues lining TMS2 and -7 were inactivated by MTSET+, indicating that these two TMSs are part of the cytoplasmic ionic pathway. The speed of the current block while NCX1.1 was held in the inward-facing state is summarized in C, whereas mean rate values during transport are shown in D. #Values represent an upper limit; they cannot be accurately measured because of our limited temporal resolution, and faster kinetics may occur. Mutant exchangers that were not modified by MTSET+ are indicated as no block (N.B.).
The structural model of inward-facing NCX_Mj (7) predicts that TMS7 contributes to the cytoplasmic permeation pathway. We, therefore, introduced cysteine at eight strategic positions, resulting in the following active mutants: A807C, L808C, G809C, V812C, P813C, F816C, A817C, and V820C (summarized in Fig. 5). Mutants A807C, L808C, F816C, and V820C were not significantly affected by MTSET+, regardless of whether NCX1.1 was cycling or in its inward-facing conformation. The results indicate that these residues either are not functionally important or inaccessible from the cytoplasm. The facts that they are located on the same side of the TMS7 helix and that the homologous positions (Ala206, Phe207, Met215, and Leu219) in NCX_Mj appear to be immersed within the lipid bilayer are consistent with their inaccessibility.
Four mutants, G809C, V812C, P813C, and A817C, were inhibited by cytoplasmic MTSET+ in both the absence and presence of Na+ (Fig. 5 A and B). Because the NCX_Mj homologous residues to Gly809, Val812, and Pro813 are found in its core, the results indicate that NCX1.1 possesses a deep cavity that penetrates into the center of the protein. As shown in Fig. 5C, the time course of inactivation of these exchangers by MTSET+ was much slower when cycling, suggesting that this region of NCX1.1 undergoes conformational changes during transport.
To define further the TMSs forming a passage for Na+ and Ca2+, we also investigated the MTSET+ sensitivity of G833C of TMS8. As predicted by the crystal structure of NCX_Mj (7), this residue is expected to face the same aqueous cavity as residues Ala106 and Leu107 and therefore, should be exposed to cytoplasmic MTSET+. As shown in Fig. 5A, MTSET+ strongly inhibited the ionic currents of G833C only when in the inward-facing state. The result indicates that this segment of TMS8, near to the cytoplasmic surface, is solvent-accessible and contributes to the formation of an aqueous cavity that undergoes rearrangement during ion translocation.
Probing the Relative Size of the Two NCX1.1 Cavities.
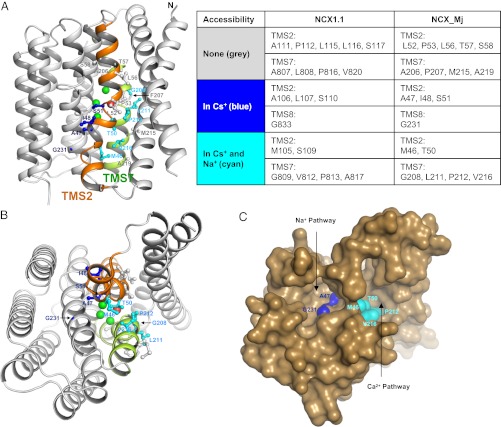
The crystal structure of NCX_Mj predicts the presence of two independent solvent-accessible pathways leading to Na+ and Ca2+ binding sites (7). Our accessibility data are consistent with the presence of these two water-filled cavities, as shown in Fig. 6 and Fig. S2, where the residues of NCX1.1 reactive with MTSET+ are indicated on the NCX_Mj inward state model. Residues of both TMS2 (Met105 and Ser109) and -7 (Gly809, Pro813, Val812, Ala817, and Val820) face the crevice inferred as the Ca2+ pathway in NCX_Mj, whereas cysteines at positions 106, 107, and 109 of TMS2 are exposed to the lumen suggested to be the Na+ passageway in NCX_Mj. To determine the relative sizes of these aqueous cavities, we tested cysteine modification by the membrane-impermeable MTS reagents 1-(Trimethylammonium)methyl Methanethiosulfonate Bromide (MTSMT+) and 2-(Tripentylammonium)ethyl Methanethiosulfonate Bromide (MTS-TPAE). MTSMT+ is slightly shorter than MTSET+ because of the presence of a methyl group instead of an ethyl chain, whereas MTS-TPAE is a larger reagent with pentyl chains (Fig. S3). Reactivity of these reagents with S110C and G809C was investigated. These positions are the deepest residues to react with MTSET+, and each lies in separate cavities, as predicted by the NCX_Mj inward-facing model. Results indicate modification of S110C currents only by the smaller reagent MTSMT+ (Fig. S3). Although the extent of block during cycling was minimal (28 ± 11%, n = 5), S110C current was extensively inhibited by MTSMT+ in the inward-facing state (82 ± 2%, n = 5) and more rapidly blocked than by MTSET+ (24 ± 2 s, n = 5 vs. 41 ± 3, n = 4, respectively) (Fig. S3E). In contrast, G809C currents were completely inhibited by both cytoplasmic MTSMT+ and MTS-TPAE independently of its state.
