Abstract
Hepatitis C virus (HCV) envelope glycoprotein E2 has been considered as a major target for vaccine design. Epitope II, mapped between residues 427–446 within the E2 protein, elicits antibodies that are either neutralizing or nonneutralizing. The fundamental mechanism of antibody-mediated neutralization at epitope II remains to be defined at the atomic level. Here we report the crystal structure of the epitope II peptide in complex with a monoclonal antibody (mAb#8) capable of neutralizing HCV. The complex structure revealed that this neutralizing antibody engages epitope II via interactions with both the C-terminal α-helix and the N-terminal loop using a bifurcated mode of action. Our structural insights into the key determinants for the antibody-mediated neutralization may contribute to the immune prophylaxis of HCV infection and the development of an effective HCV vaccine.
Hepatitis C virus (HCV) infection is a major public health problem with an estimated 170 million people infected worldwide (1). HCV is transmitted primarily through direct contact with the blood or other bodily fluids of an infected individual. Although acute hepatitis C is typically mild or even subclinical, the infection becomes chronic in more than 75% of those infected (2, 3). Patients with chronic HCV infection have a high risk of developing cirrhosis and, in some cases, hepatocellular carcinoma (2, 3).
Significant advances have been made in the treatment of hepatitis C with the recent introduction of HCV-specific protease and polymerase inhibitors; sustained virologic responses, tantamount to cure, can now be achieved in more than 70% of the most difficult to treat HCV genotype 1-infected patients (4). However, the use of such drugs for treatment is not economically or logistically feasible in most parts of the world; therefore, vaccine development remains an important goal for the global control of HCV infection. Thus far, no HCV vaccine formulation has been able to induce sterilizing immunity, but a recombinant envelope protein vaccine has significantly reduced the rate of chronic HCV infection in a chimpanzee model (5). Thus, designing a vaccine that successfully elicits neutralizing antibodies remains a practical strategy to either prevent primary HCV infection or to reduce the frequency of progression from acute to chronic HCV infection (6).
HCV envelope glycoprotein E2 has been studied extensively as a potential candidate for the immune prophylaxis of HCV infection and vaccine development. Several segments of the E2 protein have been identified as key components of conformational or linear epitopes that are critical to antibody-mediated neutralization of HCV in vitro (7–16). Interestingly, naturally evoked antibodies and those produced in vitro that are specifically directed against a short peptide located in the E2 protein between residues 427–446, also known as epitope II, displayed one of three activities: virus neutralization, E2 binding but no neutralization, or interference with virus neutralization (15, 16). To capture the full spectrum of antibody responses in hepatitis C patients, we have previously characterized biochemically a panel of murine monoclonal antibodies (mAbs) into these three categories (17). All of the mAbs we have examined bind epitope II with a distinct activity: mAbs#8 and -#41 are both neutralizing antibodies, mAbs#12 and -#50 are nonneutralizing antibodies, and mAb#12 has the additional ability to interfere with neutralization (17). We further showed that Trp437 and Leu438 are the core residues for antibody recognition, regardless of the neutralizing capability of the antibody, whereas Leu441 is required for both nonneutralizing antibodies (mAbs#12 and -#50), and Phe442 is only specific for the binding of mAb#50 (17). We thus hypothesized that the effectiveness of antibody-mediated neutralization of HCV could be deduced from the interactions between an antibody and a specific set of amino acid residues. A significant amount of information on several candidate HCV E2-binding sites has been generated in recent years by epitope-mapping techniques (7–16); however, the underlying mechanism at the atomic level is still poorly understood. Here, we present the crystal structure of the epitope II peptide complexed with a neutralizing monoclonal antibody, mAb#8.
Results
Overview of mAb#8–Epitope II Complex Structure.
