Abstract
The T box leader sequence is an RNA element that controls gene expression by binding directly to a specific tRNA and sensing its aminoacylation state. This interaction controls expression of amino acid-related genes in a negative feedback loop. The T box RNA structure is highly conserved, but its tRNA binding mechanism is only partially understood. Known sequence elements are the specifier sequence, which recognizes the tRNA anticodon, and the antiterminator bulge, which base pairs with the tRNA acceptor end. Here, we reveal the crucial function of the highly conserved stem I distal region in tRNA recognition and report its 2.65-Å crystal structure. The apex of this region contains an intricately woven loop–loop interaction between two conserved motifs, the Adenine-guanine (AG) bulge and the distal loop. This loop–loop structure presents a base triple on its surface that is optimally positioned for base-stacking interactions. Mutagenesis, cross-linking, and small-angle X-ray scattering data demonstrate that the apical base triple serves as a binding platform to dock the tRNA D- and T-loops. Strikingly, the binding platform strongly resembles the D- and T-loop binding elements from RNase P and the ribosome exit site, suggesting that this loop–loop structure may represent a widespread tRNA recognition platform. We propose a two-checkpoint molecular ruler model for tRNA decoding in which the information content of tRNA is first examined through specifier sequence–anticodon interaction, and the length of the tRNA anticodon arm is then measured by the distal loop–loop platform. When both conditions are met, tRNA is secured, and its aminoacylation state is sensed.
Keywords: riboswitch, RNA-RNA complex
Cells must maintain appropriate intracellular pools of aminoacylated transfer RNA (aa-tRNA) to survive. This is usually accomplished by tight regulation of many cellular factors, including amino acid biosynthesis and transporter genes, and tRNA synthetases (1). Most Gram-positive bacteria use the T box regulatory system to control their aa-tRNA levels (2). T box elements are a special family of regulatory RNAs located in the 5′-untranslated region of the mRNA for a regulated gene or operon. Unlike small molecule-sensing riboswitches, T box RNAs recognize tRNAs containing the cognate anticodon and discriminate between uncharged tRNA and aa-tRNA. They usually control gene expression via premature transcription termination, although there are rare examples that are predicted to regulate at the level of translation initiation (2–4). The T box motif was initially identified in the Bacillus subtilis (Bsub) tyrosyl-tRNA synthetase gene (tyrS) and was shown to respond to the aminoacylation state of tyrosine tRNA (5, 6). More than 1,000 T box elements have since been identified (3, 4).
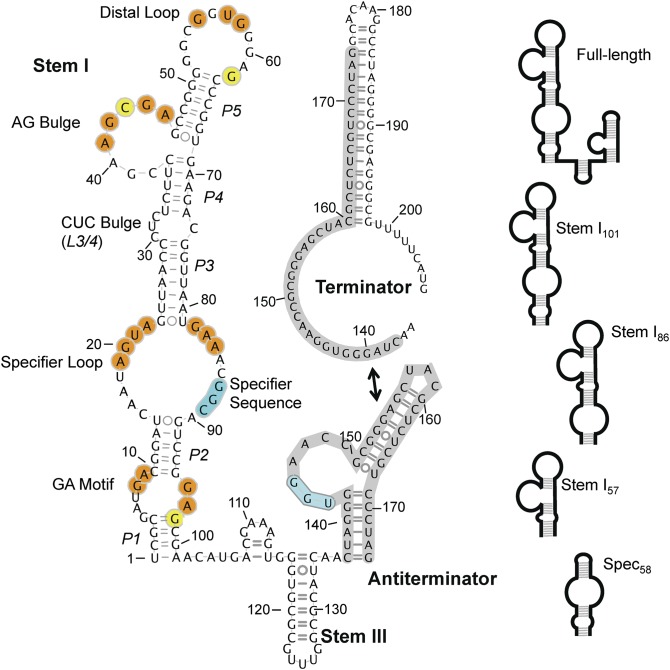
Selective tRNA recognition is achieved through a collective effort from several conserved domains in the T box RNA [Geobacillus kaustophilus (Gkau) glyQS T box sequence is shown in Fig. 1]. Stem I is the largest element. It is an ∼100-nucleotide (nt) stem–loop structure with several conserved interior features, from proximal to distal, including the Guanine-Adenine (GA) motif (K-turn), the specifier loop, a small variable bulge, an 8-nt AG bulge, and a large 11-nt distal loop. Cognate tRNA is selectively recognized by Watson–Crick (WC) base pairing between its anticodon and the specifier sequence, within the specifier loop, which is a codon that corresponds to the amino acid specificity of the regulated gene (6–10). Stem 2 and stem 3 form short, variable-length stem loops, and are usually separated by a pseudoknot element (stem 2A/B); stem 2 and the pseudoknot are absent in certain subclasses of T box RNAs, and their functions are unclear (2). Finally, the expression platform for the transcriptional T box system is its ability to form a transcription terminator or a competing antiterminator structure (11). The terminator is thermodynamically more stable; however, the alternatively base-paired antiterminator is stabilized when a conserved UGGN sequence motif in its internal bulge base pairs with the complementary NCCA sequence at the 3′-end of an uncharged tRNA, thereby preventing formation of the terminator helix and allowing the synthesis of the full-length transcript (12). The aa-tRNA is not capable of the same interaction, presumably as a result of steric hindrance. The prevailing model for the T box system envisions a stepwise recognition of tRNA. First, tRNA is recruited by anticodon–specifier sequence pairing, and next the tRNA aminoacylation state is sensed to affect terminator formation. As a consequence, the ratio of the aa-tRNA to uncharged tRNA is more important than the total level of tRNA in determining the outcome (13).