Fig. 6.
NCX1.1 appears to have two large cytoplasmic cavities. View of inward-facing model of NCX_Mj (7) from the membrane (A) and the cytoplasmic side (B). Homologous residues of NCX1.1 tested in these studies are highlighted and indicated in the table in A. Mutants unreactive to MTSET+ are shown in gray. Residues reactive to MTSET+ when NCX1.1 is held in the inward-facing state (Cs+ present) are in blue, and those residues accessible both while cycling (Na+ present) and in the inward-facing state are in cyan. The surface representation of NCX_Mj in the inward-facing configuration as seen from the cytoplasmic side is shown in C. Note the presence of two cytoplasmic passages for ions to reach their binding sites.
The data indicate that the two ion translocation pathways of NCX1.1 have different sizes. The one represented by G809C is sufficiently wide to accommodate the larger MTS-TPAE reagent, whereas the other cavity is too narrow to allow MTS-TPAE to react with S110C; however, it is reachable by the smaller MTSMT+ and MTSET+ reagents.
Discussion
X-ray crystallography of the archaebacterial NCX_Mj in its outward-facing conformation (7) revealed that NCX is organized into two sets of five TMSs with similar structure but opposite orientation. Residues from TMSs segments 2, 3, 7, and 8 contribute to one specific binding site for Ca2+ and three inferred Na+ ion binding sites in the center of the protein reachable through two extracellular cavities. Based on the symmetrical nature of NCX_Mj, a model for the inward-facing state was proposed that mirrors the outward configuration but with an inverted orientation, placing the two ion permeation pathways facing the cytoplasm (7). We sought evidence as to whether homologous regions in the eukaryotic exchanger had similar structure and important functional roles. Particularly, we assessed the presence of deep aqueous cavities and their potential rearrangements during ion transport using cysteine scanning mutagenesis and electrophysiological analyses.
Mutations at Coordination Sites for Na+ and Ca2+.
By comparison with NCX_Mj, NCX1.1 residues Ala106, Ser109, Ser110, and Glu113 within TMS2 and Ala807, Thr810, Ser811, and Asp814 within TMS7 are predicted to coordinate ion binding (Fig. 1). Mutation at residues Glu113 and Asp814 resulted in nonfunctional exchangers, consistent with earlier Na+-dependent Ca2+ uptake studies (4). These results are expected, because two homologous residues in NCX_Mj (Glu54 and Glu213) contribute to multiple ion binding sites coordinating both Na+ and Ca2+ (7). We observed a similar phenotype for mutant S811C of TMS7. Based on NCX_Mj structure, Ser811 is found in the break of helix TMS7 between segments B and C and coordinates the Na+ ion nearest to the external surface. Serines within an unwound segment are common among Na+-coupled membrane transporters, providing coordination to Na+ ions (15). The slightly larger thiol group introduced at this position may prevent Na+ binding or affect the structure of the helix. In either case, our data underline the important role of this residue in transport function of the mammalian exchanger. Mutation at the remaining sites (Ala106, Ser109, Ser110 Ala807, and Thr810) resulted in low-activity exchangers with generally decreased apparent affinity for cytoplasmic Na+ and increased affinities for intracellular Ca2+. This result supports the notion that Na+ binding destabilizes Ca2+ from its site because of their close proximity (7). Replacement of Ala807 and Ser109 with cysteine produced the most prominent increases in Ca2+ affinity. Alanine 807 of TMS7 is homologous to Ala206 of NCX_Mj, which coordinates Na+ at the site nearest to the extracellular side (7). Because this residue participates in ion chelation with its backbone carbonyls, the effect of cysteine substitution is likely because of a steric hindrance destabilizing Na+ at this site and allowing Ca2+ to bind more efficiently. In contrast, Ser109 coordinates intracellular Na+ through its hydroxyl group and Ca2+ through its backbone carbonyl. A conservative substitution at this position (Ser to Cys) decreases the apparent affinity of NCX1.1 for intracellular Na+, and this phenotype was further enhanced by introduction of the apolar and smaller alanine, emphasizing the role of the side chain at this position in Na+ coordination.
