Abstract
The methyl-binding protein gene, MECP2, is a candidate for involvement in autism through its implication as a major causative factor in Rett syndrome that has similarities to autism. Rare mutations in MECP2 have also been identified in autistic individuals. We have examined the possible broader involvement of MECP2 as a predisposing factor in the disorder. Analysis of polymorphic markers spanning the gene and comprising both microsatellites and single nucleotide polymorphisms (SNPs) by the transmission disequilibrium test in two collections of families (219 in total), one in the USA and one in the UK, has provided evidence for significant association (P = 0.009) for a three-marker SNP haplotype of MECP2 with autism/autism spectrum disorders. This association is supported by association of both Single Sequence Repeat (SSR) and SNP single markers located at the 3′ end of the MECP2 locus and flanking sequence, the most significant being that of an indel marker located in intron 2 (P = 0.001 – Bonferroni corrected P = 0.006). This suggests that one or more functional variants of MECP2 existing at significant frequencies in the population may confer increased risk of autism/autism spectrum disorders and warrants further investigation in additional independent samples.
Keywords: Autism, autism spectrum disorder, MECP2, polymorphisms, X-chromosome
Autism is a neurodevelopmental disorder characterized by social and communication impairments, coupled with stereotyped and repetitive behaviours. It is far more common in males (4–10:1 male:female ratio; Folstein & Rosen-Sheidley 2001). Linkage studies have provided only weak evidence for the involvement of X-linked loci but have limited power to detect those of small effect size (Freitag 2007). Gauthier et al. (2006) have provided some evidence for the involvement of a locus at Xq27-q28 in the disorder employing both linkage and association, and rare mutations in X-linked genes such as neuroligin 3 (NLGN3) and neuroligin 4 (NLGN4) have also been reported to be associated with autism (Jamain et al. 2003).
Rett syndrome (RTT) is an X-linked neurodevelopmental disorder, with a prevalence of around 1 per 10 000–15 000 (Hagberg 1985; Hagberg & Hagberg 1997; Leonard et al. 1997). Many symptoms, including impaired language, stereotypic behaviours, high frequency of seizures and sleep abnormalities as well as the developmental timing are common to both RTT and autism. Indeed, misdiagnosis of RTT individuals as autistic can occur (Abdul-Rahman & Hudgins 2006; Moretti et al. 2005). Both disorders are grouped under the heading of pervasive developmental disorders (PDD) in Diagnostic and Statistical Manual of Disorders-IV (DSM-IV).
The MECP2 gene (Xq28) encodes the methyl-CpG-binding protein 2 (MECP2), and mutations in the gene are reported to be responsible for around 75% of cases of classical RTT. Methyl-CpG-binding protein 2 protein binds to methylated CpGs and is believed to repress the transcription of downstream genes, e.g. Bdnf in mice (Chen et al. 2003; Lewis et al. 1992; Meehan et al. 1992; Moretti & Zoghbi 2006); however, there is evidence that the situation is more complex with opposite directions of regulation existing in different tissues (LaSalle 2007). MECP2 comprises four exons, with the coding sequence shared among exons 2, 3 and 4 and a highly conserved 3′ untranslated region (UTR) of 8.5 kb (Coy et al. 1999). Both increases and decreases in MECP2 expression have been implicated in a range of PDDs including autism suggesting that a common pathway may be involved in these disorders (Samaco et al. 2004, 2005; Van Esch et al. 2005).
Although they are rare, mutations in the coding region of MECP2 have been observed in autistic individuals (Beyer et al. 2002; Carney et al. 2003; Lam et al. 2000; Lobo-Menendez et al. 2003; Vourc’h et al. 2001). Other studies by Shibayama et al. (2004) detected one missense and two 3′ UTR variants in 24 autism patients vs. only one missense mutation in 144 ethnically matched individuals without autism. Recently, Liu and Francke (2006) showed that certain sequence motifs, distributed over a distance of 130 kb in and around the MECP2 gene, make up a ‘functional expression module’ containing enhancers and silencers. These interact with the MECP2 promoter and affect the tissue-specific, developmental stage-specific or splice-variant-specific control of MECP2 protein expression.
Thus, variations throughout the coding and non-coding regions of the MECP2 gene, as well as flanking regions, could be important factors contributing to the complex disorder of autism. This study was designed to investigate a series of polymorphic variants in the MECP2 gene, including flanking and intronic regions, as potential markers for the disorder by transmission disequilibrium tests (TDTs) in two series of autistic families.
Methods
The DNA was available from 219 families with an affected autism spectrum disorder proband, some of their siblings (n = 81) and one or both the proband’s parents (219 mothers and 196 fathers). The sample collections taken from two main sources are as follows.
