Abstract
Family, twin, and adoption studies provide convincing evidence for a genetic contribution to suicidal behavior. The heritability for suicidal behavior depends in part on the transmission of psychiatric disorders, such as mood disorders and substance use disorders, but is also partly independent of them. Three linkage studies using the attempted suicide phenotype in pedigrees with bipolar disorder, major depression, or alcoholism have provided consistent evidence that 2p11-12 harbors a susceptibility gene for attempted suicide. A microarray expression study using post-mortem brain samples has implicated a gene from the 2p11-12 candidate region, the trans-Golgi network protein 2 (TGOLN2) gene, as being consistently up-regulated in suicide cases as compared to controls. Here, we present a TGOLN2 case-control association study using nine single nucleotide polymorphisms (SNPs). These nine SNPs, which include seven tag SNPs and two coding SNPs, have been genotyped in 517 mood disorder subjects with a history of attempted suicide and 515 normal controls. Allelic and genotypic analyses of the case-control sample did not provide evidence for association with the attempted suicide phenotype. Eight of the nine SNPs provided supportive evidence for association (p-values ranging from 0.008–0.03) when we compared the attempted suicide cases with a history of alcoholism to the attempted suicide cases without a history of alcoholism. However, this association finding was not replicated in an independent sample. Taken together, these analyses do not provide support for the hypothesis that common genetic variation in TGOLN2 contributes significantly to the risk for attempted suicide in subjects with major mood disorders.
Keywords: suicidal behavior, bipolar disorder, major depression
Introduction
Suicidal behavior is a heterogeneous phenotype that includes attempted and completed suicide (for review, see Mann et al., 2009). Attempted and completed suicide are strongly associated with psychiatric disorders, especially mood disorders and substance use disorders. The majority of people who die by suicide have depression or bipolar disorder (Barraclough et al., 1974; Henriksson et al., 1993), with one study finding that the risk of suicide in those with severe depression is 78-fold higher than in those without it (Hagnell et al., 1981). Alcoholism has also been shown to increase the risk of suicide (Murphy and Wetzel, 1990), as have other substance use disorders (Fowler et al., 1986), and many who die in this way are found to be intoxicated at the time of death. Interestingly, individuals with both major depression and substance use disorders are at significantly higher risk for suicidal behavior than those with major depression and no substance use disorders (Davis et al., 2005).
Family, twin, and adoption studies provide strong evidence for a genetic component to suicidal behavior, with a heritability estimate of 30–50% for the suicidality phenotype (for review, see Brent and Mann, 2005; Brezo et al., 2008; Mann et al., 2009). The heritability appears to depend in part on psychiatric disorders, such as mood disorders and substance abuse and, importantly, to also be partly independent of them. This independent factor has been hypothesized to influence personality traits, such as impulsive-aggression (Brent et al., 2003; Melhem et al., 2007), with individuals having a psychiatric disorder and a tendency towards impulsive aggression being at greatest risk for suicidal behavior.
In the past, studies of the biologic basis for suicidality have focused mostly on the serotonergic system because of diverse evidence such as the finding of lowered levels of the serotonin metabolite 5-hydroxyindolacetic acid (5-HIAA) in the cerebrospinal fluid (CSF) of patients who attempted suicide, especially those who used violent means (Asberg et al., 1976). The most studied suicidal behavior candidate genes from the serotonergic system are the tryptophan hydroxylase genes (TPH1 and TPH2) and the serotonin transporter. However, association studies with suicidal behavior and these genes have been mixed, with some showing evidence for association while others showed no evidence for association (for review, see Brezo et al., 2008)
Four genome-wide linkage studies have been conducted using attempted suicide as a phenotype in pedigrees with alcoholism, bipolar disorder, and major depression. Three of these provided compelling evidence that the 2p11-12 candidate region contains a genetic risk factor for suicidal behavior (Hesselbrock et al., 2004, Zubenko et al., 2004, Willour et al., 2007). In addition, two of the studies implicated the 6q25-26 region (Cheng et al., 2006, Willour et al., 2007).
One of the genes in the 2p11-12 candidate region is the trans-Golgi network protein 2 (TGOLN2) gene, which encodes a transmembrane protein primarily localized to the trans-Golgi network (TGN), an important protein sorting station in the cell (Ponnambalam et al., 1996). TGOLN2 is of particular interest because it was identified in a microarray expression study as having a consistently up-regulated expression pattern in brains of suicide completers as compared to controls (Sequeira et al., 2006). We were interested in determining whether common variants in TGOLN2 confer risk for attempted suicide. Towards this end, we tested seven tag SNPs and two coding SNPs from the TGOLN2 region for association with attempted suicide in 517 cases (subjects with a major mood disorder and a history of attempted suicide) and 515 controls.
Materials and Methods
Sample
We selected our 517 cases from among the families originally ascertained as part of the Chicago, Hopkins, NIMH Intramural Program (CHIP) bipolar disorder study (Zandi et al., 2007), the Genetics of Recurrent Early-Onset Depression (GenRED) study (Levinson et al., 2003), or the National Institute of Mental Health (NIMH) Genetics Initiative Bipolar Disorder Collaborative study waves 1–4 (1997). The detailed description of the ascertainment and assessment protocols for each of these studies can be found in the initial study reports. All subjects signed IRB-approved written informed consent forms prior to enrolling in the studies.