The results show that residues participating in Na+ and Ca2+ binding in the mammalian NCX1.1 are analogous to those sites identified in NCX_Mj. The data provides strong functional evidence for the identity of the Na+ binding sites, which were only inferred in the NCX_Mj X-ray structure (7).
Previously, Na+-dependent 45Ca2+ uptake measurements indicated that mutations at sites 109, 110, and 810 lead to inactive transporters, despite their plasma membrane localization (4). Although the ionic currents generated by these mutants were low compared with WT, we could routinely measure their transport properties using the electrophysiological approach, enabling their initial characterization. Possibly, the large decrease in Na+ affinity for these mutants prevented measurements of a significant 45Ca2+ influx.
Mutagenesis Outside the Ion Coordinating Sites.
Within the α-repeats of NCX1.1 are two highly conserved regions: 108GSSAPE113 (TMS2) and 809GTSVPD814 (TMS7). As predicted by the NCX_Mj structure, the glycine and proline residues within these sequences are located deep in the protein but are not directly involved in Na+ and Ca2+ coordination (7). Our data showed that replacement of both glycines 108 and 809 and prolines 112 and 813 did not drastically alter NCX1.1 activity, suggesting that they are not essential for function. This result is striking because these amino acids are highly conserved among the exchangers, and both residues have essential structural roles in many membrane proteins. The homologous prolines in NCX_Mj participate in forming part of TMS2 and -7 helices instead of creating a helix break or kink, which may explain why they are more resistant to mutation (7). In NCX_Mj, these conserved prolines are also part of a hydrophobic patch that has been argued to allow the adjacent TMS1 and -6 to slide, facilitating the switch between the inward- and outward-facing states (7). By replacing the remaining residues of this hydrophobic patch, Ala111 and Leu115 of TMS2 and Val812 and Phe816 of TMS7, with cysteine, we showed the most substantial shifts in both Na+ and Ca2+ apparent affinities for L115C (Fig. 4 and Fig. S1), substantiating its important role in ion transport.
TMS2 and -7 Are Part of the Exchanger Cytoplasmic Ion Translocation Pathway.
By examining the effects of the membrane-impermeable thiol-reactive agent MTSET+ on mutants with cysteine substitutions within TMS2 and -7, we have mapped the residues accessible from the cytoplasm. Within TMS2, cysteines at positions 105, 106, 107, 109, and 110 were effectively modified by MTSET+ when the ion binding sites were trapped facing the cytoplasm (inward-facing state). As shown in Fig. 6 and Fig. S2, the model for the inward configuration of NCX_Mj shows a similar ionic-accessible surface of this portion of TMS2, with the exception of serine 51 (equivalent to Ser110 of NCX1.1), which is modeled as buried within the protein. However, our data clearly indicate exposure of this residue (S110C) to the cytoplasm when NCX1.1 is in the inward-facing configuration, suggesting a pocket deeper than predicted by the structural model. This result may underlie potential differences between the mammalian and archaebacterial isoforms. However, it is worth noting that the structural model represents a static picture. The protein during electrophysiological measurements is dynamic, and therefore, the cavity size likely fluctuates, making certain residues transiently accessible.
We investigated the accessibility of residues during transport initiated by the addition of Na+ at the intracellular surface. Currents generated by mutant exchangers M105C and S109C were completely blocked by MTSET+ in both the presence and absence of Na+. Cytoplasmic Na+, however, eliminated cysteine reactivity at positions 106, 107, and 110. Conformational changes occurring during transport may render these positions inaccessible, and/or a short-lived inward state may not provide sufficient time for cysteine modification. In contrast, mutants M105C and S109C seem to have greater exposure, allowing MTSET+ to also react during cycling. Note that, in the absence of Na+, MTSET+ reacted faster with these cysteines than mutations at positions 106, 107, and 110 (Fig. 5 C and D), supporting the idea that residues 105 and 109 have maximal accessibility from the cytoplasm when NCX1.1 is in the inward-facing state. An alternative explanation is that positions 105 and 109 are always exposed to the cytoplasmic aqueous environment, independent of the transport cycle. We consider the latter to be the less probable scenario, because residue 109 is part of one of the Na+ binding sites, as revealed by the NCX_Mj structure, and likely to become occluded during the transport cycle.