Molecular Genetics of Autism Study collection
These 121 families were mainly families in which the proband was diagnosed by the National Specialist service multidisciplinary team at the Michael Rutter Centre, South London and Maudsley (SLAM) NHS Trust, London, UK. Some additional probands were included in this sample from three other sources: Dr Anne O’Hare at Edinburgh University; Dr A. J. Sharma at the Mary Sheridan Child Development Centre, Camberwell and The Behaviour Genetics Clinic, SLAM NHS Trust (young people with autism spectrum disorders). Cases with an autism spectrum disorder (which was defined as a case with autistic disorder, atypical autism, Asperger’s syndrome, pervasive developmental disorder and others) were included. Diagnoses were made by experienced clinicians following multidisciplinary assessments according to ICD-10 criteria and wherever possible with the assistance of structured parent interviews and semi-standardized observational assessments [the Autism Diagnostic Interview (ADI-R; Lord et al. 1994) or an updated version of the Development and Wellbeing Assessment (Goodman et al. 2000) and/or the Autism Diagnostic Observational Schedule (ADOS); n = 70]. Cases were excluded if they had another known, possible autism-causing medical condition (e.g. fragile X syndrome or tuberous sclerosis) or if their IQ was below 35.
Autism Genetic Resource Exchange collection
This sample (n = 98 families) is a subset of the sample collected for the Autism Genetic Resource Exchange (AGRE) (Geschwind et al. 2001) with the support of the Cure Autism Now (http://www.cureautismnow.org: now merged with Autism Speaks – http://www.autism-speaks.org) under the direction of a scientific steering committee (see websites). The DNA used in this study is from trios with an affected proband and was collected for the initial purposes of screening. All probands meet the criteria for autism spectrum disorder in one of three categories: (1) autism diagnosed using the ADI-R, (2) Not Quite Autism (NQA), where individuals are not more than one point away from meeting autism criteria on any or all the three ‘content’ domains (i.e. social, communication and non-social behaviour) and meet criteria for the ‘age of onset’ domain or who meet criteria on all three ‘content’ domains but not on the ‘age of onset’ domain and (3) broad spectrum where individuals who showed patterns of impairment along the spectrum of PDD. This is a broad diagnostic category and encompasses PDDs such as Asperger’s syndrome. It is operationally defined according to domain scores on the ADI algorithm and includes cases who show (1) severe deficit in at least one domain, (2) more moderate deficits in at least two domains and (3) only minimal deficits, but in all three domains.
Diagnoses were confirmed using the ADI-R (Lord et al. 1994) and/or the ADOS in 140 cases. In total, there were 170 cases with a diagnosis of autism and 1 case with NQA, and 38 with a broad-spectrum diagnosis [13 from the AGRE sample and 25 from the Molecular Genetics of Autism Study (MGAS) sample]. Genotypic data were available on 715 individuals. This included 219 autism spectrum disorder probands, 81 siblings and 415 parents.
Genotyping
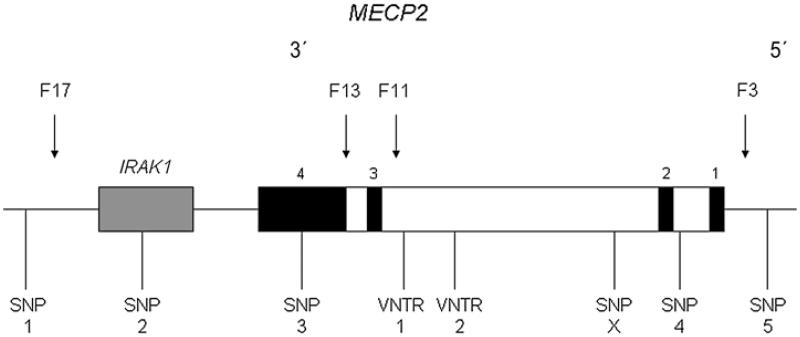
Microsatellite genotyping and analysis was set up and performed in-house. Using the University of California Santa Cruz (UCSC) Genome Browser, four microsatellite repeats were identified in the MECP2 gene whose properties suggested that they were likely candidates to be variable in the population. Primers were designed using Primo Multiplex to flank all four of these loci, and initial polymerase chain reactions (PCRs) were carried out using standard conditions to investigate heterozygosity in around 20 samples of female control DNA. Two of the four markers were extensively polymorphic and therefore informative. These markers were PCR amplified in separate 25 μl reactions using different template DNA concentrations, annealing temperatures and magnesium chloride concentrations (40 ng DNA, 3.5 mm MgCl2 and 59.3°C for marker 1 and 20 ng DNA, 2.5 mm MgCl2 and 56°C for marker 2). The PCR products from the single reactions were multiplexed for loading onto an ABI prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Microsatellite marker 1 was an AT dinucleotide repeat located at chrX:152823442. Marker 2 was a GT dinucleotide repeat located at chrX:152832198. These are both loci within the large intron 2 in the MECP2 gene (Fig. 1).