The TGOLN2 study design focused on combining subjects from all three sample collections to maximize power, with one case selected per pedigree. The cases had a history of one or more suicide attempts and a major mood disorder (bipolar I disorder (BPI), bipolar II disorder with recurrent major depression (BPII), schizoaffective disorder, manic or bipolar type (SA/BP), or recurrent major depression (MDDR)). In all three samples, phenotype data were collected using the Diagnostic Interview for Genetic Studies (DIGS) and diagnoses were made using best-estimate procedures (Nurnberger et al., 1994). We also utilized the Bipolar Disorder Phenome Database (Potash et al. 2007), which has combined the CHIP and NIMH phenotypic datasets and subjected them to a variety of quality control measures. To reduce heterogeneity, we excluded all subjects who declared themselves as being non-white and/or not of European origin.
We selected 515 normal controls from the NIMH Genetics Initiative repository (http://nimhgenetics.org/). These control subjects, collected as part of the Molecular Genetics of Schizophrenia (MGS) Part 2 (Sanders et al., 2008), were recruited by Knowledge Networks, Inc. (Menlo Park, CA), a survey research company, from participants in a nationally-representative marketing panel recruited by random digit dialing methods. Briefly, control subjects signed an informed consent for DNA and clinical information to be used for any medical research, with full anonymization of samples. Each control subject completed an online questionnaire consisting of a lifetime version of the Composite International Diagnostic Interview-Short Form (CIDI-SF), which diagnoses common mood, anxiety and substance use disorders (Kessler et al., 1998), supplemented by questions about any history of schizophrenia, psychosis or bipolar disorder. The MGS study excluded controls who endorsed or failed to answer any of these latter questions or who were outliers in the number of total questionnaire items endorsed or not answered.
We further excluded control subjects 1) if they had a history of a major depressive episode, 2) if they met criteria for alcohol or substance dependence, or 3) if their age at interview was < 21. The CIDI-SF interview did not ask specifically about prior suicide attempts, but many subjects were asked if they thought frequently about death. We excluded all subjects answering in the affirmative to this question. It is possible that some of the 56.3% who were not asked the “death question” might be positive for attempted suicide, though this number should be small because ~89.5% of attempters have had a depressive episode or alcohol abuse or dependence (Suominen et al., 1996). Control subjects were matched with case subjects for race/ethnicity and sex (80% female).
SNP selection
We used the HapMap database (Phase II) to select the SNP markers required to capture the common genetic variation across the TGOLN2 gene (2003). Tag SNPs spanning the RefSeq TGOLN2 transcript (10.2 kb ± 10kb) were chosen using the Tagger program (de Bakker et al., 2005), which allows the user to select tag SNPs based on linkage disequilibrium (LD) parameters. We required an r2 of 0.8 and a minor allele frequency (MAF) of 0.05 in the CEU sample. Eight tag SNPs were initially chosen to adequately cover the region: rs11547160, rs7428, rs1061782, rs6547611, rs2292653, rs10460585, rs6733795, and rs3923229. We also genotyped two coding SNPs: rs1044973, which is a synonymous cSNP (G357G), and rs4247303, which is a nonsynonymous cSNP (R259W). These ten SNPs were genotyped using an ABI 7900HT and TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA, USA). Marker rs11547160, which was located in the neighboring gene, TCF7L1, with a MAF of 0.058, failed to produce discrete genotypic clusters and was removed from the study prior to analysis. Thus, we included a total of nine SNPs in our analysis.
Analytic Methods
We performed SNP quality control in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), examining the genotype data for minor allele frequency, missing data rates and Hardy-Weinberg Equilibrium (HWE) (Purcell et al., 2007). We also tested whether the missing data rates were differential by case-control status. We used the program Haploview (Barrett et al., 2005) to explore the extent of LD across the region, evaluating pair-wise r2 between SNPs and assessing the region for evidence of haplotype-block structure (Gabriel et al., 2002).
We tested for disease association with single SNPs using the assoc function in PLINK to perform a basic case-control allelic association test and the logistic function to assess association under an additive genotypic model. While the case and control samples were matched by sex, age at interview differed between the two sample sets – with average age at interview of 40.9 in the cases and 52.9 in the controls (t=13.0, p<0.001). Thus, we also included a term in the logistic regression analysis to adjust for age at interview. The inclusion of this term did not significantly affect our results, so this analysis is not presented.
Haplotypes have the ability to capture LD more informatively than individual SNPs, so testing for associations with haplotypes may be a more powerful approach. Therefore, we also used the sliding window specification in PLINK to test for evidence of haplotypic association. Variable length sliding windows were determined using SNPs ordered according to physical map locations obtained from the UCSC genome database March 2006 (http://genome.ucsc.edu/).
We were also interested in whether a history of alcoholism or substance abuse might have an effect on the genetic association in this gene. Therefore, we performed a secondary case-only analysis for each of these covariates, including in the analysis only cases with a history of suicide attempts and mood disorder and comparing the genotypes of subjects with the covariate (alcoholism or substance abuse) to the genotypes of subjects without the covariate. We utilized an additive genotypic model in a logistic regression analysis as described above. We then expanded the logistic regression equation to adjust for sex, age at interview, and study (NIMH, CHIP or GenRED). Controlling for these variables did not meaningfully affect the results and therefore the results of this analysis are not presented.
