Summary
Understanding human-specific patterns of brain gene expression and regulation can provide key insights into human brain evolution and speciation. Here, we use next generation sequencing, and Illumina and Affymetrix microarray platforms, to compare the transcriptome of human, chimpanzee, and macaque telencephalon. Our analysis reveals a predominance of genes differentially expressed within human frontal lobe and a striking increase in transcriptional complexity specific to the human lineage in the frontal lobe. In contrast, caudate nucleus gene expression is highly conserved. We also identify gene co-expression signatures related to either neuronal processes or neuropsychiatric diseases, including a human-specific module with CLOCK as its hub gene and another module enriched for neuronal morphological processes and genes co-expressed with FOXP2, a gene important for language evolution. These data demonstrate that transcriptional networks have undergone evolutionary remodeling even within a given brain region, providing a new window through which to view the foundation of uniquely human cognitive capacities.
Identification of human-specific patterns of gene expression is necessary for understanding how the brain was modified in human evolution. Moreover, uncovering these human expression profiles is crucial for understanding human-specific neuropsychiatric and neurodegenerative disorders. Genetic changes resulting in changes in the amino acid sequences of proteins are likely too few to account for the phenotype differences between humans and our closest relative, the chimpanzee, prompting the suggestion that changes in gene expression are likely to drive some of the major phenotypic differences between humans and chimpanzees (Consortium, 2005; King and Wilson, 1975). Recent detailed comparisons of human and chimpanzee DNA differences have identified important differences related to gene expression including human accelerated regions (HARs) (Pollard et al., 2006a; Pollard et al., 2006b) or conserved noncoding sequences (CNSs) (Prabhakar et al., 2006) genomic neighborhood differences (De et al., 2009), copy number variations (CNVs) (Gazave et al., 2011; Perry et al., 2008), and promoter and enhancer variations (Haygood et al., 2007; Planas and Serrat, 2010) that could contribute substantially to differences in phenotype.
In addition to these DNA studies, several previous studies have directly examined human-chimpanzee differences in gene expression in the brain using microarrays to measure RNA transcript levels (Cáceres et al., 2003; Enard et al., 2002a; Khaitovich et al., 2004a). While these studies were an important first step in uncovering human-specific patterns of gene expression in the brain, microarray technology has several limitations that are especially germane to evolutionary comparisons. First, microarray analysis relies on a priori knowledge of the sequence of the sample being measured, which precludes identifying unannotated transcripts. The dynamic range of microarrays is also narrow compared to that of new sequencing technologies (Asmann et al., 2009; Feng et al., 2010). Perhaps most importantly with respect to cross-species comparisons is the tremendous loss of usable probes due to sequence divergence (Preuss et al., 2004).
To avoid these limitations, we utilized next generation sequencing (NGS) (Metzker, 2010) to compare gene expression in the brains of three primates: humans, chimpanzees, and rhesus macaques, employing 3′ digital gene expression (DGE) tag-based profiling to assess levels of mRNA expression. DGE has been shown to be both highly sensitive and reproducible when assessing gene expression from human brain (Asmann et al., 2009). Importantly, the present study included rhesus macaques as an out-group, which provides a basis for inferring whether differences between humans and chimpanzees occurred in the human lineage or the chimpanzee lineage. With a few exceptions (Brawand et al., 2011; Cáceres et al., 2003; Liu et al., 2012; Somel et al., 2009; Somel et al., 2011), previous microarray or NGS studies have not included an outgroup, or only investigated one brain region (Babbitt et al., 2010; Cáceres et al., 2003; Enard et al., 2002a; Khaitovich et al., 2005; Khaitovich et al., 2004a; Liu et al., 2011; Marvanova et al., 2003; Somel et al., 2009; Uddin et al., 2004; Xu et al., 2010a). We examine three brain regions representing different developmental origins within the telencephalon: subpallial (caudate), allocortical (hippocampus), and neocortical (frontal pole). Frontal pole is of particular interest because it was enlarged and structurally modified in human evolution (Semendeferi et al., 2001; Semendeferi et al., 2011), is involved in higher-order cognitive functions (including mental multitasking, social cognition, and planning and manipulation of abstract representations) (Dumontheil et al., 2008; Wendelken et al., 2011), has a protracted course of development extending into adolescence and beyond (Dumontheil et al., 2008; Rakic and Yakovlev, 1968; Wendelken et al., 2011), and appears to be affected in diseases that affect higher-order cognition, including autism and schizophrenia (see the review of (Dumontheil et al., 2008)).
We find many more expression changes using NGS than with microarrays, and use network biology to put the changes observed into a systems level context, showing high conservation of the caudate transcriptome, while identifying eight human specific gene co-expression modules in frontal cortex. Moreover, we discover gene co-expression signatures related to either neuronal processes or neuropsychiatric diseases, in addition to a human-specific frontal pole module that has CLOCK as its hub, and includes several psychiatric disease genes. Another frontal lobe module that underwent changes in splicing regulation on the human lineage is enriched for neuronal morphological processes and contains genes co-expressed with FOXP2, a gene important for speech and language. By using NGS, by including an outgroup, and by surveying several brain regions, these findings highlight and prioritize the human-specific gene expression patterns that may be most relevant for human brain evolution.
Results
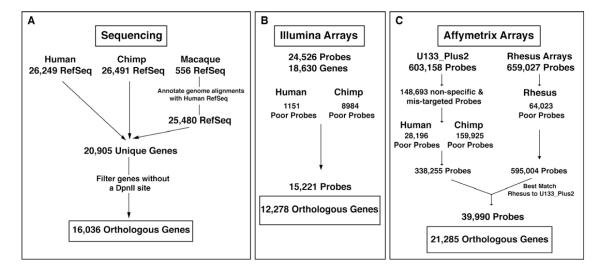
At least four individuals from each species and each brain region were assessed (Table S1) using DGE-based sequencing and two different microarray platforms, Affymetrix and Illumina (Figure 1). The total number of unique genes available for analysis among the species was 16,813 for DGE, 12,278 for Illumina arrays (ILM), and 21,285 for Affymetrix arrays (AFX) (Figure 1). Analysis of DGE data revealed an average of 50% human, 43% chimp, and 39% macaque DGE reads mapping to its respective genome with 2-3 million total reads mapping on average (Table S1); pair-wise analysis of DGE samples revealed high correlations (Table S1). Neither the total number of reads, nor the total number of mapped reads were significantly different among species for a given region, eliminating these as potential confounders in cross species comparisons (Total reads: FP (P=0.993), CN (P=0.256), HP (P=0.123); Uniquely mapped reads: FP (P=0.906), CN (P=0.216), HP (P=0.069), ANOVA). The samples primarily segregated based on species and brain region using hierarchical clustering (data not shown). We also conducted thorough outlier analysis as well as covariate analysis and do not find that factors such as postmortem interval, sex, RNA extraction, library preparation date, sequencing slide, or sequencing run are significant sample covariates (see Experimental Procedures).
Figure 1. Schematic of experimental design.
Filtering strategy for (A) DGE samples, (B) Illumina microarrays, and (C) Affymetrix microarrays. See also Table S1.
DGE identifies more expressed genes in primate brain than microarrays
On average, DGE identified 25-60% more expressed genes in the brain than either microarray platform (Figures S1A-B). Correlation plots of DGE expressed genes versus AFX expressed genes shows an average correlation of 0.470 (Pearson correlation; Figures S1C-K), and emphasize that a large number of genes are detected using DGE rather than AFX. Further, weighted Venn diagrams demonstrate that DGE captures the majority of the same genes detected by either microarray platform, but identifies more than 50% additional genes as present in human or chimp brain (Figure S2).