Figure 1. Diagram of MECP2 gene (and neighbouring IRAK1 gene in grey), with positioning of markers selected in this study shown beneath (not to scale).
The exons are filled in black and are labelled 1–4 from 5′ to 3′. SNP 1 = rs5945173; SNP 2 = rs11465839; SNP 3 = rs2734647; SNP 4 = rs5945175 and SNP 5 = rs5945397. F numbers above gene indicate regulatory regions identified by Liu and Francke (2006) – F3 and F13 are silencers and F11 and F17 are enhancers.
Six single nucleotide polymorphisms (SNPs) were selected initially, using Haploview version 3.2, on the basis of their having minimum allele frequencies predicted to be high enough to be informative (>0.05) and locations that were spread across the gene and capable of capturing variation in the single block of linkage disequilibrium (LD) across the whole gene. These SNPs (for which one assay failed) were genotyped at K Biosciences (Hoddesdon, UK; http://www.kbioscience.co.uk). The five SNPs for which data were obtained are from 3′ to 5′: (1) rs5945173 (A/G), chrX:152771019; (2) rs11465839 (C/T), chrX: 152801114; (3) rs2734647 (C/T), chrX:152813027; (4) rs5945175 (C/T), chrX:152879604 and (5) rs5945397 (A/G), chrX:152894373 (Fig. 1). Genotyping success rates achieved by the company varied between 93% and 97% for female samples and between 95% and 97% for male samples with an exception being rs5945173, where the success rate was 90%. This SNP was of low information content in transmission analysis, and although it was included in the preliminary analysis, its contribution to the final haplotype analysis was minimal. Allele frequencies were calculated separately for males and females and showed no significant differences (P > 0.6). All SNPs examined were in Hardy–Weinberg equilibrium with P > 0.05 after correction for multiple testing, apart from rs2734647 (P = 0.011). The five SNPs cover the region defined as the ‘MECP2 functional expression module’ (Liu & Francke 2006). SNPs rs2734647, rs5945175 and rs5945397 and the Single Sequence Repeat (SSR) markers are within a block of high LD covering the MECP2 locus, and SNPs rs5945173 and rs11465839 are located on either side of the 3′ enhancer region F17.
Based on May 2004 assembly release, the co-ordinates of the MECP2 region (which includes the IRAK1 locus which abuts the 3′ region of MECP2 and overlaps the enhancers examined) were chrX:152,789,145-152,902,943. The tag SNPs selected capture 94% of the known variation at r2 = 0.8 within this region (http://www.broad.mit.edu/mpg/tagger/) based on HAPMAP Build 35 release 21a/phase II jan 07 on NCBI B35 assembly dpSNP 125. The Centre d’Etude du Polymorphisme Humain Collection-European (CEPH CEU) population was examined by pairwise tagging.
Analysis
Distortion in the transmission of microsatellite and SNP alleles was tested for by the TDT implemented in tdtphase from the unphased version 2.404 (2003) suite of programs (Dudbridge 2003). Analysis was stratified by sample collection, sex and phenotypic subgroups. For microsatellite 1, alleles were clumped using the program nocom version 20 (April 2007) (Ott 1979) to estimate the best fitting normal distributions for observed allele counts.
Results
Microsatellites
Data were analysed using the TDT for deviations from expected random inheritance of alleles by offspring, which may indicate their tracking of predisposing or protective variants of the gene. Once data with any Mendelian errors or missing information had been removed, 219 affected families were available for analysis with both markers employing tdtphase.
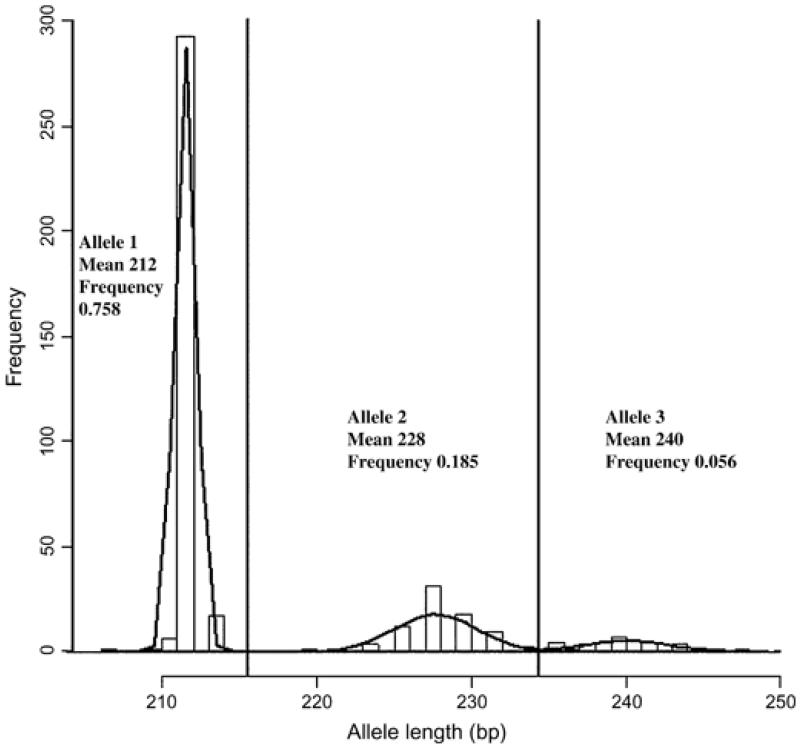
For SSR marker 1, an inspection of the pattern of allele frequencies indicated that allele sizes approximated to a trimodal distribution, suggesting that the three groups (numbered 1–3) contained alleles that may be closely related in evolutionary terms by replication slippage. Examining the fit of the allele size and frequency data to one or more normal distributions employing the program nocom (Ott 1979) indicate that two distributions fit significantly better than one and three distributions significantly better than two (P = < 0.0001) centring on mean allele sizes of 212, 228 and 240 bp with frequencies of 0.76, 0.18 and 0.06 (Fig. 2).