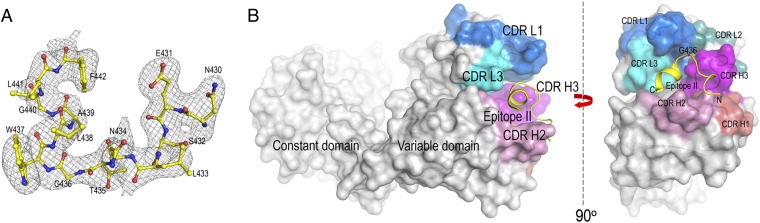
A 17-mer synthetic peptide (430NESLNTGWLAGLFYQHK446) of epitope II, whose sequence was derived from the E2 sequence of HCV genotype 1a (H77) (17), was cocrystallized with the Fab fragment of the neutralizing antibody, mAb#8. The crystal structure of the complex was determined to 2.85-Å resolution (Table 1). The first 13 amino acids of the peptide were unambiguously modeled into a difference electron density map (Fig. 1A). The last four residues of the peptide (443YQHK446) were not visible from the electron density map, indicating that they are not involved in the antigen recognition by mAb#8.
Table 1.
X-ray crystallographic statistics
| mAb#8–epitope II peptide | |
| Data processing statistics | |
| Resolution limit, Å* | 2.85 (3.00–2.85) |
| Space group | P43212 |
| Cell dimensions, Å | a = 137.2 b = 137.2 c = 140.5 |
| Unique reflections* | 30,296 (2,328) |
| Completeness, %* | 99.8 (53.8) |
| Rmerge, %* | 13.5 (37.2) |
| I/σI* | 10.8 (3.8) |
| Refinement statistics | |
| Rwork, % | 22.1 |
| Rfree, %† | 27.7 |
| rmsds from ideality | |
| Bond lengths, Å | 0.011 |
| Bond angles, ° | 1.811 |
| Ramachandran plot statistics | |
| Most favored, % | 86.8 |
| Additionally allowed, % | 11.9 |
| Generously allowed, % | 1.3 |
| Disallowed, % | 0 |
Values in parentheses are statistics of the highest resolution shell.
Rfree is calculated for a randomly selected 5.0% of reflections not included in the refinement.
Fig. 1.
(A) Composite omit electron density map of mAb#8–epitope II complex at 2.85-Å resolution showing the epitope II peptide. The density is contoured at 2.0σ. (B) Surface representation of the mAb#8–epitope II complex structure. The CDR H1 loop is colored in bronze, H2 in pink, H3 in magenta, L1 in blue, L2 in teal, and L3 in cyan. The peptide is illustrated as a yellow cartoon. The CDR loops of mAb#8 form a half-circle-shaped groove to accommodate the peptide. The location of Gly436 is indicated by a red dot.
The six complementarity-determining regions (CDRs) from the heavy (H1, H2, and H3) and the light (L1, L2, and L3) chains of mAb#8 that form the antigen-binding site were well defined. There were two mAb#8–epitope II complexes in a crystallographic asymmetric unit. The two noncrystallographic symmetry-related complexes showed nearly identical conformations as indicated by the root-mean-square deviations of 0.5 Å, 0.3 Å, and 0.08 Å in the α-carbon atoms of the variable and constant domains of the Fab molecules and the epitope II peptides. Based on this close similarity, the following description of mAb#8–epitope II interactions applies to both complexes.
The epitope II peptide consists of two major elements: an N-terminal loop (residues 430–436) and a C-terminal 1.5-turn α-helix (residues 437–442) (Fig. 1A).
Among the six CDRs of mAb#8, four CDR loops including H2, H3, L1, and L3 constitute a half-circle-shaped groove to accommodate the epitope II peptide (Fig. 1B). The arch of the groove is formed by CDR loops of H3, L1, and L3; the remaining part is made up of the CDR loop H2. The epitope II peptide is located inside the groove as if it entered the groove from one side of CDR H2, made a turn at the position Gly436, and then came out of the groove from the other side of CDR H2. In addition, the epitope II peptide, as revealed by its amino acid sequence, displays a dipolar composition; that is, the C-terminal helix is occupied exclusively by hydrophobic residues, whereas the N-terminal loop consists of mostly hydrophilic residues. Accordingly, mAb#8 engages the C-terminal and N-terminal portions of the epitope II peptide with distinct chemistry. The mAb#8–epitope II complex buried a total of 1,478 Å2 of solvent-accessible surface area, which is at the lower end of the range of 1,400–2,300 Å2 observed typically for antibody–antigen interactions (18).