To map further the aqueous cavity, we analyzed the cytoplasmic accessibility of residues within TMS7. Among the eight mutants (A807C, L808C, G809C, V812C, P813C, F816C, A817C, and V820C), only currents recorded from the exchangers G809C, V812C, P813C, and A817C were inhibited by cytoplasmic MTSET+. By analogy to NCX_Mj, residues Gly809, Val812, and Pro813 are close to the binding site for Ca2+ near the center of the protein, and their accessibility indicates the presence of a deep cavity reaching the core of NCX1.1. As seen for positions 105 and 109, the rates at which MTSET+ blocked G809C, V812C, and P813C were extremely fast in the absence of transport, but the kinetics of reactivity markedly slowed during ion exchange.
The present accessibility data provide strong evidence that TMS2 and -7 are part of the ion translocation pathway of NCX1.1, and the changes in accessibility under different states of the transport cycle show that these regions rearrange during exchange.
The Eukaryotic Exchanger Seems to Have Two Cytoplasmic Ion Permeation Pathways of Different Sizes.
The consistency between our accessibility results and the recently proposed structural model of NCX_Mj (Fig. 6 and Fig. S2) strongly supports the presence of two inferred deep aqueous cavities that may allow Na+ and Ca2+ to reach their ion coordinating sites. The first aqueous cavity is delimited by both TMS2 (Met105 and Ser109) and -7 (Gly809, Pro813, Val812, Ala817, and Val820). This ion pathway penetrates deep into the core of NCX, as demonstrated by the accessibility of residues Gly809, Pro813, and Val812, and it is quite wide to accommodate the bulky reagent MTS-TPAE. The presence of a large cavity at these positions is also supported by kinetic studies revealing the fast time course of the MTSET+ block for P813C and V812C, regardless of whether NCX1.1 is held in the inward state or cycles. The result suggests that cytoplasmic MTSET+ can rapidly reach these positions as NCX1.1 transitions through the inward state during transport. The second cavity seems to be narrower, as judged by the slow reactivity of MTSET+ with the facing side chains of cysteines at positions 106, 107, and 110 of TMS2 and 833 of TMS8, in the inward configuration and the absence of MTSET+ modification of the same residues during transport. Consistent, with this hypothesis is the faster reactivity of position 110 with the smaller MTSMT+ reagent when NCX1.1 is held in the inward state and the inability of MTS-TPAE to significantly modify S110C currents.
It has been proposed that the two ion permeation pathways allow Na+ and Ca2+ to access their respective binding sites separately (7). Whether the kinetic data and size-dependent MTS reagents accessibility confirm the notion of the wider cavity being responsible for Ca2+ access and the narrower cavity being responsible for Na+ access will require additional investigation. Nevertheless, our data provide experimental evidence of two aqueous cavities of different sizes accessible from the cytoplasmic face of NCX1.1 that undergo conformational changes during transport. Comparison between the accessibility data presented here and the structural model of the inward-facing configuration of NCX_Mj suggests that these two proteins have similar architecture within the α-repeats. The data validates the structural information and suggests that the high-resolution structure of the archaebacterial NCX provides a foundation for studying the eukaryotic exchanger.
Methods
Molecular Biology.
Mutations were generated and sequenced in 200- to 400-bp cassettes from the full-length exchanger using QuikChange Mutagenesis (Stratagene). RNA was synthesized from linearized DNA using T3 mMessage mMachine (Ambion) and injected into Xenopus laevis oocytes as described in ref. 6.
Electrophysiology.
The apparent affinities for Na+ and Ca2+ and sensitivity to MTS reagents were assessed in giant inside-out patches (6, 16). Before recording, the excised patch is briefly exposed to chymotrypsin to remove NCX1.1 Na+ and Ca2+ secondary regulation (17). Experiments are performed at VH = 0 mV and 35 °C. Intracellular solutions were rapidly changed using a computer controlled 20-channel solution switcher. Pipettes were filled with 100 mM N-methylglucamine, 20 mM Hepes, 20 mM tetraethylammonium hydroxide, 8 mM Ca(OH)2, 0.1 mM niflumic acid, and 0.15 mM ouabain, pH 7 (using methanesulfonic acid). Bath solution contained 100 mM CsOH or NaOH, 20 mM tetraethylammonium hydroxide, 20 mM Hepes, and 10 mM EGTA, pH 7 (using methanesulfonic acid). MTS reagents (Toronto Research Chemicals) (Fig. S3) solutions were prepared immediately before each experiment because of the instability of compounds. MTSET+ (5 mM), MTSMT+ (5 mM), and MTS-TPAE (100 µM) were applied cytoplasmically in presence of either cytoplasmic Cs+ or Na+ and constant 8 mM external Ca2+ (pipette).