Figure 2. Distribution of marker 1 alleles (measured in base pairs of the amplicon) and the three groups into which they were categorized for further TDT analysis of clumped alleles.
Allele lengths are in base pairs and their means within each of the three groups.
As an expedient to reduce the requirement for conservative multiple testing, we therefore analysed the frequencies of the three size groups by TDT. This indicated that allele group 1 is over-transmitted to the probands in the combined sample (P = 0.021), while allele group 2 shows a trend to be under-transmitted to the probands (P = 0.073) (Table 1).
Table 1. TDT analysis for SSR marker 1 alleles grouped into three allele categories.
| Allele | T | Freq-T | NT | Freq-NT | T (%) | Chi-square | P value |
|---|---|---|---|---|---|---|---|
| AGRE and MGAS combined (SSR marker 1 alleles grouped into three categories) | |||||||
| 1 | 169 | 0.81 | 150 | 0.71 | 52.98 | 5.3 | 0.021 |
| 2 | 32 | 0.15 | 45 | 0.21 | 41.56 | 3.22 | 0.073 |
| 3 | 9 | 0.04 | 15 | 0.07 | 37.5 | 1.52 | 0.218 |
| AGRE only (SSR marker 1 alleles grouped into three categories) | |||||||
| 1 | 77 | 0.80 | 64 | 0.67 | 54.61 | 5.62 | 0.018 |
| 2 | 14 | 0.15 | 27 | 0.28 | 34.15 | 6.53 | 0.010 |
| 3 | 5 | 0.05 | 5 | 0.05 | 50 | 0 | 1 |
| MGAS only (SSR marker 1 alleles grouped into three categories) | |||||||
| 1 | 92 | 0.81 | 86 | 0.75 | 51.69 | 0.95 | 0.329 |
| 2 | 18 | 0.16 | 18 | 0.16 | 50 | 0 | 1 |
| 3 | 4 | 0.04 | 10 | 0.09 | 28.57 | 2.66 | 0.103 |
Allele, allele category; T, number of times allele is transmitted to affected offspring, Freq-T, frequency allele is transmitted; NT, number of times allele not transmitted to affected offspring; Freq-NT, frequency allele is not transmitted; T (%), percentage of transmissions (of total transmissions and non-transmissions); Chi-squared, chi-squared statistic.
Analyses for marker 1 were also carried out on each sample (AGRE and MGAS) separately to see if one cohort was driving the pattern of results (Table 1). As shown, the strongest significance was found in the AGRE sample for the under-transmission of allele group 2 (P = 0.010), followed by over-transmission of allele group 1 (P= 0.018), with the MGAS samples following the same pattern of over-transmission for allele group 1, although the figures are not significant.
The distribution of the allele sizes was also examined for marker 2; however, there was no obvious division into allele size groupings that would have enabled simplification of the analysis. Nevertheless, it appeared that although the original sequence analysis of marker 2 indicated it to be a dinucleotide repeat (with consecutively sized products that are presumptively 2 bp different from one another), there were a substantial number (n = 101) of alleles representing nine different sizes separated by 1 bp from the dinucleotide peaks (data not shown). As the amplification method had been designed to eliminate problems with the sporadic addition of an extra nucleotide (plus-A) (Brownstein et al. 1996), the possibility that these additional alleles represented independent alleles was therefore investigated. It was subsequently found from the published sequence that the amplified region between the primers for marker 2 also contains a single base pair indel polymorphism G/− (rs11348580) in a string of G residues located 3 bp 3′ to the dinucleotide repeat, which created two allelic series separated by 1 bp. Unfortunately, the sequence surrounding the indel precluded a simple PCR-based genotyping assay. We decided, therefore, to analyse the transmittance and non-transmittance of alleles with and without a G deletion indirectly by comparing products with an odd number vs. products with an even number of base pairs (Table 2). The alleles of the indel employing this indirect assay were in Hardy–Weinberg equilibrium (P > 0.05 after correction for multiple testing). As indicated, even-sized alleles (no G deletion) were significantly over-transmitted, while odd-sized alleles (G deletion) were significantly under-transmitted (P = 0.001). A TDT was also carried out for this single base pair deletion allele in the MGAS and AGRE samples separately. As shown in Table 2, the MGAS samples showed a significant over-transmission of the even-sized (no deletion) allele (P = 0.039), and although this over-transmission was not significant for the AGRE sample (P = 0.103), the bias was again for the even-sized allele to be transmitted more often (75 transmitted vs. 65 not transmitted). Because the over-transmission of the even-sized allele was at its most significant when the MGAS and AGRE samples were combined (P = 0.001), it appears that the finding is consistent in both cohorts.