DGE identifies more differentially expressed genes in the human brain
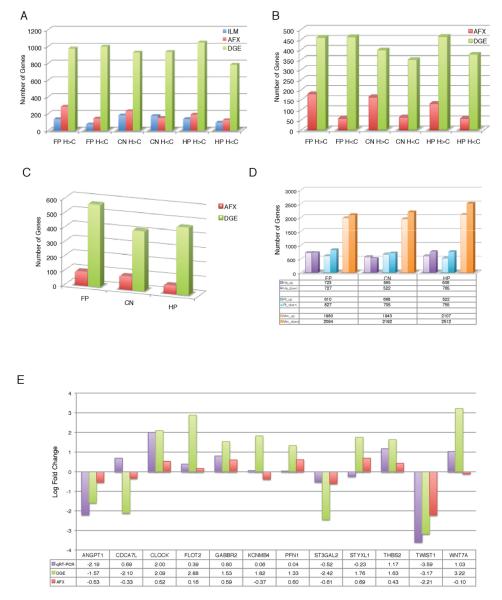
Next, we assessed differential expression, identifying more than five times as many differentially expressed (DE) genes in the brain between human and chimpanzee using DGE than AFX and almost eight times more using DGE than ILM (Figure 2A). The number of DE genes within the microarray datasets and the FP DGE was consistent with what has been previously published (Babbitt et al., 2010; Cáceres et al., 2003; Khaitovich et al., 2004a), and there was significant overlap with previous data from frontal lobe between human and chimpanzee (P=3.1E-03 (Babbitt et al., 2010) and P=2.7E-02 (Khaitovich et al., 2004b)). When we included the macaque outgroup data, using both the DGE and AFX datasets, we identified approximately five times as many human-chimp DE genes using DGE compared with AFX (Figure 2B). As expected, the total number of DE genes between humans and chimps decreased by about 50% upon inclusion of outgroup data, since many genes change in their expression levels between the chimp and macaque lineage. Correlation analysis of DE genes between platforms showed significant concordance (0.37-0.52 Spearman; P=9.6E-78-1.2E-105).
Figure 2. Identification of genes specifically expressed in human brain.
(A) Graph of number of differentially expressed genes using pairwise comparisons between humans and chimps without outgroup inclusion in each brain region (upregulated genes= H>C; downregulated genes=H<C; Bayesian t-test, FDR <0.05, absolute fold change > 2). (B) Graph of number of differentially expressed genes between humans and chimps with the inclusion of macaque outgroup data in each brain region (Bayesian t-test, FDR <0.05, absolute fold change > 2). (C) Graph of number of genes from (B) that are uniquely differentially expressed in only one brain region. (D) Graph of number of genes that are changing along a specific lineage in each brain region (ANOVA; FDR < 0.05). (E) qRT-PCR analyses of identified hDE genes in FP using DGE. See also Figures S1 and S2.
Due to the inclusion of three distinct brain regions, we were also able to identify many genes differentially expressed in only one of the regions examined (Figure 2C). Interestingly, FP had the greatest number of region-specific differentially expressed genes, even after correcting for the greater number of total differentially expressed genes in the FP (Figure 2B). Finally, we confirmed a number of specific genes using a completely independent platform, qRT-PCR (Figure 2E). These independent qRT-PCR analyses demonstrate a 67% and 58% confirmation rate with DGE and AFX respectively, in line with published high correlations between DGE and qRT-PCR (Asmann et al., 2009). Thus, the use of NGS compared with microarray produces an increased number of true positives in terms of genes differentially expressed in the human brain, reflecting the higher dynamic range and lower variance of DGE, especially at lower levels of expression, where arrays are known to suffer (Asmann et al., 2009). Together, we were able to directly confirm the validity of the DGE DE data using both an independent whole-genome method, as well as a robust gene-specific method. Thus, DGE is a more powerful method for identifying unique gene expression signatures in the primate brain, providing a real-world example demonstrating the power of next generation sequencing for analysis of a complex tissue such as brain.
Identification of genes specifically changing in the human brain
We next assessed the number of genes changing along each species’ lineage in the DGE data (Figure 2D) using two distinct methods, parsimony and an F-test, which insured robustness of our results (see Experimental Procedures). Genes with similar expression values between chimpanzee and macaques, but significantly different in humans would be indicative of those changing specifically on the human lineage (hDE). Examination of hDE genes revealed several striking findings. First, the number of hDE genes was greater in the FP than in the two other brain regions examined. For example, nearly 30% more hDE genes are detected in hFP (1450 genes) than hCN (1087 genes) (Figure 2D). This could not simply be explained by a greater number of reads in these samples, as the FP samples had fewer mapped reads on average than either CN or HP (Table S1). Moreover, the FP predominance for the lineage-specific DE genes is not observed in macaque and chimpanzee, indicating that this is truly human specific. The increase in genes changing in the frontal pole is of special interest given the recent finding of an enrichment of evolutionary new genes in the human lineage specifically within the prefrontal cortex using different methods (Zhang et al., 2011). Thus, our data identify for the first time the increasing number of genes changing specifically in the frontal cortex compared to other non-cortical regions in human brain evolution.
Gene ontology (GO) analyses identified enrichment of several key neurobiological processes. In the FP, genes involved in neuron maturation (FARP2, RND1, AGRN, CLN5, GNAQ, and PICK1) and genes implicated in Walker-Warburg syndrome (FKTN, LARGE, and POMT1), a disorder characterized by agyria, abnormal cortical lamination, and hydrocephalus (Vajsar and Schachter, 2006), were enriched. Filtering the FP list for those specifically hDE in FP and not other brain regions revealed additional categories of interest including regulation of neuron projection development (e.g. MAP1B, NEFL, PLXNB1, and PLXNB2), the KEGG category for neurotrophin signaling (e.g. BAX, CSK, CALM2, and IRAK1) and the cellular component category for axon (e.g. GRIK2, LRRTM1, NCAM2, MAP1B, NEFL, and STMN2). HP hDE GO analyses uncovered enrichment of genes involved in cell adhesion (e.g. CAV2, DSG2, SDC1, SDC4, TJP2, CDH3, and NEDD9) and HP-specific analyses demonstrated enrichment for neuron differentiation (e.g. EFNB1, MAP2, NNAT, REL2, and ROBO1) and the cellular component category for synaptosome (e.g. ALS2, DLG4, SYNPR, and VAMP3). CN-specific GO analyses identified enrichment for genes involved in dendrites and dendritic shafts (e.g. CTNNB1, EXOC4, GRM7, and SLC1A2), synapse (e.g. SYNGR3, SYT6, and CHRNA3), and sensory perception of sound (e.g. SOX2, CHRNA9, USH2A, and KCNE1).
Genes that are hDE are also enriched for genes under positive selection (dN/dS > 1 for human compared to chimpanzee) with FP and HP containing more genes under positive selection than CN: FP (56), HP (60), and CN (48). These genes are significantly enriched for the GO categories cytokine receptor activity (P=6.0E-03) and the JAK/STAT signaling pathways (P=1.0E-03) in the FP (IL11RA, IL13RA2, and GHR), for carboxylic acid catabolic process (P=3.3E-03) in the HP (ASRGL1, CYP39A1, and SULT2A1), and for synaptic transmission (P=3.5E-02) in the CN (LIN7A, MYCBPAP, and EDN1). Together, these data suggest that human-specific gene evolution is important for signaling pathways in the brain.
The human frontal pole has increased transcriptional complexity
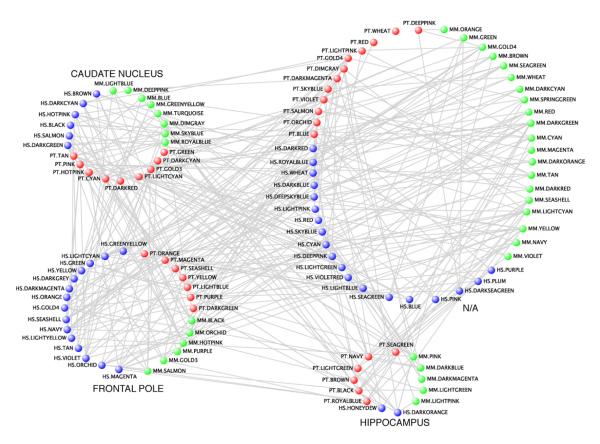
We next applied weighted gene co-expression network analysis (WGCNA) (Oldham et al., 2008) to build both combined and species-specific co-expression networks, so as to examine the systems level organization of lineage-specific gene expression differences. We constructed networks in each species separately and performed comparisons of these networks to insure a robust and systematic basis for comparison (Oldham et al., 2008). The human transcriptional network was comprised of 42 modules containing 15 FP modules, 6 CN modules, 2 HP modules, and 19 modules not representing a specific brain region (Figure 3 and Tables S2 and S3; Experimental Procedures). The FP samples correlated less with the CN and HP samples, using a composite measure of module gene expression, the module eigengene, or first principal component (Oldham et al., 2008) (data not shown).