Power
The sample of 517 cases and 515 controls had 80% power to detect evidence of association for a locus of moderate effect (genotypic relative risk = 1.6) assuming an additive model, an attempted suicide rate of 5%, and α = 0.003 (0.05÷18, representing a Bonferroni correction for the nine markers and two genetic models tested--allelic and genotypic).
Replication Sample
We sought to replicate our case-only results in an independent sample of attempted suicide cases and normal controls. For this purpose, we selected a convenience sample of cases from two sources: NIMH Genetics Initiative Bipolar Disorder Collaborative study wave five cases that were included as part of the Genetics Association Information Network (GAIN) bipolar study (Smith et al., 2009) and additional NIMH bipolar cases that were included in the Translational Genomics (TGen) bipolar study (Smith et al., manuscript in progress). None of these subjects were included in our initial sample. All subjects were assessed using the DIGS and detailed clinical information was available. The subjects from both initiatives were genotyped genome-wide on the Affymetrix Genome-Wide Human SNP Array 6.0 (http://www.affymetrix.com/). Subject and SNP quality control measures were applied to both samples as outlined in the GAIN bipolar manuscript (Smith et al., 2009). HapMap II SNPs were imputed using the program BEAGLE for the GAIN and TGen samples separately (Browning & Browning, 2009). The imputed allelic dosages from each sample were then combined into a single file for a mega-analysis. The program EIGENSTRAT was used to identify the top 10 principal components to use in adjusting for population stratification (Price et al., 2006).
We focused our subsequent attention on the nine TGOLN2 SNPs that were included in the initial case-only analyses. We tested these nine SNPs for association in the replication sample, limiting the sample to cases with a history of suicide attempt and mood disorder and testing for an association by alcoholism status. The program mach2dat was utilized to perform logistic regression using the imputed SNP dosages (Li et al, 2009). We included terms in the model to adjust for the top 10 principal components. We also expanded the model to include terms to adjust for sex, age at interview, and study (GAIN or TGen). Controlling for these additional variables did not meaningfully affect the results, and therefore the results of this analysis are not presented.
Results
Our sample contained 243 cases from GenRED, 201 cases from NIMH-BP and 73 cases from CHIP for a total of 517 attempted suicide cases with a major mood disorder. Of these, 46.8% had MDDR, 47.2% BPI, 2.9% BPII and 3.1% SA/BP. SNP names, locations and quality control measures are presented in Table I. Briefly, the minor allele frequencies ranged from 8% to 49%, all SNPs were in HWE in the control sample (p=0.14–0.85) and the missing data rates ranged from 0.4% to 1.6%. None of the SNPs showed differential missingness by case-control status (p=0.12–1). The TGOLN2 chromosomal location, gene structure, SNP locations, and LD structure are depicted in Figure 1. Using our data, the nine SNPs include two haplotype blocks as defined by the “confidence intervals” approach (Gabriel et al., 2002). The first block includes rs7428–rs10460585 and covers the 3’ end of the TGOLN2 gene. The second block includes rs6733795-rs3923229 and covers the 5’ end of the TGOLN2 and its promoter region.
Table I.
Allelic and genotypic results for attempted suicide
| SNP description and quality control measures | Allelic Test | Genotypic Test | ||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chr. 2 Location (bp) |
Minor/ major allele |
Minor allele frequency |
HWE P-valuea |
Odds Ratiob |
P- valuec |
Odds Ratiod |
P- valuec |
| rs7428 | 85399001 | C/T | 0.3855 | 0.8511 | 1.02 | 0.8071 | 1.02 | 0.8083 |
| rs1061782 | 85400126 | C/T | 0.4499 | 0.5932 | 0.98 | 0.8280 | 0.98 | 0.8284 |
| rs6547611 | 85402150 | A/G | 0.3916 | 0.5187 | 0.96 | 0.6508 | 0.96 | 0.6521 |
| rs2292653 | 85405772 | G/A | 0.0833 | 0.1377 | 1.03 | 0.8522 | 1.03 | 0.8574 |
| rs1044973 | 85407295 | C/T | 0.4545 | 0.6551 | 0.98 | 0.8336 | 0.98 | 0.8343 |
| rs4247303 | 85407591 | G/A | 0.4931 | 0.2135 | 0.96 | 0.6581 | 0.96 | 0.6631 |
| rs10460585 | 85408773 | T/G | 0.3748 | 0.3963 | 0.98 | 0.8193 | 0.98 | 0.8203 |
| rs6733795 | 85413994 | G/A | 0.4086 | 0.2406 | 0.88 | 0.1731 | 0.89 | 0.1788 |
| rs3923229 | 85415683 | C/T | 0.2614 | 0.8215 | 0.94 | 0.5280 | 0.94 | 0.5171 |
HWE=Hardy-Weinberg Equilibrium, calculated using the control sample only
Odds ratios calculated for the minor allele
P-value not corrected for multiple testing
Odds ratio calculated for the minor allele under an additive model. The odds ratio presented is for the heterozygotes versus the common homozygotes. Under the additive model, the effect size for the rare homozygotes versus the common homozygotes is assumed to be twice that of the heterozygotes versus common homozygotes.
Figure 1.