Interaction of mAb#8 with the C-Terminal α-Helix of Epitope II.
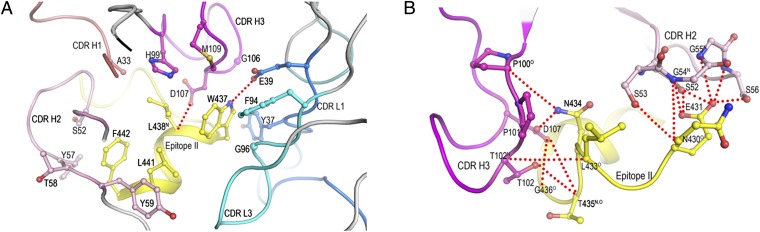
Next we examined the interactions of mAb#8 with the epitope II peptide. The complex is formed with binding contacts from both the light and heavy chains of the antibody, although the majority of interactions are provided by the heavy chain (Fig. 2 A and B). The four large residues (Trp437, Leu438, Leu441, and Phe442) of the epitope II peptide form a hydrophobic patch, which is almost completely buried in the antigen-binding groove of the antibody. Residues Trp437 and Leu438 fit snugly into a niche at the center of the binding groove, primarily through hydrophobic interactions with CDR loops of H1 (Ala33), H3 (His99, Gly106, Asp107, and Met109), L1 (Tyr37 and Glu39), and L3 (Phe94 and Gly96) (Fig. 2A). Such interactions contribute to an improved shape complementarity at this site, based on the shape correlation statistic (19). The shape correlation statistic for the interface formed by Trp437 and Leu438 with mAb#8 was 0.79 (the shape correlation statistic for a perfect fit is 1.0) (19). By contrast, the correlation statistic between the interface of the rest of the peptide and mAb#8 was 0.69. Furthermore, the hydrogen bond interaction between the side-chain atoms of Trp437 Nε1 and Glu39 Oε2 of CDR L1 further locked Trp437 into position. This combination of biophysical factors probably accounts for the strong preference for Trp437 and Leu438, especially for Trp at position 437.
Fig. 2.
Interactions of mAb#8 with epitope II. The side chains of interacting residues are drawn in a ball-and-stick representation, with nitrogen atoms in blue, oxygen atoms in red, and sulfur atoms in light brown. Hydrogen bonds are indicated by red dotted lines. (A) Close-up view of the interactions of mAb#8 with the C-terminal portion of epitope II (437WLAGLF442). (B) Close-up view of the interactions of mAb#8 with the N-terminal portion of epitope II (430NESLNTG436).
Our previous observation of HCV genotype-1a-specific neutralization by mAb#8 (17) may now be explained in terms of the complex structure. The sequence alignment of epitope II across HCV genotypes showed that Phe is substituted for Trp437 in other genotypes. In the context of the complex structure, the substitution of Trp437 by Phe could lead not only to a decrease in shape complementarity, but also a loss of the hydrogen bond. An additional effect of the change from Trp to Phe at position 437 is the reduction of buried surface at the binding site.
Residues Leu441 and Phe442 on the α-helix were identified as being particularly important for nonneutralizing antibodies mAbs#12 and -#50 (17). In the complex structure of mAb#8–epitope II, Leu441 and Phe442 are positioned near the edge of the mAb#8-binding groove, facing inward and interacting exclusively with CDR H2 via van der Waals contacts (Fig. 2A). In the presence of nonneutralizing mAbs#12 and -#50, we speculate that Leu441 and Phe442 are further buried in the antigen-binding groove. As a result, the strength of binding between epitope II and mAbs#12 and -#50 becomes more susceptible to mutations occurring at these two positions.
Interaction of mAb#8 with the N-Terminal Loop of Epitope II.