Table 2. TDT analysis for the indel associated with SSR marker 2 comparing alleles with and without G deletions (‘odd’ and ‘even’, respectively).
| Allele | T | Freq-T | NT | Freq-NT | T (%) | P value |
|---|---|---|---|---|---|---|
| AGRE and MGAS combined, SSR marker 2 (indel rs11348580) | ||||||
| Even | 174 | 0.82 | 153 | 0.72 | 53.21 | 0.001 |
| Odd | 39 | 0.18 | 60 | 0.28 | 39.39 | |
| AGRE sample only, SSR marker 2 | ||||||
| Even | 75 | 0.77 | 65 | 0.6 | 53.57 | 0.103 |
| Odd | 23 | 0.23 | 33 | 0.34 | 41.07 | |
| MGAS samples only, SSR marker 2 | ||||||
| Even | 99 | 0.86 | 88 | 0.77 | 52.94 | 0.039 |
| Odd | 16 | 0.14 | 27 | 0.23 | 37.21 | |
Allele, allele size; T, number of times allele is transmitted to affected offspring; Freq-T, frequency allele is transmitted; NT, number of times allele not transmitted to affected offspring; Freq-T, frequency allele is not transmitted, T (%), percentage of transmissions (of total transmissions and non-transmissions), Chi-square, chi-squared statistic.
Single nucleotide polymorphisms
The distribution of the five successfully genotyped SNPs across the MECP2 locus is shown in Fig. 1. As was the case for microsatellites, TDT analysis of the SNP data was carried out using tdtphase (Table 3). The data for the two samples combined indicated that SNPs rs5945173 and rs2734647 were significantly biased in transmission (P = 0.006 and P = 0.016, respectively). Evidence for the remaining SNPs was compromised by the low frequencies of the rarer alleles – in spite of our preliminary selection for minor allele frequencies to be >0.05 – the frequencies for SNPs rs11465839, rs5945175 and rs594539 that we observed were only between 0.01 and 0.04, and consequently, few informative transmissions were available for analysis, and they were not considered from the viewpoint of multiple testing. When the AGRE and MGAS cohorts were analysed separately, the same markers were significant in the AGRE cohort (rs5945173, P = 0.003 and rs2734647, P = 0.008). Analysis of MGAS data alone did not show significance for the two SNPs that were significant in the AGRE sample, although the direction of the biased transmission in the AGRE sample was weakly echoed.
Table 3. TDT results for all five MECP2 SNPs on combined sample.
| MGAS and AGRE combined | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Allele | T | Freq-T | NT | Freq-NT | T (%) | P value |
| 1 | A | 31 | 0.16 | 51 | 0.27 | 37.8 | 0.006 |
| G | 161 | 0.84 | 141 | 0.73 | 53.31 | ||
| 2 | C | 5 | 0.02 | 7 | 0.03 | 41.67 | |
| T | 199 | 0.98 | 197 | 0.97 | 50.25 | ||
| 3 | C | 179 | 0.86 | 162 | 0.78 | 52.49 | 0.016 |
| T | 30 | 0.14 | 47 | 0.22 | 38.96 | ||
| 4 | C | 2 | 0.01 | 3 | 0.02 | 40 | |
| T | 187 | 0.99 | 186 | 0.98 | 50.13 | ||
| 5 | A | 5 | 0.03 | 9 | 0.05 | 35.71 | |
| T | 183 | 0.97 | 179 | 0.95 | 47.5 | ||
T, number of times allele is transmitted to affected offspring; Freq-T, frequency allele is transmitted; NT, number of times allele not transmitted to affected offspring; Freq-T, frequency allele is not transmitted; T (%), percentage of transmissions (of total transmissions and non-transmissions); Chi-square, chi-squared statistic.