Fig. 1.
The Gkau glyQS T box RNA secondary structure. Nucleotides in stem I that are conserved in >70% of transcriptional T box RNAs (4) are highlighted in orange or yellow, indicating their presence or absence in the Gkau glyQS T box RNA, respectively. Stem II and the pseudoknot are absent from several glyQS T box RNAs. Regions previously shown to bind tRNA are highlighted in cyan. (Top Right) The alternatively paired terminator is shown with the common region highlighted in gray. (Right) Schematic representations of construct used in this study.
Outside of specifier sequence–anticodon and antiterminator–acceptor pairing, the specifics of T box–tRNA binding are unclear. For example, several regions within the stem I AG bulge and distal loop are highly conserved, and mutational analysis demonstrated their importance for antitermination (14); however, their functions are unknown (3, 4). In this study, we show that stem I forms a stable complex with tRNA that requires the distal portion of stem I, in addition to the specifier loop. We further solve the X-ray crystal structure of the distal region of stem I, which reveals a tightly woven loop–loop structure between the highly conserved AG bulge and distal loop. The RNA conformation in this region resembles the tRNA recognition elements in RNase P and the 70S ribosome, all of which include base stacking against the tRNA elbow region (D- and T-loops). Follow-up experiments with the use of several techniques support the existence of this tRNA contact. With the aid of small-angle X-ray scattering (SAXS), we propose a double-checkpoint tRNA recognition model that examines the information content and the geometry of tRNA to ensure the recruitment of a cognate ligand.
Results
In Vitro Reconstitution to Dissect T Box–tRNA Interactions.
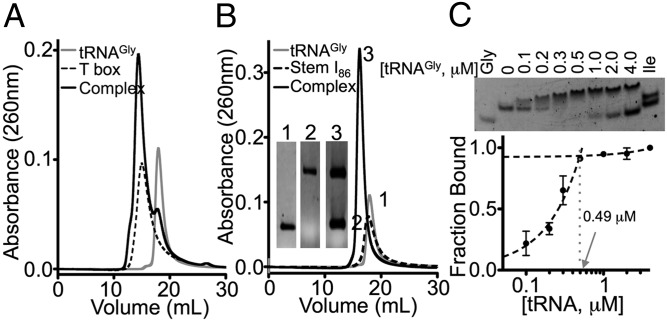
The Gkau glyQS T box sequence contains all notable secondary structural features of the well-studied Bsub glyQS T box RNA, but its folding is predicted to be thermodynamically more stable (Fig. S1 shows T box constructs used in this work) (15). Indeed, the full-length Gkau glyQS T box RNA refolded into a single species, whereas the Bsub glyQS T box RNA folded into two conformers under a variety of refolding conditions (Fig. S2A). Size-exclusion chromatography (SEC) confirmed that purified T box RNAs bind glycine tRNA (tRNAGly), but the Gkau RNA was notably more homogeneous, and therefore was chosen as the model system for this work (Fig. 2A and Fig. S2B). Regulatory RNAs are typically modular in nature, so to determine whether stem I alone is capable of binding tRNA, stem I101 (nucleotides 1–99, with an additional P1 G⋅C pair, Fig. S1), was produced. It formed an equimolar complex with tRNA, with a calculated mass of ∼58 kDa and ∼50 kDa (hydrodynamic radius, 4.7 nm), determined by using RNA standard curves for SEC and dynamic light scattering, respectively (Fig. S2 C and D). These values were only slightly larger than the expected 47-kDa molecular weight for a 1:1 complex.
Fig. 2.
In vitro reconstitution of T box RNA–tRNAGly complexes. SEC elution profiles for equimolar (A) full-length T box RNA and (B) stem I86 and tRNAGly. (Inset) Denaturing PAGE for labeled peaks. (C) EMSA for stem I86 (0.5 μM) titrated with tRNAGly (0–4 μM, shown on log scale). Isoleucine tRNA (tRNAIle, Ile) and tRNAGly alone (Gly) controls are shown. Dashed lines represent a linear fit to data between 0 and 0.5 μM and between 0.5 and 4 μM, demonstrating that binding is approximately linear to saturation (1:1) under these conditions.
To identify tRNA-binding requirements, a series of stem I deletion mutants was generated: (i) stem I86 (nucleotides 10–95) contains everything above the GA motif; (ii) stem I57 (nucleotides 24–80) encompasses the distal portion above the specifier loop; and (iii) spec58 (nucleotides 2–28, GAAA, nucleotides 75–100) contains the base of stem I to just beyond the specifier loop (Fig. 1 and Fig. S1). Stem I86 formed a robust 1:1 complex with tRNA (Fig. 2B), but no stable complexes were observed for the other truncations. Electrophoretic mobility shift assays (EMSA) were used to estimate binding affinities for tRNA. Stem I101 and stem I86 bound tRNA with apparent dissociation constants (Kd < 0.25 μM; Fig. 2C and Fig. S2 E and F); however, complexes were unstable in the EMSA conditions at concentrations low enough to accurately determine binding affinities, and therefore are presented as an upper limit for the affinity. tRNA binding was completely abolished in stem I57 and significantly impaired in spec58 (Fig. S2G). Titrating tRNA into spec58 caused the band to smear and disappear with increasing tRNA, but did not result in a homogeneous shift. Data for the disappearance of spec58 could not be modeled by a single binding site. Instead, the data more closely resembled multiple binding sites that reached half saturation at ∼12 μM (Fig. S2H), a >50-fold increase in Kd relative to the upper limit for stem I101. Overall, our deletion mapping data demonstrate that, in addition to the previously recognized specifier loop, the distal portion of stem I is essential for robust tRNA binding.