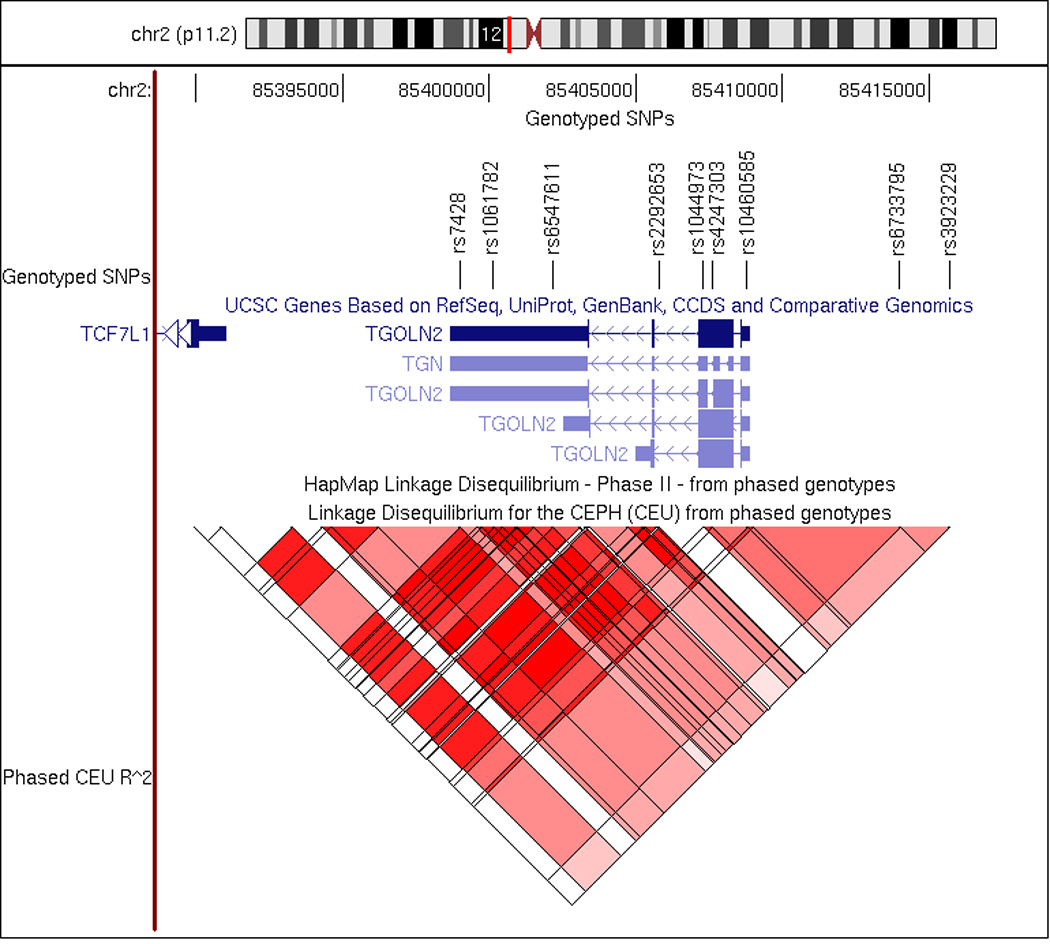
Location and structure of TGOLN2. Shown are the chromosomal location genotyped SNPs, gene structure and LD structure in the HapMap CEU population for TGOLN2.
We tested for evidence of association between the alleles and genotypes of the nine SNPs and the attempted suicide phenotype (Table I), but neither of these analyses provided supportive evidence for association. Haplotype analysis using a sliding window approach with a variable window size identified a six SNP haplotype with evidence for association with attempted suicide (p= 0.011) (Table II). However, this haplotype is very rare, occurring in only 1.9% of cases and 0.6% of controls, and the global test of association did not provide evidence for association (p=0.19).
Table II.
Results for the six SNP haplotype rs6547611|rs2292653|rs1044973|rs4247303|rs10460585|rs6733795
| Haplotype | Frequency Cases | Frequency Controls | χ2 | df | p-valueb |
|---|---|---|---|---|---|
| Globala | NA | NA | 9.90 | 7 | 0.1941 |
| AACGTG | 0.3096 | 0.3260 | 0.63 | 1 | 0.4262 |
| GACGGG | 0.0417 | 0.0470 | 0.34 | 1 | 0.5587 |
| GATGGG | 0.0354 | 0.0423 | 0.65 | 1 | 0.4186 |
| AACGTA | 0.0697 | 0.0559 | 1.64 | 1 | 0.1999 |
| AACGGA | 0.0168 | 0.0208 | 0.45 | 1 | 0.5040 |
| GACGGA | 0.0186 | 0.0060 | 6.55 | 1 | 0.0105 |
| GGTAGA | 0.0798 | 0.0768 | 0.07 | 1 | 0.7986 |
| GATAGA | 0.4285 | 0.4251 | 0.02 | 1 | 0.8780 |
Omnibus test for a trend across all possible haplotypes
P-values are not corrected for multiple testing
We also wanted to determine whether the presence of alcoholism influenced the evidence for association. To do this, we conducted a secondary analysis using a case-only approach under an additive genotypic model. We compared the genotypes from 182 attempted suicide cases with a history of alcoholism to the genotypes from 335 attempted suicide cases without a history of alcoholism. Under these conditions, eight of the nine SNPs provided at least nominal evidence for association (Table III). The most significant SNP was rs1044973 with a p-value of 0.008. We also conducted a comparable analysis using history of drug abuse and dependence and found no evidence for association with any of the nine SNPs (p> 0.1; data not shown).