Supplementary Material
Acknowledgments
We thank Drs. Kenneth D. Philipson and Debora Nicoll for critical review of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218751110/-/DCSupplemental.
References
- 1.Philipson KD, et al. The Na+/Ca2+ exchange molecule: An overview. Ann N Y Acad Sci. 2002;976:1–10. doi: 10.1111/j.1749-6632.2002.tb04708.x. [DOI] [PubMed] [Google Scholar]
- 2.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 3.Lytton J. Na+/Ca2+ exchangers: Three mammalian gene families control Ca2+ transport. Biochem J. 2007;406(3):365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 4.Nicoll DA, Hryshko LV, Matsuoka S, Frank JS, Philipson KD. Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem. 1996;271(23):13385–13391. doi: 10.1074/jbc.271.23.13385. [DOI] [PubMed] [Google Scholar]
- 5.Shigekawa M, Iwamoto T, Uehara A, Kita S. Probing ion binding sites in the Na+/Ca2+ exchanger. Ann N Y Acad Sci. 2002;976:19–30. doi: 10.1111/j.1749-6632.2002.tb04710.x. [DOI] [PubMed] [Google Scholar]
- 6.Ottolia M, Nicoll DA, Philipson KD. Mutational analysis of the alpha-1 repeat of the cardiac Na+-Ca2+ exchanger. J Biol Chem. 2005;280(2):1061–1069. doi: 10.1074/jbc.M411899200. [DOI] [PubMed] [Google Scholar]
- 7.Liao J, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz EM, Benzer S. Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc Natl Acad Sci USA. 1997;94(19):10249–10254. doi: 10.1073/pnas.94.19.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doering AE, et al. Topology of a functionally important region of the cardiac Na+/Ca2+ exchanger. J Biol Chem. 1998;273(2):778–783. doi: 10.1074/jbc.273.2.778. [DOI] [PubMed] [Google Scholar]
- 10.Altimimi HF, Fung EH, Winkfein RJ, Schnetkamp PP. Residues contributing to the Na(+)-binding pocket of the SLC24 Na+/Ca2+-K+ Exchanger NCKX2. J Biol Chem. 2010;285(20):15245–15255. doi: 10.1074/jbc.M109.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicoll DA, Longoni S, Philipson KD. Molecular cloning and functional expression of the cardiac sarcolemmal Na+-Ca2+ exchanger. Science. 1990;250(4980):562–565. doi: 10.1126/science.1700476. [DOI] [PubMed] [Google Scholar]
- 12.Matsuoka S, Hilgemann DW. Steady-state and dynamic properties of cardiac sodium-calcium exchange. Ion and voltage dependencies of the transport cycle. J Gen Physiol. 1992;100(6):963–1001. doi: 10.1085/jgp.100.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilgemann DW, Nicoll DA, Philipson KD. Charge movement during Na+ translocation by native and cloned cardiac Na+/Ca2+ exchanger. Nature. 1991;352(6337):715–718. doi: 10.1038/352715a0. [DOI] [PubMed] [Google Scholar]
- 14.Iwamoto T, Uehara A, Imanaga I, Shigekawa M. The Na+/Ca2+ exchanger NCX1 has oppositely oriented reentrant loop domains that contain conserved aspartic acids whose mutation alters its apparent Ca2+ affinity. J Biol Chem. 2000;275(49):38571–38580. doi: 10.1074/jbc.M003788200. [DOI] [PubMed] [Google Scholar]
- 15.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilgemann DW, Lu CC. Giant membrane patches: Improvements and applications. Methods Enzymol. 1998;293:267–280. doi: 10.1016/s0076-6879(98)93018-x. [DOI] [PubMed] [Google Scholar]
- 17.Hilgemann DW. Regulation and deregulation of cardiac Na+-Ca2+ exchange in giant excised sarcolemmal membrane patches. Nature. 1990;344(6263):242–245. doi: 10.1038/344242a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.