Distal Portion of Stem I Contains a Loop–Loop Structure Resembling Key tRNA Recognition Element in RNase P.
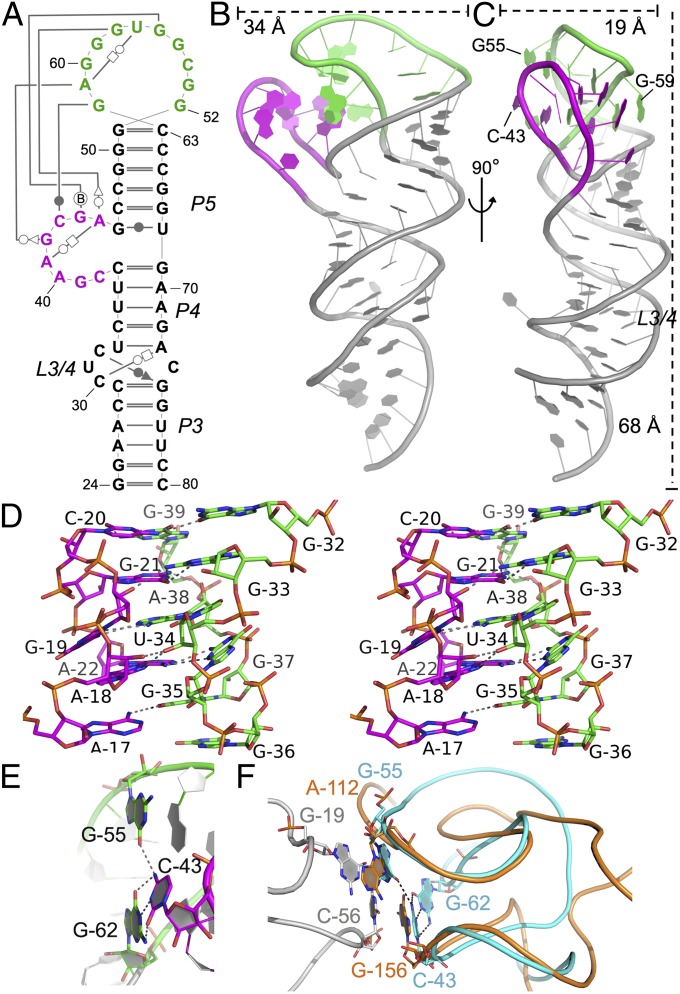
We determined the 2.65-Å crystal structure of the distal region of stem I to gain insight into its potential tRNA-binding function (stem I57, nucleotides 24–80; Fig. 3A). The overall arrangement of this portion of the T box resembles an upper limb. The three helical segments (P3, P4, and P5) in stem I57 are coaxially stacked, resembling the lower (P3) and upper arms (P4/5; Fig. 3 B and C), bent by ∼45° at an internal bulge, L3/4 (i.e., elbow). This bulge is extensively stabilized through tertiary contacts; C30 forms a C30⋅A73-U33 base triple via trans-WC–Hoogsteen pairing and C32 forms a C33⋅G75-C29 base triple via a cis-WC–sugar pair, leaving C74 as a nucleotide flip-out poorly resolved in the electron density (Fig. 3A; Fig. S3 shows hydrogen bond distances).
Fig. 3.
Stem I57 crystal structure. (A) Secondary structure of the stem I57 crystallization construct with tertiary contacts drawn by using the Leontis–Westhof notation (43), with the AG bulge (magenta) and distal loop (green) highlighted. (B and C) Orthogonal views of the stem I57 crystal structure shown as a cartoon, colored as in A. (D) Stereo image of the AG bulge–distal loop interaction. (E) The apical base triple, highlighted in sticks, colored as in B and C. Oxygen, nitrogen, and phosphorus are shown in red, blue, and orange, respectively. Hydrogen bonds are shown as dashed lines. Stem I57 is in the same orientation as in C. (F) Superposition of the stem I apical loops (cyan) and the RNase P specificity domain (orange), which is in complex with tRNA (gray) [Protein Data Bank (PDB) ID 3Q1Q]. Structures are shown as ribbons with interacting bases highlighted in sticks. Hydrogen bonds in the stem I apical base triple are highlighted as dashed lines.