Figure 3. Connectivity among module eigengenes.
Visualization of the module eigengene overlaps. Nodes for human modules are blue, nodes for chimpanzee modules are red, and nodes for macaque modules are green. See also Table S2.
The chimp network analysis yielded 34 modules, including 7 FP modules, 9 CN modules, 7 HP modules, and 11 modules that were unrelated to a specific brain region (Figure 3, Tables S2 and S3). The macaque analysis yielded 39 modules with 6 FP modules, 8 CN modules, 5 HP modules, and 20 modules not related to a specific brain region (Figure 3, Tables S2 and S3). Thus, only in human brain were more modules related to FP than either of the other regions, consistent with increased cellular and hence transcriptional complexity in FP relative to the other regions. While the smaller number of chimpanzee (n=15) and macaque (n=12) samples compared to human (n=17) samples could potentially affect the outcome of the network analysis, we used the same thresholding parameters, and there were equivalent numbers of human and chimpanzee FP samples (n=6), similar numbers of total modules in human and macaque samples (42 and 39, respectively), and proportionally more FP modules compared to total modules in human samples (18/42=43%) compared to chimpanzee (8/34=23%) or macaque (5/39=13%), mitigating this concern. This indicates that even within a single region of human frontal lobe, transcriptome complexity is increased with regards to other primates.
We next determined the conservation of the modules defined in humans in the other species (see Experimental Procedures; Table S3). Interestingly, we found that four out of six of the human CN modules were highly preserved in chimps and macaques whereas only four of the FP and none of the human HP modules were highly preserved in the other primates. Consistent with these data, the Hs_brown module, which is a highly preserved caudate module, has high overlap with a previously documented conserved caudate module between humans and chimpanzees (Oldham et al., 2006). In addition, the conserved CN module, Hs_darkcyan, has significant overlap with a recently identified module in layer 6 of the rhesus macaque parietal lobe (P=2.12E-54; hypergeometric overlap; Bernard et al., 2012) annotated as containing genes important for oligodendrocytes (Oldham et al., 2008). The conserved CN module, Hs_hotpink, also has significant overlap with layers 2/3 in the macaque (P=1.92E-04; hypergeometric overlap; Bernard et al., 2012) annotated as an astrocyte module (Oldham et al., 2008). In fact, only conserved modules from our dataset had high overlap with these recently described rhesus macaque cortical modules. Together, these data highlight the power of our systems approach to identify conserved cell-type-related networks among primate brains. Interestingly, the gene CUX2 in the Hs_hotpink module demonstrated conserved laminar expression by in situ hybridization in both primate and mouse cortex (Bernard et al., 2012), providing additional evidence for confirmation of our network findings.
In contrast to our findings in the caudate, at least two of the conserved human FP modules (Hs_orchid and Hs_magenta) overlapped with two cortical modules denoted as non-conserved in an earlier microarray-based dataset (Oldham et al., 2006), and the Hs_magenta module also significantly overlapped an additional rhesus macaque frontal region module (P=1.18E-13; hypergeometric overlap) (Bernard et al., 2012), again highlighting the increased power of the DGE analysis over microarrays. In addition, two of the genes in the conserved Hs_tan FP module, RORB and RXFP1, have conserved laminar expression between primates and mice (Bernard et al., 2012).
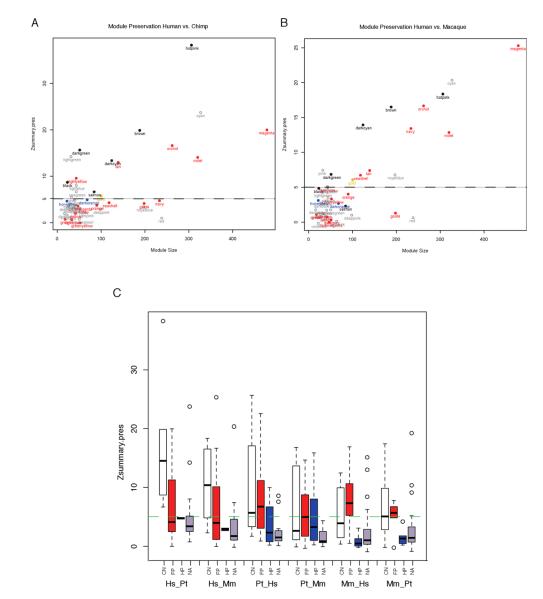
Most remarkably, eight out of 15 of the human FP modules were human-specific and were not preserved in either chimpanzee or macaque (Figures 4A-B), whereas only three out of seven FP modules in chimpanzee and one of six macaque FP modules were species specific (Table S3), suggesting increased transcriptional complexity in human frontal pole. In contrast, highly preserved modules among the three primate brains include the modules with the strongest eigengene for CN: Hs_brown and Hs_hotpink (Figures 4A-C and Tables S2 and S3). After controlling for module size using a MedianRank function (Langfelder et al., 2011; see Experimental Procedures), we found similar module preservation results across species as above (Table S3).
Figure 4. Module preservation among primate brain regions.
Graph of the module preservation score (Zsummary.pres) compared to the number of genes in a module (module size) for the human network as reference compared to (A) chimpanzee and (B) macaque. CN modules are in black, FP in red, HP in blue, and modules not correlated to region in grey. Modules below the dashed line at 5 in both comparisons are human-specific whereas those at or above 5 are conserved. (C) Distribution of preservation scores using each species for the reference network. CN networks are conserved in all cases whereas FP networks are only specific when human is used as the reference genome. See also Figure S3 and Table S3.
The conserved CN modules are very interesting as they highlight a robust set of key conserved regulatory networks across primates and likely other mammals. We explored the function of the hub genes in these modules, as these genes are a primary indicator of the module network function. The hub genes of the Hs_hotpink module are significantly enriched for genes involved in CNS development (P=1.0E-04; SERPINF1, PRDM8, NEUROD2, RTN4R, CA10, and MEF2C). The hub genes of the Hs_brown module are significantly enriched for genes involved in regulation of G-protein coupled receptor protein signaling (P=3.0E-07; RGS9, RGS14, RGS20, and GNG7). The most highly connected gene in the Hs_brown module is PPP1R1B, or DARPP-32, which is a critical mediator of dopamine signaling in medium spiny neurons in the striatum (Walaas and Greengard, 1984). In addition, five other hubs in the Hs_brown module (ADORA2A, GNG7, PDE10A, PRKCH, and RXRG) overlap with the top 25 cell type specific proteins in Drd1 or Drd2 striatal neurons in mouse characterized by translational profiling (Doyle et al., 2008). The hub gene ADORA2A also overlaps with the top differentially expressed genes from microarray profiling of striatal neurons in mouse (Lobo et al., 2006). When considering all of the genes in the conserved CN modules, we also find a high level of confirmation: six genes overlap with striatal microarrays (ADORA2A, CALB1, HBEGF, NRXN1, STMN2, and SYT6), eight genes overlap with Drd1 translational profiling ADORA2A, BCL11B, GNG7, GPR6, GPR88, MN1, PDE10A, and RXRG), and nine genes overlap with Drd2 translational profiling (ADRA2C, ERC2, EYA1, KCNIP2, MYO5B, PDE10A, PDYN, PRKCH, and WNT2). Interestingly, four CN hub genes have been implicated in addiction, three are involved in alcohol addiction (MEF2C, RGS9, and VSNL1), one is involved in nicotine addiction (GABBR2) (Li et al., 2008), and two CN hub genes have been linked to obsessive compulsive disorder (HTR1D and HTR2C) (Grados, 2010). Taken together, this cross species conservation and link to disease has implications for pharmacotherapeutics of neuropsychiatric diseases being developed in rodent models because these data showing conservation between primates and mice further validate rodents as appropriate models for striatal function in humans.