The N-terminal residues (430NESLNT435) of the epitope II peptide adopt an extended loop conformation in the complex structure (Fig. 2B). Residues Thr102 and Asp107 of CDR H3 and Ser53 of CDR H2 are directed toward the main-chain atoms of Asn430, Leu433, and Thr435 of the epitope II peptide. By contrast, residues Ser52, Gly54, and Ser56 of CDR H2 and residues Pro100 and Asp107 of CDR H3 engage in a network of hydrogen bonds with the side chains of residues Glu431 and Asn434 of the epitope II peptide, indicating that Glu431 and Asn434 in the N-terminal loop serve as a previously unrecognized anchor for antibody binding.
To further determine the contribution of these newly identified anchor residues to antibody binding, a series of competitive ELISA experiments was performed (Fig. 3). A biotinylated epitope II peptide termed epitope II–long (427LNCNESLNTGWLAGLFYQHK446) was coated on a 96-well streptavidin-coated plate. Before the addition to the coated ELISA plate, mAb#8 was incubated with different concentrations of competitive peptides, the wild-type epitope II (430NESLNTGWLAGLFYQHK446) that was used for producing the complex crystal, or mutants of epitope II containing either a single site replacement of Glu431 > Ala or double site replacements of Glu431 > Ala and Asn434 > Glu. The curve produced by the wild-type epitope II peptide was used as a reference for comparison with those generated by the epitope II mutants. The substitution of Glu431 > Ala reduced its binding to mAb#8 as shown by the EC50 values that shifted from 25 ng/mL (epitope II) to 134 ng/mL (epitope II Glu431 > Ala). The introduction of the additional change of Asn434 > Glu abolished the binding despite the substitution of Glu at position 434 in an attempt to compensate for the loss of Glu at 431. These results demonstrated that the specific interactions occurring at Glu431 and Asn434 contribute significantly to the antibody binding to epitope II. These results implicate that Glu431 and Asn434 not only contribute to antibody-binding affinity, but also are involved in antibody-binding specificity through the engagement of their side chains.
Fig. 3.
Binding of mAb#8 to the epitope II peptide mutants in a competitive ELISA. The ELISA plate was coated with epitope II–long peptide. The antibody mAb#8 was incubated with different concentrations of a competitor peptide, as indicated on the X-axis: epitope II, epitope II Glu431 > Ala, or epitope II Glu431 > Ala, Asn434 > Glu before addition to the wells of the ELISA plate.
We then surveyed the prevalence of Glu431 and Asn434 across HCV strains (Table 2) (20). The alignment of 1,957 HCV E2 protein sequences deposited in the National Institute of Allergy and Infectious Diseases (NIAID) Virus Pathogen Database and Analysis Resource (ViPR, www.viprbrc.org) showed that over 52% (1,031 of 1,957) of the sequences contain either Glu or Asp at position 431 and Asn at position 434. Interestingly, when a substitution occurs at position 431 (n = 926), position 434 is frequently taken over by either Glu or Asp (n = 299), suggesting a preference for an acidic residue at this location. If simultaneous mutations occur at positions 431 and 434, as seen during HCV infection (i.e., the condition under which mAb#8 loses its binding to epitope II), the virus may be able to avoid neutralization by mAb#8-like antibodies in vivo.
Table 2.
Prevalence of residues of epitope II associated with antibody binding
| Pattern | % of sequences examined (n = 1,957*) |
| D/E431–N434–GW437LAGL | 52.7 |
| X431–D/E434–GW437LAGL | 15.3 |
| X431–X434–GW437LAGL | 32.1 |
X represents any individual amino acids other than D/E. Italics indicate sequences derived from HCV H77 strain.
ViPR, www.viprbrc.org.
Pivot Point for the Epitope II Peptide Structure.
Gly436 within epitope II is known to be a highly conserved residue across HCV genotypes and has been implicated in virus entry (21). In the complex structure, Gly436 is located at the junction between the C-terminal α-helix and the N-terminal loop, where an ∼65-degree turn was observed (Fig. 2B). The peculiar location of Gly436 in epitope II makes it a possible pivot point connecting the α-helix with the rest of the peptide, thus providing epitope II the necessary flexibility to accommodate the antibody binding to the two anchor sites.