The most prominent structural feature is found in the “hand” region of distal stem I, where the highly conserved AG bulge (thumb; nucleotides 38–45) and the 11-nt G-rich distal loop (fingers; nucleotides 52–62) form extensive loop–loop tertiary interactions. The AG bulge (thumb) folds into a narrow flap via a tight turn at C43 and multiple base–ribose contacts, and pinches toward the G-rich distal loop (palm and fingers) through five layers of tertiary interactions (three base triples and two base–ribose interactions; Fig. 3D and Fig. S3). The central axis of the loop–loop interface is formed by stacked bases in a zipper-like arrangement by alternating bases from each element, in the sequence of G62-G44-A61-A45-G60 from top to bottom. The network of contacts starts from the G62-C43⋅G55 base triple at the apex, and ends with a trinucleotide internal turn in the distal loop (G58–G60), stabilized from the minor groove side by an adenosine-platform–like contact from the AG bulge (G58⋅A45⋅A41 base triple; Fig. 3 D and E and Fig. S3). The intricate loop–loop hydrogen-binding network explains the sequence conservation in these two loops (3, 4). Additional tertiary contacts are found outside the loop–loop interface, stabilizing the rest of the structure. These are primarily base to sugar–phosphate backbone hydrogen bonds within the AG-bulge and distal loop as well (Fig. S3). The extensive tertiary contacts in the hand region alter the base stacking direction in the loop–loop region to be approximately perpendicular to the P3–P4–P5 Superhelix, exposing stacking and hydrogen bonding surfaces on both sides to form crystal packing interactions (Fig. 3C). In the front side (minor groove side of the P5 helix), the G62-C43⋅G55 base triple stacks vertically on top of the equivalent bases in the pseudo-twofold related dimer (Fig. S4 A–C). The backside of the loop–loop structure forms another, less extensive crystal contact, although the residues involved are not highly conserved (Fig. S4 D and E).
The elaborate loop–loop structure and its ability to mediate RNA–RNA contacts prompted us to investigate the likelihood of its direct involvement in tRNA recognition. Known tRNA-binding RNA structures were surveyed for a similar theme. tRNA recognition in the RNase P involves a stacking interaction from A112 and G156 of its specificity domain to the tRNA D- (G19) and T- (C56) loops, respectively (16). Although the specificity domain loops are formed by an entirely different supporting structure, the resulting conformation at their surface closely resembles the conformation of the T box stem I apex (Fig. 3F). The structures overlay (17, 18) with an rmsd of ∼2.4 Å over all sugar–phosphate backbone atoms in the corresponding loops (15 nt, 180 atoms from each molecule). Importantly, in this alignment, C43 and G55 from the stem I apical base triple directly overlay with the tRNA-binding bases in RNase P, G156, and A112, respectively. Recognition of tRNA D-/T-loops through base stacking is also found in the 70S ribosome structures, suggesting that this may be a conserved tRNA recognition mechanism (Discussion).
Mutagenesis Revealed Importance of G55 in Apical Base Triple.
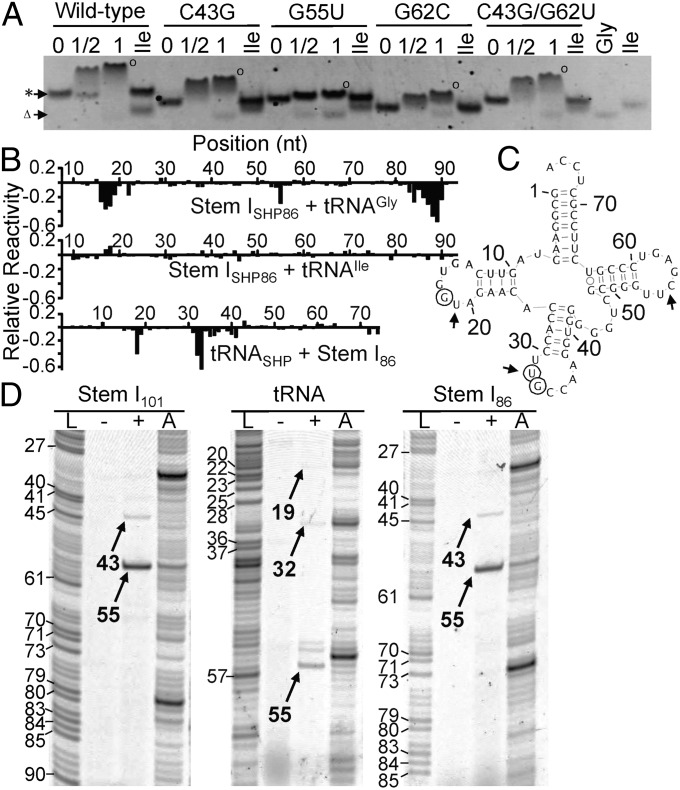
The structural resemblance to the RNase P tRNA-binding platform prompted us to further evaluate the function of the loop–loop structure by using mutagenesis. In particular, we focused on the G62-C43⋅G55 base triple that we hypothesize to stack directly against the D-/T-loops of tRNA. Interestingly, G55 is highly conserved among T box RNAs, but the G62–C43 base pair is not conserved [A43 and A62 in >70% of sequences (4); Fig. 1]. C43G and G62C variants had increased mobility in native PAGE relative to wild type, but, under these conditions, neither mutant significantly impaired stem I86–tRNAGly complex stability (Fig. 4A). C43G/G62C restored the WC base pair and restored the migration behavior of the stem I86-tRNAGly complex to the wild-type level; this is consistent with the prediction from lack of sequence conservation that the identity of these bases in Gkau stem I86 is not essential for tRNAGly binding (Fig. 4A). Replacing the highly conserved G55 (>80%) (4) with a U residue almost completely disrupted complex formation. The stem I86 G55U band is only slightly retarded, and uncomplexed tRNAGly was observed, even with G55U in twofold excess (Fig. 4A). Overall, our mutagenesis results clearly indicate that G55 in the apical base triple is essential for tRNA binding.
Fig. 4.