GO analysis of FP hub genes reveals an enrichment of genes involved in neural tube development (FZD3, PAX7, PSEN2, and SMO), and regulation of synaptic plasticity (ARC, KRAS, and STAR). However, the majority of FP and HP hub genes are not enriched for specific ontological categories. These results emphasize the importance of these human-specific modules as it suggests that due to their unique expression patterns in the human brain, very little is known about the coordinated function of these genes. Finally, at least one of the conserved modules that was not associated with a particular brain region, Hs_cyan, does overlap with a previously identified module containing an enrichment of genes involved in ATP synthesis and the mitochondrion (Oldham et al., 2008). These data suggest that genes important for subcellular components important in all brain regions throughout evolution may drive some of the network eigengenes.
We next sought to integrate these findings showing regional differences in network module conservation with changes occurring at the DNA level in specific genes. Four out of eight of the human-specific FP modules (6 genes: BBS10, MTFR1, TCP10L, FKBP15, KIAA1731, and TRIM22) and both of the human-specific HP modules (2 genes: CP110 and DFFA) contain hub genes (genes ranked in the top 20 for connectivity) under strong positive selection (Experimental Procedures). In addition, six genes from human FP modules with some level of conservation are under positive selection (C15orf23, C20orf96, CYP8B1, GSDMB, REEP1, and UACA). In contrast, only one hub gene from a CN module conserved between humans and chimpanzees is under positive selection (APTX). Even considering all of the genes in a module for each brain region, both FP (4.3%) and HP (6.9%) have more genes under positive selection than CN (3.5%). Therefore, overall, non-conserved modules tend to have more genes evolving faster. These data again highlight the biological importance of the network preservation findings: human-specific FP and HP modules contain genes with fewer constraints to allow for new cognitive functions, whereas highly preserved CN modules contain genes with more constraint in order to participate in essential brain functions necessary for all primates. These DNA sequence data indicating positive selection of specific genes more preferentially in the frontal lobe, supports the network data based on gene expression, indicating that this region is most divergent, and highlights specific hub genes with multiple levels of evidence for their evolutionary importance.
Human-specific co-expression networks are enriched for genes involved in neuronal processes and neuropsychiatric diseases
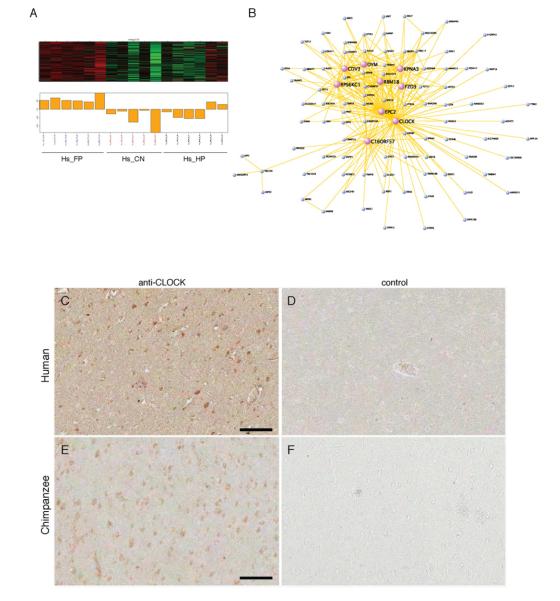
Further functional annotation of the human-specific FP modules revealed several important findings relevant to evolution of human brain function. One of the non-preserved FP modules was the human orange module (Hs_orange). Visualization of the co-expressed genes in this module revealed that CLOCK, a circadian rhythm gene implicated in neuropsychiatric disorders such as bipolar disorder (Coque et al., 2011; Menet and Rosbash, 2011), is a major hub and the most central gene in the module (Figure 5 and Table S2). CLOCK is also differentially expressed and is increased in human FP (Figure 2E). We therefore asked whether the CLOCK protein was increased in human FP and confirmed increased CLOCK protein expression in human FP compared to chimpanzee FP using immunohistochemistry (Figures 5C-F). In addition, the Hs_orange module is significantly enriched for other genes involved in neuropsychiatric disorders, such as seasonal affective disorder (P=2.5E-02), depression (P=2.1E-02), schizophrenia (P=4.7E-02), and autism (P=4.0E-02) (e.g. HTR2A, FZD3, HSPA1L, KPNA3, and AGAP1; Table S2). To determine whether this might reflect differences in typical circadian genes due to differences in time of death or other factors, we compared the genes in the Hs_orange module to genes annotated as circadian rhythm genes based on gene expression and other functional studies (See Experimental Procedures). Interestingly, none of the genes in the Hs_orange module overlap with previously identified circadian rhythm genes in the liver or brain of rodents, suggesting that we may have identified unique targets of CLOCK in human brain. This is especially interesting, as the histone acetyltransferase function of CLOCK is conserved from viruses to human (Kalamvoki and Roizman, 2010, 2011). The hub role of CLOCK in this module suggests potential transcriptional regulatory relationships with other module genes.
Figure 5. Visualization of a human-specific gene co-expression network: the Hs_orange module.
(A-B) Visualization of the top 300 connections among the co-expressed genes in the module. Hub genes are denoted using purple circles. (C-F) Immunohistochemistry for CLOCK in human FP (C) and chimpanzee FP (E). Corresponding negative control sections are shown in (D) and (F). Scalebars are 100 microns.
Another FP module not preserved in chimp or macaque is the Hs_darkmagenta module. Hs_darkmagenta is enriched for genes involved in CNS development (e.g. BMP4, ADAM22, KIF2A NRP1, NCOA6, PEX5, PCDHB9, SEMA7A, SDHA, TWIST1), growth cones (FKBP15), axon growth (KIF2A), cell adhesion (ADAM22), and actin dynamics (EIF5A2) (Figure S3 and Table S2). These data are congruent with the finding that human neurons have unique morphological properties in terms of the number and density of spines (Duan et al., 2003; Elston et al., 2001), providing a potential molecular basis for these ultrastructural differences for the first time. Additionally, the combination of these molecular data with the previous morphological data support the hypothesis that in addition to the expansion of cortical regions, the human brain has been modified by evolution to support higher rates of synaptic modification in terms of growth, plasticity, and turnover (Cáceres et al., 2007; Preuss, 2011).
An exon-level analysis of gene connectivity reveals accelerated evolution in the human brain
We next examined each unique read individually to determine whether there was information about the expression of alternative isoforms. Among the 22,761 Refseq genes detected, 86% of those genes had more than one read aligning to it, demonstrating that most transcripts had alternative forms detected. Although some genes (about 40%) had a dominant variant that accounted for more than 90% of the reads aligning to a specific gene, more than half (57.3%) of genes had a dominant variant that accounted for less than 90% of the expression detected. We then examined the expression of these alternative variants by calculating the Pearson correlation between all reads that align to the same gene. We found that most pairs were slightly negatively correlated and that the average correlation between all pairs aligned to the same gene was zero (data not shown), suggesting that these reads do indeed represent differentially regulated variants.