Table III.
Genotypic results for attempted suicide cases with and without alcoholism
| SNP | Odds Ratioa | P-valueb |
|---|---|---|
| rs7428 | 0.75 | 0.0305 |
| rs1061782 | 1.37 | 0.0181 |
| rs6547611 | 1.33 | 0.0334 |
| rs2292653 | 0.83 | 0.4209 |
| rs1044973 | 1.42 | 0.0080 |
| rs4247303 | 1.35 | 0.0246 |
| rs10460585 | 1.40 | 0.0146 |
| rs6733795 | 1.39 | 0.0138 |
| rs3923229 | 1.42 | 0.0231 |
Odds ratio calculated for the minor allele under an additive model. The odds ratio presented is for the heterozygotes versus the common homozygotes. Under the additive model, the effect size for the rare homozygotes versus the common homozygotes is assumed to be twice that of the heterozygotes versus common homozygotes.
P-value not corrected for multiple testing
We next attempted to replicate the alcoholism finding in an independent sample of cases with mood disorder and suicide attempt. We utilized a case-only approach comparing the genotypes of 430 attempted suicide cases with a history of alcoholism to the genotypes of 335 attempted suicide cases without a history of alcoholism. None of the nine TGOLN2 SNPs tested showed statistically significant evidence of association (p>0.1; Table IV).
Table IV.
Replication of genotypic results for attempted suicide cases with and without alcoholism
| SNP | Odds Ratioa | P-valueb |
|---|---|---|
| rs7428c | 0.97 | 0.8062 |
| rs1061782c | 1.01 | 0.9151 |
| rs6547611c | 1.03 | 0.7623 |
| rs2292653 | 1.26 | 0.2817 |
| rs1044973 | 1.01 | 0.9107 |
| rs4247303 | 1.02 | 0.8213 |
| rs10460585 | 1.02 | 0.8727 |
| rs6733795 | 0.91 | 0.3890 |
| rs3923229 | 0.98 | 0.8509 |
Odds ratio calculated for the minor allele, adjusted for the top 10 principal components to control for population stratification.
P-value not corrected for multiple testing
SNP was genotyped in the replication sample. All other SNPs were imputed.
Discussion
The 2p11-12 candidate region, which contains the TGOLN2 gene, was initially identified in linkage analyses using the attempted suicide phenotype (Hesselbrock et al. 2004; Zubenko et al. 2004; Willour et al. 2007). However, we did not detect evidence of association for the TGOLN2 SNPs using either the allelic or the genotypic approach and the attempted suicide phenotype in this TGOLN2 association study. While we did identify modest evidence for association of eight SNPs from the TGOLN2 region when we compared attempted suicide cases with and without alcoholism, this finding was not replicated in an independent mood disorder sample set.
The TGOLN2 gene is widely expressed, with several alternatively spliced transcripts (each of which localizes to the genomic region genotyped in this experiment). Expression studies support a connection between TGOLN2 and suicidal behavior. Sequeira et al., conducted a microarray expression study that identified TGOLN2 as up-regulated in suicide completers as compared to controls (Sequeira et al., 2006), and the TGOLN2 gene was identified in the Stanley Medical Research Institute brain collection as being up-regulated in bipolar disorder samples from subjects who died by suicide (https://www.stanleygenomics.org/). Since our study was designed to identify only common variants conferring risk for the attempted suicide phenotype, we did not test for the impact of rare variants or copy number variants. It is possible that these expression results could be explained by these types of variants or by regulatory elements outside of the genomic region that we screened.
We selected our case samples primarily from the CHIP sample set and GenRED sample set, both of which show evidence for linkage to the 2p11-12 candidate region (Willour et al., 2007; data not shown), with the goal of increasing our ability to detect association signals in TGOLN2. We felt that combining these two samples, along with the NIMB-BP sample, was appropriate as the recruitment and ascertainment of each sample was similar. Furthermore, we compared the allele counts for each SNP across studies using a χ2 test with two degrees of freedom and found the counts to be very similar across studies (all p>0.3). However, it is still possible that combining subjects from multiple samples may have introduced small levels of heterogeneity into our study and reduced our ability to detect evidence for association with the attempted suicide phenotype.
The Psychiatric GWAS Consortium (PGC) is currently conducting meta-analyses for bipolar disorder and for major depression, and these sample sets along with others, such as those from schizophrenia, will be used to test for evidence of association with the attempted suicide phenotype (Psychiatric GWAS Consortium Coordinating Committee, 2009). These analyses may provide further evidence for association between genetic variants in the TGOLN2 gene and the risk for suicidal behavior.
Our findings need to be interpreted cautiously and in light of several study limitations. Our sample size was such that we would have missed associations with low relative risk in the main analyses (GRR < 1.6). Also, we did not include environmental risk factors in our study, such as parental abuse and early parental loss, which may interact with genetic risk factors and thereby increase the risk for suicidal behavior at this locus. For the alcoholism case-only analysis, the initial case samples were drawn from bipolar disorder and major depression pedigrees with evidence for linkage to the candidate region, while the replication sample was not. It should also be noted that our replication sample consisted of primarily bipolar I disorder cases, while the initial sample contained about half major depression and half bipolar disorder cases. However, in the initial sample the results were consistent across mood disorder categories (interaction p-value 0.05–0.70) and thus the inclusion of mainly BPI cases in the replication sample should be appropriate.