SNPs are labelled 1–5 from 3′ to 5′ as follows: 1: rs5945173 (A/G); 2: rs11465839 (C/T); 3: rs2734647 (C/T); 4: rs5945175 (C/T), chrX:152879604; 5: rs5945397 (A/G), chrX:152894373. P values not included for MECP2 SNPs 2, 4 and 5 as there is less than 50% power to detect a variant of 5% frequency conferring an allele-specific risk of 1.7; this rising to >80% for alleles with frequencies >12% (http://pngu.mgh.harvard.edu/~purcell/gpc/).
Application of a Bonferroni correction for multiple testing of the indel and the two usefully informative SNPs together with the microsatellite SSR marker 1 reduces the probability for association in the combined samples as follows: indel rs11348580 (0.004), rs5945173 (0.024) and rs2734647 (0.064) leaving the indel and rs5945173 significant and rs2734647 showing a trend to significance.
To investigate the effect of using narrower phenotypic criteria, TDT analysis was repeated on the combined sample after removing all families in which the proband was allotted a broad-spectrum diagnosis. This left only families where the proband had a pure ‘autism’ or an ‘Asperger’s syndrome’ diagnosis. Asperger’s individuals were still included in the narrow phenotype sample both to maintain a relatively large sample size and because their social interactions are as severely impaired as they are for autistic individuals. We have previously indicated the importance of X-linked loci to the social substrate of the autistic phenotype (Loat et al. 2004) and, therefore, all probands for whom the diagnosis implied a level of deficit in social functioning equivalent to autism were analysed. Analysis of the data after the removal of the less severe phenotypic group of probands (which included 39 individuals, most of whom had a PDD nitric oxide synthase diagnosis) marginally increased the significance of rs5945173 (P = 0.002) and rs2734647 (P = 0.006).
Haplotype analysis for rs5945173 and rs2734647 (i.e. those with appreciable heterozygosity, c. 30%) indicated that the G-C haplotype was significantly over-transmitted (P = 0.011) and the A-T haplotype was significantly under-transmitted (P = 0.021). When the LD patterns at the rare SNPs are also included to determine if this can subdivide the common G-C haplotype using the whole panel of five SNPs (numbered in order from 3′ to 5′ – Fig. 1) this showed that the G-T-C-T-G haplotype was significantly over-transmitted (P = 0.004), while the A-T-T-T-G haplotype was under-transmitted P = 0.032) from parents to autistic offspring (Table 4). If the usefully informative SNP markers (rs5945173 and rs2734647) are combined with the indel within SSR marker 2, the haplotype significance for over-transmission for G-C odd is 0.009. Given the patterns of significance for the SSR marker 1, the indel and SNPs, it appears that any functional variant associated with autism and tracked by these may be located towards the 3′ end of the gene.
Table 4. Haplotype TDT results for all five SNPs.
| Haplotype | T | Freq-T | NT | Freq-NT | T (%) | Chi-square | P value |
|---|---|---|---|---|---|---|---|
| A-T-C-T-G | 4 | 0.07 | 8 | 0.15 | 33.33 | 1.359 | 0.244 |
| A-T-T-C-A | 1 | 0.02 | 3 | 0.05 | 25 | 1.046 | 0.306 |
| A-T-T-T-A | 2 | 0.04 | 4 | 0.07 | 33.33 | 0.6796 | 0.410 |
| A-T-T-T-G | 8 | 0.15 | 19 | 0.76 | 29.63 | 4.615 | 0.032 |
| G-C-C-T-G | 3 | 0.05 | 5 | 0.09 | 37.5 | 0.5053 | 0.477 |
| G-T-C-T-G | 35 | 0.64 | 15 | 0.27 | 70 | 8.228 | 0.004 |
| G-T-T-T-G | 2 | 0.04 | 1 | 0.02 | 66.67 | 0.3398 | 0.560 |
T, number of times allele is transmitted to affected offspring; Freq-T, frequency allele is transmitted; NT, number of times allele not transmitted to affected offspring; Freq-T, frequency allele is not transmitted; T (%), percentage of transmissions (of totaltransmissions and non-transmissions); Chi-square, chi-squared statistic.
Alleles are listed in order for SNPs 1–5 in a 3′ to 5′ direction.
Discussion
The possibility that MECP2 variants may be involved in autism has been suggested by previous research identifying a clear association between mutations in MECP2 and RTT and by the acknowledgement of an overlap in behavioural phenotypes and gene expression profiles in brain tissue from RTT and autistic individuals. Furthermore, there is some evidence suggesting that mutations in the coding, and 3′ UTRs of MECP2 may be associated with the disorder (Carney et al. 2003; Lam et al. 2000; Shibayama et al. 2004). Recent evidence additionally suggests that regions of the genome within and near to the MECP2 locus harbour regulatory elements affecting its transcription (Liu & Francke 2006). The study reported here aimed to investigate whether there is evidence for a bias in the transmission, from parents to autistic offspring in two different sample collections, of particular alleles at several polymorphic sites in and around MECP2. Data from two microsatellites located in intron 2, and five SNPs within, upstream and downstream of the gene were analysed. Our results indicate that one or more relatively common variants in MECP2 may play some part in altering the risk for autism. Of the two highly polymorphic microsatellite markers we investigated, one gave a significant bias in transmission to probands when alleles were grouped based on clusters of sizes closely approximating to three normal distributions, the groups themselves being distinct from each other. The other microsatellite, while not significant itself, was located in close proximity to a single base pair deletion (rs11348580), which could be detected using the same microsatellite assay, and this single base pair insertion/deletion showed a significant association with autism in the combined sample.