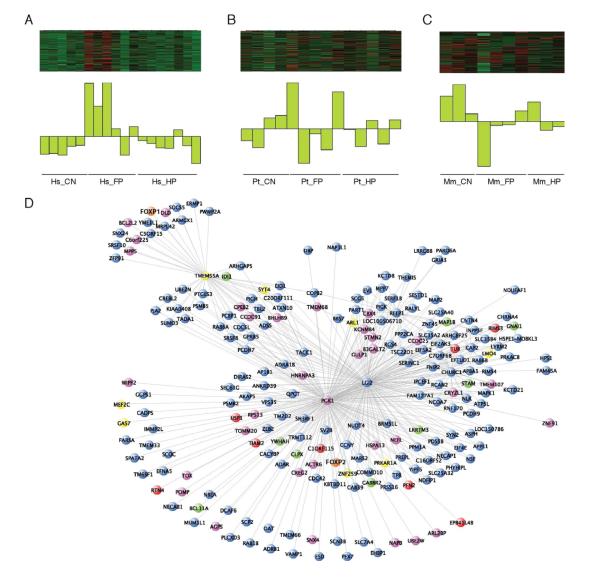
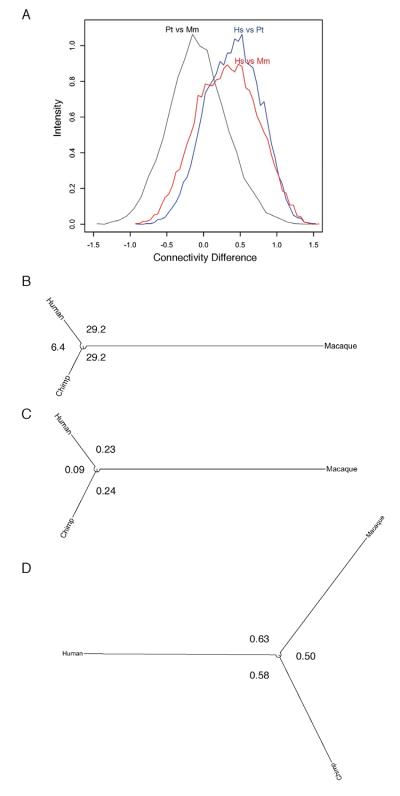
Based on these data that unique reads likely contained information about alternative variants, we built a co-expression network based upon aligning reads to specific exons rather than only to whole genes to potentially uncover an enrichment of gene co-expression patterns based on alternative splicing (See Experimental Procedures and Table S4). This analysis also resulted in the identification of several modules whose module eigengene corresponded to the human frontal pole. One of these, the olivedrab2 module (Figure 6) is a FP module enriched for genes involved in neuron projections, neurotransmitter transport, synapses, axons, and dendrites, as well as, genes implicated in schizophrenia. In addition, this module contains many of the highly connected genes within the Hs_darkmagenta module from the whole gene WGCNA (n=9, including the top five hubs out of a total of 62 genes in the Hs_darkmagenta module). Although this module is preserved in the other species, 73% and 78% of exons exhibit higher connectivity within the human data than the chimpanzee or macaque datasets, respectively, demonstrating that these genes have enhanced connectivity within the human brain (Figure S4). Connectivity is a measure of the extent to which the expression of a gene (or exon) is co-expressed with all other genes (or exons) that we and others have shown to be highly preserved in human brain networks and is related to functional properties of genes such as brain region, cellular types, and disease states (Miller et al., 2010; Oldham et al., 2008). Therefore, changes in this measure of connectivity among primate species may indicate changes in the function of these genes in evolution. Strikingly, the conservation of connectivity of genes within this human network and the other primates is less (r=0.281, human-chimp; r=0.182, human-macaque) than the connectivity preservation between chimp and macaque networks (r=0.421) (Figure 7A). In view of the very close evolutionary relationship of humans and chimpanzees, these results indicate that patterns of gene connectivity in this module underwent dramatic reorganization in the human lineage, after it diverged from the chimpanzee lineage, without, it should be noted, major changes in overall gene expression within the module (Figure 7B-D). Therefore, this new dimension of gene expression analysis, only made possible using NGS, has revealed a striking pattern of accelerated evolution of gene connectivity in the human brain.
Figure 6. Visualization of a human-specific gene co-expression network: the olivedrab2 module.
Visualization of the module eigengene in the olivedrab2 module in the (A) human samples and the expression of the same genes in the (B) chimpanzee and (C) macaque samples. (D) Visualization of the top 500 connections among the co-expressed genes in the module. FOXP2 and FOXP1 are highlighted in orange. Genes in purple are specifically expressed along the human lineage in FP, genes in red are FOXP2 targets identified in human neural progenitors, genes in yellow are FOXP2 targets identified in human fetal brain (Spiteri et al., 2007), human transformed neuronal cell lines (Konopka et al., 2009), or mouse brain (Vernes et al., 2011) and genes in green are both human-specific and FOXP2 targets. See also Figure S4 and Table S4.
Figure 7. Brain network connectivity differences among primates.
The human olivedrab2 FP module as an example of differential connectivity in the primate brain. (A) Histogram of the difference in connectivity among genes in the olivedrab2 module between each species. Human networks compared to chimpanzee (blue) and macaque (red) networks have greater differences than between chimpanzee and macaque (grey). Unrooted distance trees demonstrate that while human and chimpanzee are more similar as (B) species overall and (C) absolute gene expression, while the (D) connectivity of genes in the human FP is greater.
Human-specific co-expressed gene isoforms are enriched for a differentially expressed splicing factor
Because we hypothesized that multiple reads targeting the same gene corresponded to transcript variants of the gene, we attempted to gain insight into the potential mechanism of regulation of these transcripts by examining the exon to which the read aligned for known RNA binding motifs. We calculated an enrichment score for each module for the known RNA binding motifs, and we then clustered modules based on their motif enrichment patterns. We found that olivedrab2 clustered with lavenderblush1 and thistle4 modules, which are among the top scoring neuronal modules when compared to a previous annotation of the human brain transcriptome (Oldham et al., 2008). Moreover, the exons represented by the olivedrab2 module showed an enrichment of ELAVL2 binding motifs within the module gene membership (Table S4). ELAVL2 (alias HuB) is also hDE in FP, increasing on the human lineage, consistent with the parallel changes in splice isoforms observed in this module. The adult CNS function of this splicing factor, ELAVL2, is mostly unknown, but recent work indicates that it interacts with microRNAs to regulate cortical neurogenesis via de-repression of Foxg1 (Shibata et al., 2011). The evolutionary significance of this pathway is highlighted by the recent finding that Foxg1 mutations in humans lead to a syndrome of microcephaly, and social and language impairment (Kortum et al., 2011). Table S4 provides a list of the significantly enriched ELAVL2 targets (P<2.2E-05) within this module in human frontal lobe (637 exons in 521 genes). Importantly, we identify ELAVL1 as an enriched target. Since ELAVL2 has already been shown to regulate ELAVL1 (Mansfield and Keene, 2011), this provides validation of our computational approach. These targets are enriched in genes involved in calcium channels (P=3.3E-02; CACNB4, CACNG2, and RYR2), synaptic vesicles (P=4.4E-02; APBA1, RAB3B, SCAMP1, SYN2, and SYT11), postsynaptic density proteins (P=3.23E-02; CAMK2N1, DLG2, DLG4, GRIN2A, and MAP1B), as well as many other intrinsic CNS properties. There is also an enrichment of genes involved in Alzheimer’s disease (P=4.2E-02; BACE1, CYCS, GRIN2A, GSK3B, and SDHC) and autism spectrum disorders (P=1.1E-04; APC, CNTN4, CNTNAP2, DLX1, EIF4E, FBXO33, FOXP2, GABRB3, GALNT13, GRIN2A, HS3ST5, MAP2, MDGA2, MECP2, MEF2C, MKL2, NRXN1, SLC9A6, and TSN). These data provide a starting point for further mechanistic studies of the molecular function of neurons in the frontal pole, especially how they may relate to human cognitive disorders. Therefore, examination of gene co-expression in primate brain has revealed new biological connections and insights into the evolved human brain: gene connectivity as evidenced by modified transcriptional programs together with alternative splicing are likely critical for human-specific frontal pole cognitive functions.
FOXP2 is co-expressed with genes involved in neuronal projections in a human-specific frontal pole module
Further inspection revealed that the olivedrab2 module contains FOXP2 among its more differentially connected genes specifically in the human brain (kMEHuman=0.91, kMEChimp=0.67, kMEMacaque=0.46; P=1.24E-05) (Figure 6 and Table S4). FOXP2 is a transcription factor implicated in language and cognition that has undergone accelerated evolution and has human-specific functions (Enard et al., 2009; Enard et al., 2002b; Konopka et al., 2009; Lai et al., 2001).