Probing the stem I–tRNAGly interaction. (A) EMSA for stem I86 point mutants (0.5 μM; asterisk) with tRNAGly (Gly; Δ) at molar ratios of 1:0, 1:0.5, and 1:1. tRNAile (Ile; 1:1) was used as a negative control. (B) SHAPE analysis showing tRNAGly-induced protection of stem I86 with no significant protection by tRNAIle and stem I86–induced protection of tRNAGly. Values represent SHAPE reactivity of each nucleotide in the presence of excess tRNAGly/stem I86 minus stem ISHP86/tRNASHP alone. Raw SHAPE data are shown in Fig. S5. (C) tRNAGly secondary structure with nucleotides significantly protected upon complex formation indicated by open circles. Arrows indicate cross-link sites identified in D. (D) Denaturing PAGE of reverse transcribed cross-linked samples (stem ISHP101, stem ISHP86, and tRNASHP) in the presence (+) or absence (−) of excess unlabeled binding partner. A, reverse transcription of the cross-linked sample before gel purification; L, ddTTP ladder. Cross-linking specific products are numbered after adjusting for RTase blockage (−1 base).
Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension Further Established New tRNA Contact Through Stem I Apex.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (19) was used to identify structural effects of tRNA binding. Additional sequences were introduced into stem I101, stem I86, or tRNAGly to allow SHAPE readout through reverse transcription (19); these modifications did not affect their ability to form a complex (Fig. S6A). In the absence of tRNA, the SHAPE reactivity profiles of stem I101 and stem I86 were consistent with their structure models (Fig. S5) the GA motif and specifier loop were highly reactive, whereas the AG bulge and distal loop were poorly reactive as a result of the formation of the loop–loop structure. These findings suggest that the addition of the 3′ SHAPE primers did not affect complex formation. In the presence of excess tRNA, stem I was significantly less reactive at three main regions: G55 (the most reactive base in the apo-state distal loop); A16 to A19 (5′ side of the specifier loop); and A84 to A90 (the specifier sequence and flanking bases; Fig. 4B and Fig. S5). Whereas protection in the specifier loop region was anticipated because this region is known to interact with the tRNA anticodon loop, protection at G55 in the apical base triple was not previously recognized, and strongly supports our hypothesis that the loop–loop structure plays a direct role in tRNA binding, although, because of preferential reactivity of 1-methyl-7-nitroisatoic anhydride with flexible nucleotides, tRNA binding at any region within the distal loop–loop structure could induce the observed changes.
To identify the complimentary binding interface, tRNA SHAPE analysis was performed with excess stem I101 or stem I86. Consistent with tRNA crystal structures, Gkau tRNAGly alone was most reactive at its D-loop, anticodon loop, variable loop, and the CCA tail, and only moderately reactive at the T-loop (Fig. S5). The presence of stem I101 or stem I86 significantly reduced the tRNA reactivity at U32–G33 in the anticodon and G18 in the D-loop (Fig. 4B). The expected anticodon protection confirms that both stem I constructs specifically recognize this region. G18 protection in the D-loop is consistent with a previous studies that used inline probing of tNRAGly in complex with full-length Bsub glyQS T box RNA (20, 21). Further, the protection mirrors our stem I SHAPE data and mutagenesis in suggesting that these regions form the additional T box RNA–tRNA contact.
UV Cross-Linking Identified Distal Stem I–tRNA Contact.
UV cross-linking was used to establish direct contact points between the T box RNA and tRNA. UV cross-links can form between nucleotides that are in close proximity and achieve appropriate geometry, and can be used to identify potential RNA–RNA contacts (22). Even after short UV exposure (<5 min), the stem I101, stem I86, or tRNA constructs form a unique species with dramatically decreased mobility by denaturing PAGE, as expected for an X-shaped RNA molecule, only in the presence of tRNAGly or stem I, respectively (Fig. S6B). The cross-linked species is most likely the 1:1 stem I–tRNA complex, as its migration on native PAGE was indistinguishable from that of the un–cross-linked 1:1 complex (Fig. S6 C and D). The cross-linked species were excised and analyzed by reverse transcriptase (RTase). RTase is blocked 1 nt upstream from an RNA cross-link (23), which allowed us to locate potential cross-linking sites. Stem I101 and stem I86 had two clear RTase blockages, a strong site at G55 and a weak one at C43 (Fig. 4D). Interestingly, these nucleotides are members of the apical base triple, and although it is possible that they are cross-linked to one another, two details argue for intermolecular cross-links instead: (i) RTase will be blocked by the 3′-most cross-link in any given RNA, so C43 would not be observed if it formed an intramolecular cross-link to G55; and (ii) these are the only cross-links observed in the purified tRNA cross-linked complex, so at least one must be forming the intermolecular bond. Notably, no cross-links were observed at the specifier loop, which may be a result of more constrained geometry. Reverse transcription of tRNA revealed potential cross-links at C55, U53, and G52 from the T-loop, U32 from the anticodon loop, and U19 in the D-loop (Fig. 4 C and D). It is unclear whether the U32 band represents an intra- or intermolecular cross-link, as the corresponding band was not detected in stem I; however, the D- and T-loop cross-links correlate well with the distal stem I cross-links. These data, in concert with the distal base-triple mutagenesis and SHAPE analysis, clearly establish a direct physical contact between the distal portion of stem I and the D-/T-loop of its ligand tRNA.
SAXS Supports in Silico-Generated T Box–tRNA Structure Model.