Our SNP analyses also generated interesting results. While the most significant findings arose when one of the samples (AGRE) was considered in isolation, the two apparently associated SNPs (rs5945173 and rs2734647) continued to be significantly associated in the combined sample. While the data for the other SNPs examined did not suggest significant association, the low frequencies of the minor alleles resulted in relatively few informative transmissions. The reason for the difference between the strength of findings in the AGRE and MGAS samples is not clear but may be influenced by the fact that they were collected in different countries and diagnosed against slightly different criteria. Both samples were primarily made up from cases meeting clinical/ADI-R criteria for autism (79% MGAS and 86% AGRE). The slightly higher proportion of cases meeting the criteria for autism in the AGRE sample may be relevant, but the numbers in the samples were too small to permit meaningful subgroup analyses.
The most significant of the SNP haplotypes made up of varying numbers of SNPs was the five-marker G-T-C-T-G haplotype, which was transmitted to autistic probands on 35 occasions and not transmitted on only 15 for the combined sample. This may reflect the fact that there is strong LD in the MECP2 gene and that any true association of a functional variant with autism may be detected at some distance from the real locus of interest. The results of SNP analysis based on rs5945173 and rs2734647, which had relatively frequent minor alleles, indicated that they were significantly associated with autism both individually and as a haplotype. Furthermore, the combined haplotype for rs5945173 and rs2734647 and the indel within SSR marker 2 was also significantly associated (P = 0.009). This might imply that the 3′ end of the gene is more likely to track the truly functional variant. However, given the low number of informative transmissions for the other SNPs, it is conceivable that this concentration of significant findings at the 3′ end simply results from greater power to detect significance at these more informative loci.
The data from the MECP2 microsatellites and five SNPs were not incorporated into the same haplotype analyses because the nature of LD between these disparate types of markers is unknown and so the validity of such an approach to identify which specific region of a gene is of most interest is questionable. Nevertheless, it is reassuring that significant associations were observed with both SNPs and microsatellite/indel, and associations observed in one sample group were generally supported by trends in the same direction observed for the other group. It is reasonable to assume that true associations will be detected by single marker analysis, and this would suggest that the 3′ end of the MECP2 gene might be playing a role in vulnerability to autism. Given the concentration of Liu and Francke’s regulatory regions in this area, it is entirely possible that one or more functional polymorphisms in and around this end of the gene might be of importance. Furthermore, the observation of the functional importance of microRNA recognition elements located in the 3′ UTR of the long transcript of the gene, which is the predominant form in brain, adds support to this interpretation (Klein et al. 2007).
Further studies using other large autistic samples with clear inclusion criteria would help to clarify these findings. Overall, our results suggest that MECP2 may well be a predisposing factor for autism. Certainly the gene has an impressive list of credentials for involvement in behavioural disorders. An intriguing recent report suggests, for example, that MECP2 interacts with ATRX (X linked) and that point mutations within the methylated DNA-binding domain of MECP2 can disrupt this interaction in vitro and in vivo (Nan et al. 2007). ATRX has been implicated in a range of X-linked mental retardation syndromes (OMIM 300032). Furthermore, the involvement of the key neuronal growth factor gene, Bdnf, as a target (Chen et al. 2003) further points to MECP2’s potential involvement in important aspects of brain development. Most recently, the introduction of a conditional MECP2 allele with only 50% activity into mice (MECP2Flox/y) was observed to result in a spectrum of abnormalities including learning and motor deficiencies and altered social behaviour, indicating that more subtle changes in MECP2 activity may have a significant behavioural impact (Samaco et al. 2008).
Clearly, autism is a disorder with complex genetic aetiology, and MECP2 is a gene with wide-ranging interactions and mechanisms of effect. It is vital, therefore, that we incorporate sufficient complexity into our models for understanding in detail how MECP2 and its gene partners might be involved in vulnerability to autism.
Acknowledgments
C.S.L. was funded by a Medical Research Council (MRC) PhD studentship. Part of this work was funded by the National Institute for Health Research (NIHR) through the Biomedical Research Centre at the South London and Maudsley NHS Trust and Institute of Psychiatry. The authors also thank Ben Williams for his assistance with bioinformatics.