The 2p11-12 candidate region spans 29 Mb and is well-annotated, with 187 unique genes, per the UCSC Genome Browser’s RefSeq track (March 2006). While this number is more than can be thoroughly reviewed here, we note that several of these represent biologically plausible candidate genes for suicidal behavior susceptibility. Beyond TGOLN2, there is one additional strong functional candidate--TACR1. The TACR1 gene encodes the substance P receptor, which is of interest because antagonists of the receptor are reported to have antidepressant and anxiolytic properties (McLean, 2005), and Tacr1 knock-out mice are markedly less aggressive than their wild-type counterparts (De Felipe et al., 1998). Giegling et al., genotyped four SNPs from the 150 kb TACR1 gene in 167 suicide attempters, 92 suicide completers, and 312 controls and identified an association with reactive aggression (Giegling et al., 2007). Additional functional candidates include: 1) NPAS2, or neuronal pas-domain protein 2, a transcription factor expressed predominantly in brain. It controls circadian oscillation in the mammalian forebrain (Reick et al., 2001) and influences behavioral adaptability (Dudley et al., 2003); and 2) ADRA2B, the alpha-2B adrenergic receptor, which has a critical role in regulating neurotransmitter release from adrenergic neurons in the central nervous system.
To our knowledge, this is the first study to test whether common variants in the TGOLN2 gene are associated with suicidal behavior. While we were not able to detect any evidence of association for variants in TGOLN2 with suicidal behavior, our association results provided modest support for TGOLN2 influencing the risk for attempted suicide in combination with mood disorders and alcoholism. However, this finding was not replicated in an independent sample set. These analyses do not provide support for the hypothesis that common variants in the TGOLN2 gene increase the risk for attempted suicide. Analysis of the attempted suicide phenotype in densely genotyped sample sets might allow for the identification of loci influencing this phenotype on 2p11-12 and throughout the genome.
Acknowledgments
This work was supported by grants from the National Institute of Mental Health (MH079240 to V.L.W.) and the American Foundation for Suicide Prevention (V.L.W.). Drs. Willour and Potash were also supported by Margaret Price Investigatorships. Some DNA samples were prepared and distributed by Rutgers University under a contract from the NIMH. We are grateful to the many interviewers and diagnosticians who contributed to this project, and to the families who devoted their time and effort to the study.
Appendix
The Bipolar Disorder Phenome Group consists of Francis McMahon, Jo Steele, Justin Pearl, Layla Kassem, Victor Lopez from the Genetic Basis of Mood and Anxiety Disorders Unit, Mood and Anxiety Program, National Institute of Mental Health, National Institutes of Health, Bethesda, MD; James Potash, Dean MacKinnon, Erin Miller, Jennifer Toolan from the Department of Psychiatry, Johns Hopkins School of Medicine, Baltimore, MD; Peter Zandi from the Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD; Thomas Schulze from the Division of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Ruprecht-Karls-University of Heidelberg, Mannheim, Germany; Evaristus Nwulia from the Department of Psychiatry, Howard University Hospital, Washington, DC; Sylvia Simpson from the Department of Psychiatry, University of Colorado at Denver, Denver, CO.
Acknowledgement for Bipolar Disorder Biomaterials and Clinical Data
Data and biomaterials were collected in four projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1991–98, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, U01 MH46282, John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., and Elizabeth Bowman, M.D.; Washington University, St. Louis, MO, U01 MH46280, Theodore Reich, M.D., Allison Goate, Ph.D., and John Rice, Ph.D.; Johns Hopkins University, Baltimore, MD U01 MH46274, J. Raymond DePaulo, Jr., M.D., Sylvia Simpson, M.D., MPH, and Colin Stine, Ph.D.; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot Gershon, M.D., Diane Kazuba, B.A., and Elizabeth Maxwell, M.S.W.
Data and biomaterials were collected as part of ten projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1999–03, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Wayne State University), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, M.S., Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D., Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D, Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D. ; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini M.D.,Ph.D. ; University of California at Irvine, CA, R01 MH60068, William Byerley M.D., and Mark Vawter M.D. ; University of Iowa, IA, R01 MH059548, William Coryell M.D., and Raymond Crowe M.D. ; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner Ph.D., Francis McMahon M.D., Chunyu Liu Ph.D., Alan Sanders M.D., Maria Caserta, Steven Dinwiddie M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, PsyD, Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.
Acknowledgement for Depression Sample Biomaterials and Clinical Data
Data and biomaterials were collected in six projects that participated in the National Institute of Mental Health (NIMH) Genetics of Recurrent Early-Onset Depression (GenRED) project. From 1999–2003, the Principal Investigators and Co-Investigators were: New York State Psychiatric Institute, New York, NY, R01 MH060912, Myrna M. Weissman, Ph.D. and James K. Knowles, M.D., Ph.D.; University of Pittsburgh, Pittsburgh, PA, R01 MH060866, George S. Zubenko, M.D., Ph.D. and Wendy N. Zubenko, Ed.D., R.N., C.S.; Johns Hopkins University, Baltimore, R01 MH059552, J. Raymond DePaulo, M.D., Melvin G. McInnis, M.D. and Dean MacKinnon, M.D.; University of Pennsylvania, Philadelphia, PA, RO1 MH61686, Douglas F. Levinson, M.D. (GenRED coordinator), Madeleine M. Gladis, Ph.D., Kathleen Murphy-Eberenz, Ph.D. and Peter Holmans, Ph.D. (University of Wales College of Medicine); University of Iowa, Iowa City, IW, R01 MH059542, Raymond R. Crowe, M.D. and William H. Coryell, M.D.; Rush University Medical Center, Chicago, IL, R01 MH059541-05, William A. Scheftner, M.D. Rush-Presbyterian.