Discussion
The 3D structure of the mAb#8–epitope II complex revealed a bifurcated mode of action at epitope II, involving antibody interactions with two primary anchor sites of epitope II, i.e., the hydrophobic interactions with residues Trp437Leu438 in the α-helix and the hydrophilic interactions with residues Glu431/Asn434 in the extended loop, contributing to the binding affinity. Because residues Trp437Leu438 in the α-helix are critical for the binding of epitope-II-specific antibodies regardless of their neutralizing ability, we propose that antibody binding to the α-helical structure of epitope II is necessary, but may not by itself be sufficient for HCV neutralization. This result provides an explanation why some other antibodies have failed, during chronic HCV infection, to neutralize the virus even though they are able to bind to epitope II (15, 16). The virus may exploit this mechanism for its escape from neutralization if the host immune system simply produces the antibodies that bind preferentially to the α-helical structure of epitope II. On the other hand, neutralization may be achieved through antibody binding to Glu431/Asn434 in addition to the binding to residues of Trp437 and Leu438. This proposal could be investigated further by introducing specific mutations within the helix of epitope II that will selectively dismantle the residues (i.e., Leu441 and Phe442) specific for nonneutralizing antibodies to minimize the possibility of generating nonneutralizing antibodies, while maintaining the critical residues (i.e., hydrophobic residues Trp437 and Leu438) and the N-terminal hydrophilic residues (i.e., Glu431 and Asn434) of epitope II needed to elicit neutralizing antibodies such as mAb#8. By doing so, the neutralizing antibody is able to orient itself in a way that disrupts, directly or indirectly, the virus entry process. The structural insights from our studies should be considered in developing an effective HCV vaccine. Specific antigens can be designed to elicit neutralizing antibodies that optimally engage both anchor sites of epitope II, and to avoid nonneutralizing antibody production.
Materials and Methods
Peptide Synthesis and Protein Preparation.
All peptides were chemically synthesized by the Core Laboratory of the Center for Biologics Evaluation and Research at the US Food and Drug Administration, with an Applied Biosystems model 433A peptide synthesizer as described (15). Ascites containing mAb#8 IgG was produced by Harlan Bioproducts for Science as described (17). Fab fragment of mAb#8 was prepared with the Fab preparation kit from Thermo Scientific by following the instructions from the manufacturer.
Competitive ELISA.
Biotin-conjugated peptide (400 ng per well) was added to a streptavidin-coated 96-well Maxisorp plate (Thermo Scientific), followed by incubation at room temperature for 1 h in Super Block blocking buffer and at 37 °C for another hour. The plate was washed four times with PBS (pH 7.4) solution containing 0.05% Tween 20 (PBS-T) to remove unbound peptides. A diluted HCV antibody solution (mAb#8 in ascites fluid) was incubated with different concentrations of a competitive peptide for 1 h at room temperature before adding to the plate, followed by incubation at 37 °C for 1 h. After the removal of unbound antibodies by four washes with PBS-T, a goat anti-mouse peroxidase-conjugated IgG (KPL) at 1:5,000 dilution was added to the wells and incubated for 30 min at 37 °C. The plate was washed four times with PBS-T to remove unbound IgG conjugate. Tetramethylbenzidine substrate was then added, and the plates were incubated at room temperature in the dark for 10 min. The color reaction was terminated by adding 2 M sulfuric acid, and the absorbance was measured at 450 nm with a Spectramax M2e microplate reader (Molecular Devices). The data were plotted using the Prism software (Prism Software).
Crystallization and Data Collection.
Crystallization screenings were carried out robotically with a Mosquito liquid dispenser (TTP LabTech) by the hanging-drop vapor diffusion method. Crystals of the mAb#8–epitope II peptide complex were grown in 0.1 M Tris⋅HCl buffer (pH 8.0) containing 16% (wt/vol) polyethylene glycol 10,000, 0.2 M ammonium acetate. Glycerol (25% vol/vol) was used as a cryoprotectant. X-ray diffraction data were collected at beamline ×29 of the Brookhaven National Synchrotron Light Source with an ADSC Quantum-315 CCD detector. All data were indexed, integrated, and scaled with the program HKL2000 (22).