To put our developing model for stem I–tRNA binding into a more concrete structural context, a molecular model for stem I was assembled from our stem I57 crystal structure and a previous NMR structure of the specifier loop region of Bsub tyrS (9, 10). The 3-bp overlap in P3 enabled alignment of these two models (Fig. 5A). The CUC bulge between helix P3 and P4 caused a large arch in the stem to position the specifier codon WC edges and the apical base triple (G55⋅C43-G62) ∼60 Å apart on the same face (Fig. 5A). Notably, the distance between the tRNA anticodon and U19 in its D-loop is also ∼60 Å. tRNA was modeled by extracting mRNA and petidyl site tRNA from the Thermus thermophilus ribosome structure (24) and overlaying the mRNA codon with the specifier sequence. This projects tRNA up from the specifier loop, along the length of stem I such that the D- and T-loops (U19 and G55) are within bonding distance to the apical base triple (Fig. 6).
Fig. 5.
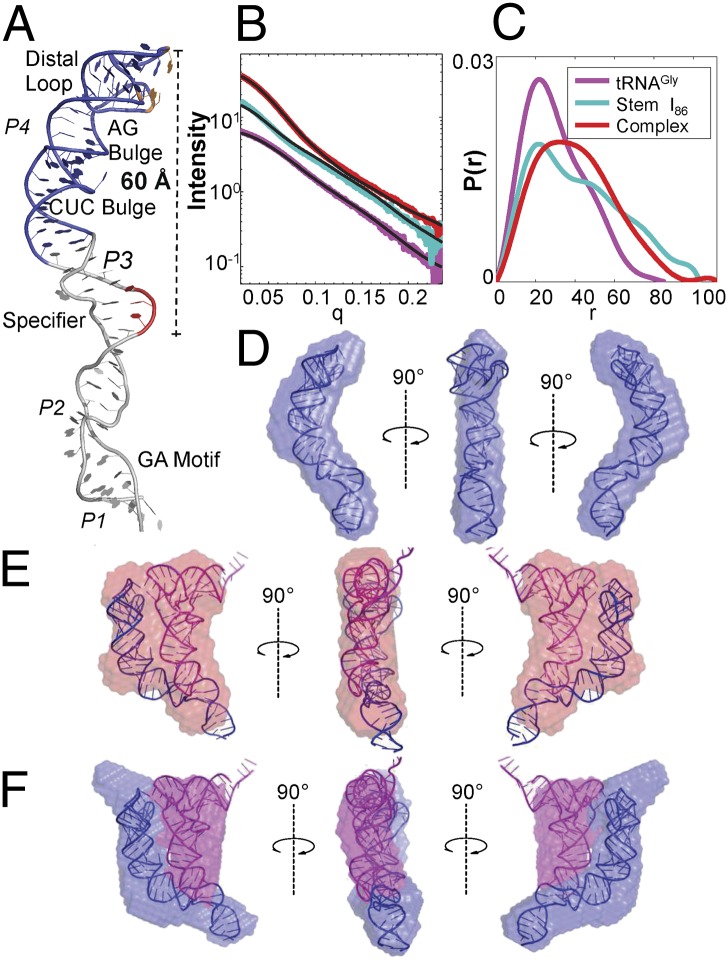
Model for stem I–tRNA interaction. (A) Hypothetical model of stem I constructed from the stem I57 crystal structure (blue) and stem I base NMR structure (gray) (PDB ID 2KZL). The specifier sequence and apical base triple are highlighted in red and orange, respectively. (B) Experimental SAXS curves and (C) pair distance distribution function P(r) of the tRNA (magenta), T box–stem I86 (cyan), and stem I86–tRNA complex (red). Black lines represent the theoretical scattering profiles calculated by CRYSOL from the tRNA crystal structure (PDB ID 2J00) and our predicted structures of T box stem I86 and its complex. (D) Docking of the T box stem I86 model into the SAXS-reconstructed envelope from DAMMIF. (E) Docking of the stem I86–tRNA complex model into the SAXS-reconstructed envelope from DAMMIF. (F) Docking of the stem I86–tRNA complex model into the averaged two-phase MONSA-reconstructed model.
Fig. 6.
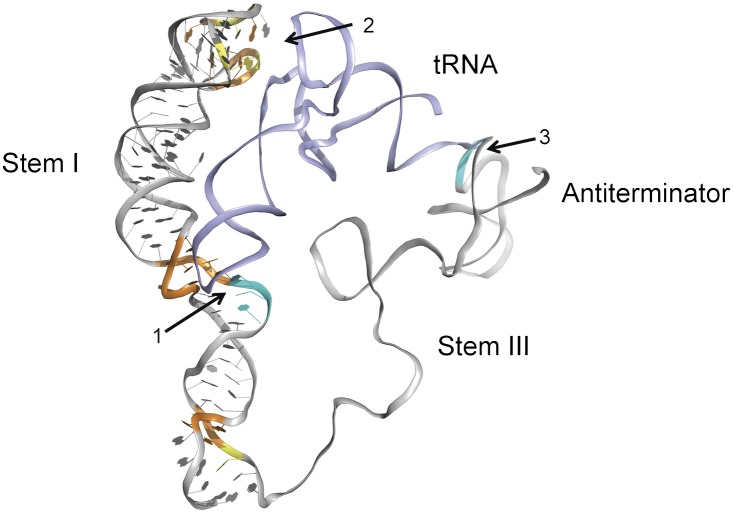
A two-checkpoint molecular ruler model for tRNA recognition by the T box RNA. The T box RNA is shown in the antitermination (“ON”) state. The RNA is shown as ribbons with tRNA (light blue) docked against stem I and the antiterminator [based on the published NMR structure (44)]. Sequence conservation and known tRNA contacts are colored as in Fig 1. The information checkpoint (1 represents the specifier sequence for tRNA anticodon recognition), geometry checkpoint (2 represents the apical region for recognition of tRNA D-/T-loops 60 Å away), and the antiterminator contact (3) are highlighted with arrows.