Acknowledgement for Control Sample Biomaterials and Clinical Data
Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials are being collected by the "Molecular Genetics of Schizophrenia II" (MGS-2) collaboration. The investigators and coinvestigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA,MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA,MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA,MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY,MH59586, Jeremy Silverman, Ph.D. (PI).
In addition, cord blood samples were collected by V L Nimgaonkar's group at the University of Pittsburgh, as part of a multi-institutional collaborative research project with J Smoller, MD DSc and P Sklar, MD PhD (Massachusetts General Hospital) (grant MH 63420).
References
- Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: a preliminary report. Am J Med Genet. 1997;74:227–237. doi: 10.1002/(sici)1096-8628(19970531)74:3<227::aid-ajmg1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Asberg M, Traskman L, Thoren P. 5-HIAA in the cerebrospinal fluid. A biochemical suicide predictor? Arch Gen Psychiatry. 1976;33:1193–1197. doi: 10.1001/archpsyc.1976.01770100055005. [DOI] [PubMed] [Google Scholar]
- Barraclough B, Bunch J, Nelson B, Sainsbury P. A hundred cases of suicide: clinical aspects. Br J Psychiatry. 1974;125:355–373. doi: 10.1192/bjp.125.4.355. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Brent DA, Mann JJ. Family genetic studies, suicide, and suicidal behavior. Am J Med Genet C Semin Med Genet. 2005;133:13–24. doi: 10.1002/ajmg.c.30042. [DOI] [PubMed] [Google Scholar]
- Brent DA, Oquendo M, Birmaher B, Greenhill L, Kolko D, Stanley B, Zelazny J, Brodsky B, Firinciogullari S, Ellis SP, Mann JJ. Peripubertal suicide attempts in offspring of suicide attempters with siblings concordant for suicidal behavior. Am J Psychiatry. 2003;160:1486–1493. doi: 10.1176/appi.ajp.160.8.1486. [DOI] [PubMed] [Google Scholar]
- Brezo J, Klempan T, Turecki G. The Genetics of Suicide: A Critical Review of Molecular Studies. Psychiatr Clin North Am. 2008;31(2):179–203. doi: 10.1016/j.psc.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Juo SH, Loth JE, Nee J, Iossifov I, Blumenthal R, Sharpe L, Kanyas K, Lerer B, Lilliston B, Smith M, Trautman K, Gilliam TC, Endicott J, Baron M. Genome-wide linkage scan in a large bipolar disorder sample from the National Institute of Mental Health genetics initiative suggests putative loci for bipolar disorder, psychosis, suicide, and panic disorder. Mol Psychiatry. 2006;11:252–260. doi: 10.1038/sj.mp.4001778. [DOI] [PubMed] [Google Scholar]
- Davis LL, Rush JA, Wisniewski SR, Rice K, Cassano P, Jewell ME, Biggs MM, Shores-Wilson K, Balasubramani GK, Husain MM, Quitkin FM, McGrath PJ. Substance use disorder comorbidity in major depressive disorder: an exploratory analysis of the Sequenced Treatment Alternatives to Relieve Depression cohort. Compr Psychiatry. 2005;46(2):81–89. doi: 10.1016/j.comppsych.2004.07.025. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- De Felipe C, Herrero JF, O'Brien JA, Palmer JA, Doyle CA, Smith AJ, Laird JM, Belmonte C, Cervero F, Hunt SP. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature. 1998;392:394–397. doi: 10.1038/32904. [DOI] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- Fowler RC, Rich CL, Young D. San Diego Suicide Study. II. Substance abuse in young cases. Arch Gen Psychiatry. 1986;43:962–965. doi: 10.1001/archpsyc.1986.01800100056008. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Giegling I, Rujescu D, Mandelli L, Schneider B, Hartmann AM, Schnabel A, Maurer K, De Ronchi D, Moeller H-J, m Serretti A. Tachykinin receptor 1 variants associated with aggression in suicidal behavior. Am J Med Genet. 2007;144B:757–761. doi: 10.1002/ajmg.b.30506. [DOI] [PubMed] [Google Scholar]
- Hagnell O, Lanke J, Rorsman B. Suicide rates in the Lundby study: mental illness as a risk factor for suicide. Neuropsychobiology. 1981;7:248–253. doi: 10.1159/000117857. [DOI] [PubMed] [Google Scholar]
- Henriksson MM, Aro HM, Marttunen MJ, Heikkinen ME, Isometsa ET, Kuoppasalmi KI, Lonnqvist JK. Mental disorders and comorbidity in suicide. Am J Psychiatry. 1993;150:935–940. doi: 10.1176/ajp.150.6.935. [DOI] [PubMed] [Google Scholar]
- Hesselbrock V, Dick D, Hesselbrock M, Foroud T, Schuckit M, Edenberg H, Bucholz K, Kramer J, Reich T, Goate A, Bierut L, Rice JP, Nurnberger JI., Jr The search for genetic risk factors associated with suicidal behavior. Alcohol Clin Exp Res. 2004;28:70S–76S. doi: 10.1097/01.alc.0000127416.92128.b0. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Andrews G, Mroczek D, Ustun TB, Wittchen H-U. The World Health Organization Composite International Diagnostic Interview Short Form (CIDI-SF) Int J Methods in Psychiatr Res. 1998;7(4):171–185. [Google Scholar]