References
- Abdul-Rahman OA, Hudgins L. The diagnostic utility of a genetics evaluation in children with pervasive developmental disorders. Genet Med. 2006;8:50–54. doi: 10.1097/01.gim.0000195304.45116.96. [DOI] [PubMed] [Google Scholar]
- Beyer KS, Blasi F, Bacchelli E, Klauck SM, Maestrini E, Poustka A. Mutation analysis of the coding sequence of the MECP2 gene in infantile autism. Hum Genet. 2002;111:305–309. doi: 10.1007/s00439-002-0786-3. [DOI] [PubMed] [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Coy JF, Sedlacek Z, Bächner D, Delius H, Poustka A. A complex pattern of evolutionary conservation and alternative polyadenylation within the long 3″-untranslated region of the methyl–CpG-binding protein 2 gene (MeCP2) suggests a regulatory role in gene expression. Hum Mol Genet. 1999;8:1253–1262. doi: 10.1093/hmg/8.7.1253. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Joober R, Dube MP, St Onge J, Bonnel A, Gariepy D, Laurent S, Najafee R, Lacasse H, St-Charles L, Fombonne E, Mottron L, Rouleau GA. Autism spectrum disorders associated with X chromosome markers in French-Canadian males. Mol Psychiatry. 2006;11:206–213. doi: 10.1038/sj.mp.4001756. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, AGRE Steering Committee The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R, Ford T, Richards H, Gatward R, Meltzer H. The development and well-being assessment: description and initial validation of an integrated assessment of child and adolescent psychopathology. J Child Psychol Psychiatry. 2000;41:645–655. [PubMed] [Google Scholar]
- Hagberg B. Rett’s syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand. 1985;74:405–408. doi: 10.1111/j.1651-2227.1985.tb10993.x. [DOI] [PubMed] [Google Scholar]
- Hagberg B, Hagberg G. Rett syndrome: epidemiology and geographical variability. Eur Child Adolesc Psychiatry. 1997;6:5–7. [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by CREB-induced microRNA. Nat Neurosci. 2007;12:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Lam CW, Yeung WL, Ko CH, Poon PM, Tong SF, Chan KY, Lo IFM, Chan LYS, Hui J, Wong V, Pang CP, Lo YMD, Fok TF. Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet. 2000;37:E41. doi: 10.1136/jmg.37.12.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM. The odyssey of MeCP2 and parental imprinting. Epigenetics. 2007;2:5–10. doi: 10.4161/epi.2.1.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard H, Bower C, English D. The prevalence and incidence of Rett syndrome in Australia. Eur Child Adolesc Psychiatry. 1997;6:8–10. [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Liu J, Francke U. Identification of cis-regulatory elements for MECP2 expression. Hum Mol Genet. 2006;15:1769–1782. doi: 10.1093/hmg/ddl099. [DOI] [PubMed] [Google Scholar]
- Loat CS, Asbury K, Galsworthy MJ, Plomin R, Craig IW. X inactivation as a source of behavioural differences in monozygotic female twins. Twin Res. 2004;7:54–61. doi: 10.1375/13690520460741444. [DOI] [PubMed] [Google Scholar]
- Lobo-Menendez F, Sossey-Alaoui K, Bell JM, Copeland-Yates SA, Plank SM, Sanford SO, Skinner C, Simensen RJ, Schroer RJ, Michaelis RC. Absence of MeCP2 mutations in patients from the South Carolina autism project. Am J Med Genet B Neuropsychiatr Genet. 2003;117:97–101. doi: 10.1002/ajmg.b.10016. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic zInterview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J. Detection of rare major genes in lipid levels. Hum Genet. 1979;51:79–91. doi: 10.1007/BF00278296. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Nagarajan RP, Braunschweig D, Lasalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet. 2004;13:629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, Lasalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Fryer JD, Ren J, Fyffe S, Chao H, Sun Y, Greer JJ, Zoghbi HY, New JL. A partial loss of function allele of methyl-CpG-binding protein predicts a human neurodevelopmental syndrome. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn062. Advanced Access 4 March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama A, Cook EH, Jr, Feng J, Glanzmann C, Yan J, Craddock N, Jones IR, Goldman D, Heston LL, Sommer SS. MECP2 structural and 3′-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. Am J Med Genet B Neuropsychiatr Genet. 2004;128:50–53. doi: 10.1002/ajmg.b.30016. [DOI] [PubMed] [Google Scholar]
- Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, Lugtenberg D, Bienvenu T, Jensen LR, Geoz J, Moraine C, Marynen P, Fryns JP, Froyen G. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc’h P, Bienvenu T, Beldjord C, Chelly J, Barthelemy C, Muh JP, Andres C. No mutations in the coding region of the Rett syndrome gene MECP2 in 59 autistic patients. Eur J Hum Genet. 2001;9:556–558. doi: 10.1038/sj.ejhg.5200660. [DOI] [PubMed] [Google Scholar]