SAXS was used to validate the proposed structural models of the stem I–tRNA complex. The experimental SAXS profiles (Fig. 5B; Fig. S7B provides Kratky representations) were superimposed with the CRYSOL-calculated (25) theoretical scattering profiles of tRNA (using P-site Escherichia coli phenylalanine tRNA) (24), T box stem I86, and the binary complex (both based on our hypothetical structure models). The radius of gyration calculated by a linear fit through the Guinier regime yielded 32.72 ± 0.20 Å for T box stem I86 and 31.62 ± 0.11 Å for the complex. These values agreed well with the CRYSOL-calculated values from our hypothetical models of 31.61 Å and 32.08 Å, respectively. The shape of the pair distance distribution function P(r) (Fig. 5C) calculated by GNOM (26) indicated that T box stem I86 adopted a kinked, rod-like structure whereas the tRNA-bound complex was similar to a flat disk (27). These distributions were also consistent with the overall shapes predicted by our models. Ab initio SAXS reconstruction was carried out to investigate the structural and geometrical properties of the three RNA samples in real space, with the use of the programs DAMMIF and MONSA (28, 29). Our hypothetical stem I86 and stem I86–tRNA structures fit tightly inside the density envelope of their unbiased SAXS reconstructions (Fig. 5 D and E and Fig. S7 C–E). Two-phase reconstructions were further carried out using by MONSA to delineate the contributions from the two distinct RNA components. These reconstructions revealed two points of contact between tRNA and T box stem I86 (Fig. 5F and Fig. S8 D–F), consistent with a two-point recognition model involving the concept of a “molecular ruler”: (i) the specifier loop examines the tRNA anticodon loop and (ii) the distal loop 60 Å away stacks against the tRNA D-/T-loops.
Discussion
The function of T box RNA in tRNA-dependent gene regulation is well established; however, a complete physical model describing detailed T box RNA–tRNA interactions is lacking. The prevailing model depicts base-pairing interactions between the T box and tRNA at the specifier sequence–anticodon and antiterminator–acceptor regions. The former interaction is responsible for recruiting specific tRNA, and the latter is responsible for sensing its aminoacylation state (6, 7). In this work, we identified a conserved region in stem I that is required for robust tRNA binding. Deletion of this element (spec58), resulted in >50-fold reduction in tRNA binding affinity. The reduced affinity is similar to that of the truncated Bsub tyrS T box stem I and the tRNA anticodon arm (8, 10, 21). This provided experimental evidence that, in addition to the specifier loop, the distal region is important for tRNA binding.
The X-ray crystal structure provided substantial insight into the potential tRNA-binding functions for the uncharacterized distal region of stem I. The intricate distal loop–loop structure explains the high sequence conservation in the AG bulge and distal loop (3, 4). Rollins et al. (14) previously determined that mutation of highly conserved regions in the AG bulge and distal loop impaired antitermination. These disruptive mutations all map to nucleotides forming loop–loop interactions (Fig. 3D), clearly implicating the biological importance of the intricate structure (14). Despite lacking sequence or gross structural conservation, the stem I apex resembles the tRNA D-/T-loop binding platform in RNase P architecturally (Fig. 3F) (16). This inspired us to analyze stem I–tRNA contacts by SHAPE, in which we identified high protection at G55 in the loop-loop region upon tRNA binding. Previous inline cleavage of the Bsub glyQS T box RNA did not detect distal stem I contacts, although the importance of the tRNA D- and T-loops for binding was noted (21). As the apical sequence is nearly identical in the Bsub and Gkau glyQS T box RNAs, the interaction was likely missed as a result of the lack of reactivity of this region to inline attack, or the tendency of Bsub T box RNA to misfold (Fig. S2A). Together, the SHAPE data, mutagenesis and cross-linking provide cross-validated evidence that in addition to the specifier sequence–anticodon pairing, the apical base triple forms an essential binding platform that base stacks against the tRNA D- and T-loops. With strong biochemical evidence pointing toward this two-point interaction, we turned to structural modeling and SAXS to test the hypothesis. Both techniques strongly support a role for the distal base triple in tRNA binding. Our in silico T box–tRNA complex model is in agreement with several previous findings by Yousef et al. that extending the tRNA anticodon arm by more than a single base pair or mutating the D- and T-loop base pair G19 and C56 significantly impaired its regulatory function (20, 21). Either of these changes would be expected to impair distance and structure-specific interactions at both the specifier loop and the stem I apex and were sufficient to significantly impair function.
A broad survey of tRNA recognition by structured RNAs reveals similar recognition not only in the RNase P–tRNA complex, as described earlier, but also in the bacterial ribosome E-site–tRNA complex. tRNA is anchored by the D- and T-loops and along the acceptor arm (Fig. 3F and Fig. S4G) by highly conserved bases in RNase P (16). In the ribosome, the E-site anchors tRNA by stacking of the D- and T-loop bases against G2112 and G2168 from separate loops of 23S ribosomal RNA in the classic and hybrid states (Fig. S4 H and I) (24, 30). The similar binding platforms in RNase P and the ribosome, in the absence of sequence or gross structural conservation, suggests that variations to the stem I loop–loop structure could represent a widespread tRNA binding platform. Perhaps more intriguingly, the ribosome contacts provide similar multilevel specificity for tRNA observed in the T box system, i.e., sequence-specific contacts at the mRNA and structure-specific contacts ∼60 Å away at the D- and T-loops. T box RNA features that enable this two-point molecular ruler are highly conserved (3, 4).