- Levinson DF, Zubenko GS, Crowe RR, DePaulo RJ, Scheftner WS, Weissman MM, Holmans P, Zubenko WN, Boutelle S, Murphy-Eberenz K, MacKinnon D, McInnis MG, Marta DH, Adams P, Sassoon S, Knowles JA, Thomas J, Chellis J. Genetics of recurrent early-onset depression (GenRED): design and preliminary clinical characteristics of a repository sample for genetic linkage studies. Am J Med Genet B Neuropsychiatr Genet. 2003;119:118–130. doi: 10.1002/ajmg.b.20009. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Sanna S, Abecasis GR. Genotype Imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, Currier D, Dougherty DM, Haghighi F, Hodge SE, Kleinman J, Lehner T, McMahon F, Mościcki EK, Oquendo MA, Pandey GN, Pearson J, Stanley B, Terwilliger J, Wenzel A. Candidate Endophenotypes for Genetic Studies of Suicidal Behavior. Biol Psychiatry. 2009;65:556–563. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean S. Do substance P and the NK1 receptor have a role in depression and anxiety? Curr Pharm Des. 2005;11:1529–1547. doi: 10.2174/1381612053764779. [DOI] [PubMed] [Google Scholar]
- Melhem NM, Brent DA, Ziegler M, Iyengar S, Kolko D, Oquendo M, Birmaher B, Burke A, Zelazny J, Stanley B, Mann JJ. Familial pathways to early-onset suicidal behavior: familial and individual antecedents of suicidal behavior. Am J Psychiatry. 2007;164(9):1364–1370. doi: 10.1176/appi.ajp.2007.06091522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE, Wetzel RD. The lifetime risk of suicide in alcoholism. Arch Gen Psychiatry. 1990;47:383–392. doi: 10.1001/archpsyc.1990.01810160083012. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S, Girotti M, Yaspo ML, Owen CE, Perry AC, Suganuma T, Nilsson T, Fried M, Banting G, Warren G. Primate homologues of rat TGN38: primary structure, expression and functional implications. J Cell Sci. 1996;109(Pt 3):675–685. doi: 10.1242/jcs.109.3.675. [DOI] [PubMed] [Google Scholar]
- Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, Schulze TG, Kassem L, Simpson SG, Lopez V, MacKinnon DF, McMahon FJ NIMH Genetics Initiative Bipolar Disorder Consortium. The bipolar disorder phenome database: a resource for genetic studies. Am J Psychiatry. 2007;8:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Coordinating Committee. Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, Lehner T, Levinson DF, Moran A, Sklar P, Sullivan PF. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;3:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165(4):497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Gwadry FG, Ffrench-Mullen JM, Canetti L, Gingras Y, Casero RA, Jr, Rouleau G, Benkelfat C, Turecki G. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Arch Gen Psychiatry. 2006;63:35–48. doi: 10.1001/archpsyc.63.1.35. [DOI] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, Eskin E, Foroud T, Gershon E, Greenwood TA, Hipolito M, Koller DL, Lawson WB, Liu C, Lohoff F, McInnis MG, McMahon FJ, Mirel DB, Murray SS, Nievergelt C, Nurnberger J, Nwulia EA, Paschall J, Potash JB, Rice J, Schulze TG, Scheftner W, Panganiban C, Zaitlen N, Zandi PP, Zöllner S, Schork NJ, Kelsoe JR. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.43. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suominen K, Henriksson M, Suokas J, Isometsa E, Ostamo A, Lonnqvist J. Mental disorders and comorbidity in attempted suicide. Acta Psychiatr Scand. 1996;94:234–240. doi: 10.1111/j.1600-0447.1996.tb09855.x. [DOI] [PubMed] [Google Scholar]
- Willour VL, Zandi PP, Badner JA, Steele J, Miao K, Lopez V, Mackinnon DF, Mondimore FM, Schweizer B, McInnis MG, Miller EB, Depaulo JR, Jr, Gershon ES, McMahon FJ, Potash JB. Attempted Suicide in Bipolar Disorder Pedigrees: Evidence for Linkage to 2p12. Biol Psychiatry. 2007;61(5):725–727. doi: 10.1016/j.biopsych.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Zandi PP, Badner JA, Steele J, Willour VL, Miao K, MacKinnon DF, Mondimore FM, Schweizer B, McInnis MG, DePaulo JR, Jr, Gershon E, McMahon FJ, Potash JB. Genome-wide linkage scan of 98 bipolar pedigrees and analysis of clinical covariates. Mol Psychiatry. 2007;12(7):630–639. doi: 10.1038/sj.mp.4002027. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Maher BS, Hughes HB, 3rd, Zubenko WN, Scott Stiffler J, Marazita ML. Genome-wide linkage survey for genetic loci that affect the risk of suicide attempts in families with recurrent, early-onset, major depression. Am J Med Genet B Neuropsychiatr Genet. 2004;129:47–54. doi: 10.1002/ajmg.b.30092. [DOI] [PubMed] [Google Scholar]