To validate the co-expression relationships in this module, we assessed enrichment of known FOXP2 targets. We identified 13 genes that overlap with previously published targets of FOXP2 from human brain, human cells, or mouse brain (Figure 6) (Konopka et al., 2009; Spiteri et al., 2007; Vernes et al., 2011; Vernes et al., 2007) with the genes in the olivedrab2 module including one of the hub genes, TMEM55A (Figure 6D). We noted that FOXP1 is also among the most connected genes in this module (Figure 6D). Not only can FOXP1 heterodimerize with FOXP2 to regulate transcription (Li et al., 2004), FOXP1 has been implicated in language impairment, intellectual disability, and autism (Carr et al., 2010; Hamdan et al., 2010; Horn et al., 2010; O’Roak et al., 2011).
We further examined FOXP2 targets in human neuronal cell lines previously shown to exhibit patterns of gene expression similar to those of forebrain neurons (Konopka et al., 2012). We manipulated FOXP2 expression during the normal four-week period of differentiation of these human cells by either forcing expression of FOXP2 or knocking down expression of FOXP2 using RNA interference (Experimental Procedures). Using Illumina microarrays, we identified over 600 target genes with expression going in the opposite direction with FOXP2 forced expression compared to FOXP2 knockdown (Figure S4). Upon comparing this list of experimentally identified FOXP2 targets in human neural progenitors using microarrays with the genes in the olivedrab2 module identified by DGE, we found a significant overlap (13 overlapping genes, P=4.0E-04; Figure 6D). Interestingly, nine FOXP2 target genes overlap with hDE genes in this module (Figure 6D). Strikingly, the FOXP2 targets in the olivedrab2 module are enriched for genes involved in neuron projections, synapse, and axonogenesis. These data fit with work showing modulation of neurite outgrowth in mouse models of Foxp2 (Enard et al., 2009; Vernes et al., 2011). Thus, while regulation of neurite outgrowth by FOXP2 may be a conserved mammalian function of FOXP2, the contribution of human FOXP2 to modulation of this critical neuronal process may be enhanced as evidenced by increased neurite length in humanized Foxp2 mice (Enard et al., 2009). Together, these data identify a human-specific FP gene co-expression network that is enriched in both genes involved in neurite outgrowth, binding sites for a differentially expressed splicing factor on the human lineage, and genes regulated by FOXP2.
Discussion
Since the sequencing of the human genome, a major goal of evolutionary neuroscience has been to identify human-specific patterns of gene expression and regulation in the brain. While several studies have addressed gene expression in primate brain (Babbitt et al., 2010; Brawand et al., 2011; Cáceres et al., 2003; Enard et al., 2002a; Khaitovich et al., 2005; Khaitovich et al., 2004a; Liu et al., 2011; Marvanova et al., 2003; Somel et al., 2009; Somel et al., 2011; Uddin et al., 2004; Xu et al., 2010a), our study is the first of its kind to ascertain human-specific patterns using multiple platforms, multiple brain regions, and sufficient sample sizes in multiple species. Moreover, ours is the first study to identify human-specific gene co-expression networks with the inclusion of an outgroup. By including these data, we find that gene co-expression or connectivity has rapidly evolved in the neocortex of the human brain. In addition, the genes with changing patterns of connectivity are important for neuronal process formation, the structures that underlie neuronal functional activity and plasticity. Therefore, the evolution of gene connectivity in the human brain may have been critical for the proposed unique abilities of human neurons for synaptic integration (Cáceres et al., 2007; Preuss, 2011).
Using this systems-level approach, we identify several human-specific FP gene co-expression modules. Since FP is a region of the neocortex that was recently enlarged and modified in human evolution (Dumontheil et al., 2008; Semendeferi et al., 2011) human-specific FP networks may provide particular insight into human brain evolution. Previous work has highlighted the evolution of prefrontal cortex in terms of its expansion, enlargement of select subdivisions, its cellular organization, and its connectivity (Rakic, 2009; Semendeferi et al., 2011). In fact, strong evidence supports the protomap model, which by connecting neuronal progenitor cell division and cortical expansion, provides a molecular basis for the evolutionary addition of new brain regions (Donoghue and Rakic, 1999; Rakic et al., 2009). Here, we demonstrate for the first time that even within a single specific cortical region, transcriptional regulation and complexity have dramatically increased on the human lineage. These changes may not be specific to the frontal lobe; it is possible that profiling of additional cortical areas will uncover a general trend for increased transcriptional connectivity in human cortex overall relative to non-neocortex. This novel network connectivity may reflect elaboration of signaling pathways within neurons, neuronal and synaptic ultrastructural elements, or even new cell types. For example, within these human FP networks, there is an enrichment of genes critical for neuronal processes, such as spines, dendrites, and axons.
These findings are striking in light of data demonstrating that human neurons contain a greater number and density of spines compared to other primates (Duan et al., 2003; Elston et al., 2001). A number of the genes identified in the Hs_olivedrab2 module support the hypothesis that our network approach is useful for prioritizing large-scale comparative genomics datasets as well as potentially providing insight into human-specific neuronal processes. STMN2 (or SCG10) has previously been shown to an important regulator of NGF-induced neurite outgrowth (Xu et al., 2010b). Thus, the human-specific increase in STMN2 may be involved in the human-specific increase in spine number. In addition, STMN2 also acts to retard the multipolar transition of neurons and subsequent migration of neurons (Westerlund et al., 2011), suggesting a potential role for increased expression of this gene in the human brain for regulating human cortical expansion. MAP1B is both increasing on the human lineage in the FP as well as a FOXP2 target in human neural progenitors. MAP1B has primarily been associated with axon growth and guidance, and was recently shown to be necessary for the maturation of spines, since loss of MAP1B causes a deficiency in mature spines (Tortosa et al., 2011). Therefore, MAP1B increased expression in human brain may also be involved in the increased number or density of spines in human neurons. The transcription factor LMO4, a previously identified FOXP2 target (Vernes et al., 2011), is also co-expressed in the olivedrab2 module. LMO4 has preferential increased expression in the right human fetal cortex (Sun et al., 2005), perhaps due to repression by FOXP2 in the left cortex. Moreover, co-expression in this human FP module, the distinct expression pattern in the right cortex, and potential regulation by FOXP2, together suggest an important role for LMO4 regulation of genes involved in asymmetrically developed cognitive processes such as language. Several other hub genes in the Hs_darkmagenta have also been directly implicated in neuronal processes such as axons and dendrites. FKBP15 (or FKBP133), which is increased in the human FP, promotes growth cone filipodia (Nakajima et al., 2006). In contrast, KIF2A is an example of a hub gene that is not differentially expressed along the human lineage, yet is highly co-expressed in a human-specific FP module. KIF2A negatively regulates growth cones (Noda et al., 2012). Together, these data suggest that human-specific expression of genes leads to positive growth and maturation of neuronal processes, while those highly co-expressed but not showing human-specific expression may have either negative or refining effects on neuronal process formation. Thus, our data provide a molecular basis for connecting anatomical changes to their underlying genomic origins, furthering our understanding of human brain evolution, and providing predictions that can be tested in model systems. Moreover, our data support the hypothesis that human brain evolution has not only relied upon the expansion and modification of cortical areas, but also on increasing molecular and cellular complexity within a given region. Such complexity is exemplified in findings of neuronal subtypes like the von Economo neurons that have evolved in animals of complex cognition such as primates and expanded in the human brain (Allman et al., 2010; Stimpson et al., 2011).