Structure Determination and Refinement.
The structure of the mAb#8–epitope II complex was determined by molecular replacement with the program Phaser (23) in the CCP4 program suite (24), using the anti-cholera toxin antibody (Protein Data Bank ID code 1ZEA) as the search model. Solutions were found in the cross-rotation and translation functions for the two complex molecules in the asymmetric unit. The epitope II peptide was omitted in the initial refinement. After refinement with RefMac 5.0 (25), the resulting difference electron-density map clearly showed the peptide at the mAb#8 antigen-binding site. The peptide was built into electron density using the program Coot (26). The CDR loops were deleted in the initial refinement and built into where an unambiguous electron density was shown. Contact residues in the mAb#8–epitope II complex were identified with the program Contact in CCP4 and were defined as residues containing an atom ≤ 4.0 Å from a residue of the binding partner. Buried surface areas were calculated by the program ArealMol in CCP4 with a 1.4-Å probe radius. PyMOL (www.pymol.org) was used to prepare the structural figures.
Acknowledgments
We thank Drs. John Finlayson and Shan-Lu Liu for their comments on the manuscript and Drs. Tsan Xiao and Tengchuan Jin (National Institute of Allergy and Infectious Diseases) for providing the instrument for crystallization screening. We thank Howard Robinson (Brookhaven National Synchrotron Light Source) for X-ray data collection. Beamline X29 is supported by the Department of Energy Offices of Biological and Environmental Research and Basic Energy Sciences and by the National Center for Research Resources of the National Institutes of Health. This study was funded by a Modernizing Science grant from the Center for Biologics Evaluation and Research, Food and Drug Administration.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4HZL).
References
- 1.Centers for Disease Control and Prevention . CDC Health Information for International Travel. New York: Oxford Univ Press; 2012. [Google Scholar]
- 2.Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: A perspective on long-term outcome. Semin Liver Dis. 2000;20(1):17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 3.Ghany MG, Strader DB, Thomas DL, Seeff LB. American Association for the Study of Liver Diseases Diagnosis, management, and treatment of hepatitis C: An update. Hepatology. 2009;49(4):1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter HJ, Liang TJ. Hepatitis C: The end of the beginning and possibly the beginning of the end. Ann Intern Med. 2012;156(4):317–318. doi: 10.1059/0003-4819-156-4-201202210-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton M. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev. 2011;239(1):99–108. doi: 10.1111/j.1600-065X.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 6.Puig M, Major ME, Mihalik K, Feinstone SM. Immunization of chimpanzees with an envelope protein-based vaccine enhances specific humoral and cellular immune responses that delay hepatitis C virus infection. Vaccine. 2004;22(8):991–1000. doi: 10.1016/j.vaccine.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Ray R, et al. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis. 2010;202(6):862–866. doi: 10.1086/655902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14(1):25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 9.Mancini N, et al. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PLoS ONE. 2009;4(12):e8254. doi: 10.1371/journal.pone.0008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong L, et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci USA. 2012;109(24):9499–9504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter JA, et al. Toward a hepatitis C virus vaccine: The structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol. 2012;86(23):12923–12932. doi: 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin TJ, et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 2012;8(8):e1002895. doi: 10.1371/journal.ppat.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci USA. 2012;109(16):6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabo MC, et al. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol. 2011;85(14):7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, et al. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104(20):8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, et al. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106(18):7537–7541. doi: 10.1073/pnas.0902749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan H, et al. Amino acid residue-specific neutralization and nonneutralization of hepatitis C virus by monoclonal antibodies to the E2 protein. J Virol. 2012;86(23):12686–12694. doi: 10.1128/JVI.00994-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundberg EJ, Mariuzza RA. Molecular recognition in antibody-antigen complexes. Adv Protein Chem. 2002;61:119–160. doi: 10.1016/s0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234(4):946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 20.Pickett BE, et al. ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(Database issue):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummer HE, Boo I, Maerz AL, Poumbourios P. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol. 2006;80(16):7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276(Pt A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 3):432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]