Overall, our data revealed an important function for the highly conserved structure motif in stem I of the T box RNA. We propose a revised two-checkpoint molecular ruler model for tRNA recognition in vivo that involves two levels of decoding. First, as RNA polymerase transcribes stem I, the information content of the tRNA ligand is sensed through the specifier sequence–anticodon base pairing interaction. Next, the length of the tRNA anticodon arm is measured by the stem I apical base triple. Only when tRNA passes both quality controls is it firmly anchored against stem I. When it has been anchored, as the antiterminator sequence is transcribed, the tRNA aminoacylation state is monitored by interaction with the antiterminator bulge to determine antiterminator/terminator formation and downstream gene expression (Fig. 6).
Materials and Methods
Cloning and RNA Transcription.
RNA constructs were designed and produced as previously described (31) with minor modifications, as described in SI Materials and Methods.
Complex Formation.
tRNA binding by SEC, dynamic light scattering, and EMSA were performed as described in SI Materials and Methods.
Crystallization, X-Ray Data Collection, and Structure Solution.
RNA was crystallized as described in SI Materials and Methods. Data were collected at Cornell High Energy Synchrotron Source on beam line A1 and were processed and scaled by using HKL2000 (32). The structure was phased by using ShelxCD (33, 34) in the HKL2Map interface (35) and AutoSol (36) from the Phenix suite (37). The structure was built and refined by using Coot (38), Rosetta ERRASER (39) and Phenix.Refine (40). Full details are described in SI Materials and Methods.
SHAPE Analysis.
All SHAPE experiments were performed as previously described (19, 41) with minor modifications, as detailed in SI Materials and Methods. Data were analyzed by using ShapeFinder software (42).
UV Cross-Linking.
Experiments are described in SI Materials and Methods.
SAXS Data Collection, Processing, and Reconstructions.
SAXS data were collected on the Cornell High Energy Synchrotron Source G1 beam line, and reconstructions were produced as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Arthur Woll [Cornell High Energy Synchrotron Source (CHESS) G1 station] for experimental assistance, Steve Meisburger for beam-stop development, Huimin Chen and Joshua Tokuda for small-angle X-ray scattering data collection, Fang Ding for technical help, and Ki Hyun Nam for helpful discussion. This work was supported by National Institutes of Health Operating Grants GM-086766 (to A.K.), GM-088645 (to L.P.), and GM-047823 (to T.M.H.); and a Canadian Institutes of Health Research Postdoctoral Fellowship (to J.C.G.). This work is based on research conducted at CHESS, which is funded by National Science Foundation Division of Materials Research Grant 0936384, using the Macromolecular Diffraction at CHESS facility, which is funded by National Institute of General Medical Sciences Grant GM103485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinate and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 4JRC (T box distal domain)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222214110/-/DCSupplemental.
References
- 1.Henkin TM. Regulation of aminoacyl-tRNA synthetase gene expression in bacteria. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Austin, TX: Landes Bioscience; 2005. pp. 309–313. [Google Scholar]
- 2.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584(2):318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14(4):717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73(1):36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: Conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174(4):1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 7.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: Codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99(17):11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson AR, Henkin TM, Agris PF. tRNA regulation of gene expression: Interactions of an mRNA 5′-UTR with a regulatory tRNA. RNA. 2006;12(7):1254–1261. doi: 10.1261/rna.29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Henkin TM, Nikonowicz EP. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res. 2010;38(10):3388–3398. doi: 10.1093/nar/gkq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Nikonowicz EP. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J Mol Biol. 2011;408(1):99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: Disparity between conservation and functional requirements. Nucleic Acids Res. 2002;30(7):1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy FJ, Collins JA, Rollins SM, Henkin TM. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA. 2000;6(8):1131–1141. doi: 10.1017/s1355838200992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol. 2005;346(1):73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 14.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25(2):411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiter NJ, et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468(7325):784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1(3):1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 20.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: A new twist on tRNA. RNA. 2003;9(9):1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349(2):273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 22.Behlen LS, Sampson JR, Uhlenbeck OC. An ultraviolet light-induced crosslink in yeast tRNA(Phe) Nucleic Acids Res. 1992;20(15):4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris ME, Christian EL. RNA crosslinking methods. Methods Enzymol. 2009;468:127–146. doi: 10.1016/S0076-6879(09)68007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 25.Svergun D, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28(6):768–773. [Google Scholar]
- 26.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25(4):495–503. [Google Scholar]
- 27.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66(10):1735–1782. [Google Scholar]
- 28.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999 doi: 10.1016/S0006-3495(99)77443-6. 76(5):2879-2886, and erratum (1999) 77:2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34(3):408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, ed Carter, Jr. CW (Academic, San Diego), Vol 276, pp 307–326. [DOI] [PubMed] [Google Scholar]
- 33.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(pt 10 pt 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 35.Pape T, Schneider TR. HKL2MAP: A graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 36.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat Methods. 2013;10(1):74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu C, et al. Variable sequences outside the SAM-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. J Mol Biol. 2011;409(5):786–799. doi: 10.1016/j.jmb.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14(10):1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7(4):499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: Seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol. 2003;326(1):189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.