Previous attempts to identify unique properties of the human brain have focused on changes in brain size, anatomy, regional connectivity, and gene expression (Preuss, 2011; Sherwood et al., 2008). Consistent with recent findings (Brawand et al., 2011), our study finds patterns of gene expression differences across species are generally consistent with known species phylogeny (Figures 7B-C). However, there are some remarkable differences between the gene co-expression connectivity tree and the species tree: the relative distance of human genes to chimpanzee and macaque genes is much larger in the connectivity tree (Figure 7D), indicating a faster evolution of gene connectivity, and hence gene regulation, in the human brain. Previously, we have found that connectivity is a more sensitive measure of evolutionary divergence than gene expression (Miller et al., 2010; Oldham et al., 2006). Therefore, by using new technology and multiple primate species, we have shown for the first time a rapidly evolving mechanism for the coordination of gene expression patterns in the human brain. This is the first demonstration at a genomic level that increased transcriptional diversity of a single brain region accompanies the cortical expansion known to occur in human evolution.
Of particular note in this regard is the olivedrab2 human FP-specific co-expression module, which is enriched in genes involved in neurite outgrowth and has as a hub, the gene for FOXP2, a transcription factor involved in human language and cognition (Lai et al., 2001). Whereas FOXP2 levels themselves are low in the adult brain and FOXP2 is not an hDE gene, FOXP2 is enriched in frontal cortex in developing human brain (Johnson et al., 2009) and it underwent sequence evolution (Enard et al., 2002b) so that it binds a number of new human-specific transcriptional targets (Konopka et al., 2009). Importantly, we experimentally validate an enrichment of human FOXP2 target genes identified during progenitor development in vitro in this human FP module in adults. Thus, the significant overlap with FOXP2 targets in the olivedrab2 module is consistent with a human-specific transcriptional program for FOXP2 in frontal pole (Table S4), which is supported by the graded reduction in FOXP2′s centrality in this network from human to chimp to macaque. So, although FOXP2 is highly expressed in the striatum, these data suggest that the key evolutionary changes are most relevant in the cerebral cortex. These data provide the first strong in vivo evidence for FOXP2 evolution in human cognition, complementing previous in vitro analyses (Konopka et al., 2009).
Another important observation is the enrichment of ELAVL2 binding sites within this module. ELAVL2 has been shown to promote a neuronal phenotype (Akamatsu et al., 1999), and has been modestly associated with schizophrenia (Yamada et al., 2011). Indeed, we find that the ELAVL2 target genes in the olivedrab2 module are enriched for genes involved in nervous system function and disease. For example, numerous genes involved in neuronal function such as ion channels as well as genes critical for synapses, dendrites, and axons are among the genes with ELAVL2 binding motif enrichment. There are also a significant number of autism candidate genes among these potential binding targets (see Results). Therefore, these data have uncovered potential new mechanisms for linking alternative splicing, gene co-expression, and neuropsychiatric disorders.
To date, most research on human brain evolution has focused on changes in brain size, although the past decade has seen contributions from comparative neuroimaging (e.g. (Rilling et al., 2008)), revealing human specializations of fiber-tract organization, and from comparative histology, revealing human specializations of cell and tissue organization (e.g. (Preuss and Coleman, 2002)). However, the number of well-documented human-specific brain phenotypes is currently quite small (Preuss, 2011). The present study demonstrates evolutionary changes at another level of organization, the level of gene interactions. Understanding changes in gene expression and interactions can help us understand how evolution crafted changes in brain morphology and physiology manifested at the levels of cells and tissues. What is more, the discovery of human-specific gene co-expression networks, such as the ones in the cerebral cortex that are described here, can drive “phenotype discovery providing information about changes in patterns of molecular expression that can be used to uncover human specializations of human brain structure and function” (Preuss, 2012; Preuss et al., 2004). In addition, the enrichment of genes associated with neuropsychiatric diseases within these networks provides affirmation of the relevancy of human-specific gene expression patterns providing insight into these cognitive disorders.
While we recognize that due to the inherent methodology of this study (profiling from tissue pieces), we are unable to fully determine the anatomical expression of transcripts within a particular brain region. For example, while we attempted to only use grey matter, we still find a number of gene co-expression modules driven by astrocyte or oligodendrocyte genes. Therefore, these data provide a road map for future immunohistochemical work that will be needed to ascertain the expression of these highlighted genes within different cell types in the brain. Additionally, tissue level expression profiling may miss low abundance transcripts expressed in small subsets of cells. The use of NGS provides significantly improved sensitivity in this regard over microarrays, yet still could miss very low abundance transcripts. We apply WGCNA, which permits in silico dissection of whole tissue into cell level expression patterns (Oldham et al., 2008). Therefore, some of the frontal pole modules may indeed correspond to specific subpopulations of neurons that may be unique to humans. However, future work using laser capture micro-dissection will be useful to uncover transcriptional profiles of additional human-specific gene expression changes at a cellular level. Nevertheless, this work provides a key foundation for connecting human-specific phenotypes to evolved molecular mechanisms at the level of new signaling pathways and genomic complexity in the human brain. Application of the approaches introduced here to other brain regions has the potential to greatly enrich our understanding of human brain organization and evolution.
Supplementary Material
Highlights.
NGS identifies more human-specific expressed genes than microarrays.
The frontal pole has increased in transcriptional complexity in humans.
Human-specific frontal pole networks contain genes involved in cognitive disorders.
Human frontal pole gene connectivity has undergone accelerated evolution.
Acknowledgements
We thank Dr. Giovanni Coppola for providing code for microarray and WGCNA analyses and Lauren Kawaguchi for lab management. This work is supported by grants from the NIMH (R37MH060233) (DHG) and (R00MH090238) (GK), a NARSAD Young Investigator Award (GK), the National Center for Research Resources (RR00165) and Office of Research Infrastructure Programs/OD (P51OD11132), and a James S. McDonnell Foundation grant (JSMF 21002093) (TMP, DHG). Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (NICHD Contract numbers N01-HD-4-3368 and N01-HD-4-3383). The role of the NICHD Brain and Tissue Bank is to distribute tissue, and therefore cannot endorse the studies performed or the interpretation of results. Gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible using GEO series accession number GSE33588.
Footnotes
Contributions G.K., M.O., T.M.P., and D.H.G. conceived the project. G.K. and L.C. conducted experiments. G.K., T.F., J.D-T., K.W., M.O., F.G., G.-Z.W., and R.L. analyzed data. T.M.P. performed IHC and tissue dissections and provided non-human primate samples. G.K. and D.H.G. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci U S A. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K., Erwin, J., Park S, Goubert V, Hof PR. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct Funct. 2010;214:495–517. doi: 10.1007/s00429-010-0254-0. [DOI] [PubMed] [Google Scholar]
- Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, Kocher JP. 3′ tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genomics. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MME, Serikawa K, Lemon T, Morgan R, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb Cortex. 2007;17:2312–2321. doi: 10.1093/cercor/bhl140. [DOI] [PubMed] [Google Scholar]
- Carr CW, Moreno-De-Luca D, Parker C, Zimmerman HH, Ledbetter N, Martin CL, Dobyns WB, Abdul-Rahman OA. Chiari I malformation, delayed gross motor skills, severe speech delay, and epileptiform discharges in a child with FOXP1 haploinsufficiency. Eur J Hum Genet. 2010;18:1216–1220. doi: 10.1038/ejhg.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium C.S.a.A. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Coque L, Mukherjee S, Cao JL, Spencer S, Marvin M, Falcon E, Sidor MM, Birnbaum SG, Graham A, Neve RL, et al. Specific role of VTA dopamine neuronal firing rates and morphology in the reversal of anxiety-related, but not depression-related behavior in the ClockDelta19 mouse model of mania. Neuropsychopharmacology. 2011;36:1478–1488. doi: 10.1038/npp.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Teichmann SA, Babu MM. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 2009;19:785–794. doi: 10.1101/gr.086165.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MJ, Rakic P. Molecular gradients and compartments in the embryonic primate cerebral cortex. Cereb Cortex. 1999;9:586–600. doi: 10.1093/cercor/9.6.586. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wearne SL, Rocher AB, Macedo A, Morrison JH, Hof PR. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb Cortex. 2003;13:950–961. doi: 10.1093/cercor/13.9.950. [DOI] [PubMed] [Google Scholar]
- Dumontheil I, Burgess PW, Blakemore SJ. Development of rostral prefrontal cortex and cognitive and behavioural disorders. Dev Med Child Neurol. 2008;50:168–181. doi: 10.1111/j.1469-8749.2008.02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J. The pyramidal cell in cognition: a comparative study in human and monkey. J Neurosci. 2001;21:RC163. doi: 10.1523/JNEUROSCI.21-17-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Holter SM, Blass T, Somel M, Bruckner MK, Schreiweis C, Winter C, Sohr R, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002a;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002b;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Feng L, Liu H, Liu Y, Lu Z, Guo G, Guo S, Zheng H, Gao Y, Cheng S, Wang J, et al. Power of deep sequencing and agilent microarray for gene expression profiling study. Mol Biotechnol. 2010;45:101–110. doi: 10.1007/s12033-010-9249-6. [DOI] [PubMed] [Google Scholar]
- Gazave E, Darre F, Morcillo-Suarez C, Petit-Marty N, Carreno A, Marigorta UM, Ryder OA, Blancher A, Rocchi M, Bosch E, et al. Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res. 2011;21:1626–1639. doi: 10.1101/gr.117242.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grados MA. The genetics of obsessive-compulsive disorder and Tourette syndrome: an epidemiological and pathway-based approach for gene discovery. J Am Acad Child Adolesc Psychiatry. 2010;49:810–819. 819, e811–812. doi: 10.1016/j.jaac.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Daoud H, Rochefort D, Piton A, Gauthier J, Langlois M, Foomani G, Dobrzeniecka S, Krebs MO, Joober R, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87:671–678. doi: 10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Horn D, Kapeller J, Rivera-Brugues N, Moog U, Lorenz-Depiereux B, Eck S, Hempel M, Wagenstaller J, Gawthrope A, Monaco AP, et al. Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum Mutat. 2010;31:E1851–1860. doi: 10.1002/humu.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A. 2010;107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol. 2011;85:9472–9477. doi: 10.1128/JVI.00876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004a;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. A neutral model of transcriptome evolution. PLoS Biol. 2004b;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Wexler E, Rosen E, Mukamel Z, Osborn GE, Chen L, Lu D, Gao F, Gao K, Lowe JK, Geschwind DH. Modeling the functional genomics of autism using human neurons. Mol Psychiatry. 2012;17:202–214. doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, Goldstein A, Horn D, Klopocki E, Kluger G, Martin P, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011;48:396–406. doi: 10.1136/jmg.2010.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Mao X, Wei L. Genes and (common) pathways underlying drug addiction. PLoS Comput Biol. 2008;4:e2. doi: 10.1371/journal.pcbi.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACSarray profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 2011;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvanova M, Menager J, Bezard E, Bontrop RE, Pradier L, Wong G. Microarray analysis of nonhuman primates: validation of experimental models in neurological disorders. FASEB J. 2003;17:929–931. doi: 10.1096/fj.02-0681fje. [DOI] [PubMed] [Google Scholar]
- Menet JS, Rosbash M. When brain clocks lose track of time: cause or consequence of neuropsychiatric disorders. Curr Opin Neurobiol. 2011;21:849–857. doi: 10.1016/j.conb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies -the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima O, Nakamura F, Yamashita N, Tomita Y, Suto F, Okada T, Iwamatsu A, Kondo E, Fujisawa H, Takei K, Goshima Y. FKBP133: a novel mouse FK506-binding protein homolog alters growth cone morphology. Biochem Biophys Res Commun. 2006;346:140–149. doi: 10.1016/j.bbrc.2006.05.113. [DOI] [PubMed] [Google Scholar]
- Noda Y, Niwa S, Homma N, Fukuda H, Imajo-Ohmi S, Hirokawa N. Phosphatidylinositol 4-phosphate 5-kinase alpha (PIPKalpha) regulates neuronal microtubule depolymerase kinesin, KIF2A and suppresses elongation of axon branches. Proc Natl Acad Sci U S A. 2012;109:1725–1730. doi: 10.1073/pnas.1107808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas J, Serrat JM. Gene promoter evolution targets the center of the human protein interaction network. PLoS One. 2010;5:e11476. doi: 10.1371/journal.pone.0011476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces Shaping the Fastest Evolving Regions in the Human Genome. PLoS Genet. 2006a;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006b;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Preuss TM. The human brain: rewired and running hot. Ann N Y Acad Sci. 2011;1225(Suppl 1):E182–191. doi: 10.1111/j.1749-6632.2011.06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM. Human brain evolution: from gene discovery to phenotype discovery. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1201894109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM, Cáceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Coleman GQ. Human-specific organization of primary visual cortex: alternating compartments of dense Cat-301 and calbindin immunoreactivity in layer 4A. Cereb Cortex. 2002;12:671–691. doi: 10.1093/cercor/12.7.671. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: making cortical maps. Trends Neurosci. 2009;32:291–301. doi: 10.1016/j.tins.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Yakovlev PI. Development of the corpus callosum and cavum septi in man. J Comp Neurol. 1968;132:45–72. doi: 10.1002/cne.901320103. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Armstrong E, Schleicher A, Zilles K, Van Hoesen GW. Prefrontal cortex in humans and apes: a comparative study of area 10. Am J Phys Anthropol. 2001;114:224–241. doi: 10.1002/1096-8644(200103)114:3<224::AID-AJPA1022>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex. 2011;21:1485–1497. doi: 10.1093/cercor/bhq191. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: tracing evolutionary changes in brain and cognition. J Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, et al. MicroRNA-Driven Developmental Remodeling in the Brain Distinguishes Humans from Other Primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am Hum Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson CD, Tetreault NA, Allman JM, Jacobs B, Butti C, Hof PR, Sherwood CC. Biochemical specificity of von Economo neurons in hominoids. Am J Hum Biol. 2011;23:22–28. doi: 10.1002/ajhb.21135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa E, Montenegro-Venegas C, Benoist M, Hartel S, Gonzalez-Billault C, Esteban JA, Avila J. Microtubule-associated protein 1B (MAP1B) is required for dendritic spine development and synaptic maturation. J Biol Chem. 2011;286:40638–40648. doi: 10.1074/jbc.M111.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci U S A. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajsar J, Schachter H. Walker-Warburg syndrome. Orphanet J Rare Dis. 2006;1:29. doi: 10.1186/1750-1172-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendelken C, O’Hare ED, Whitaker KJ, Ferrer E, Bunge SA. Increased Functional Selectivity over Development in Rostrolateral Prefrontal Cortex. J Neurosci. 2011;31:17260–17268. doi: 10.1523/JNEUROSCI.1193-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerlund N, Zdrojewska J, Padzik A, Komulainen E, Bjorkblom B, Rannikko E, Tararuk T, Garcia-Frigola C, Sandholm J, Nguyen L, et al. Phosphorylation of SCG10/stathmin-2 determines multipolar stage exit and neuronal migration rate. Nat Neurosci. 2011;14:305–313. doi: 10.1038/nn.2755. [DOI] [PubMed] [Google Scholar]
- Xu AG, He L, Li Z, Xu Y, Li M, Fu X, Yan Z, Yuan Y, Menzel C, Li N, et al. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput Biol. 2010a;6:e1000843. doi: 10.1371/journal.pcbi.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Dhanasekaran DN, Lee CM, Reddy EP. Regulation of neurite outgrowth by interactions between the scaffolding protein, JNK-associated leucine zipper protein, and neuronal growth-associated protein superior cervical ganglia clone 10. J Biol Chem. 2010b;285:3548–3553. doi: 10.1074/jbc.M109.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Iwayama Y, Hattori E, Iwamoto K, Toyota T, Ohnishi T, Ohba H, Maekawa M, Kato T, Yoshikawa T. Genome-wide association study of schizophrenia in Japanese population. PLoS One. 2011;6:e20468. doi: 10.1371/journal.pone.0020468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.