Abstract
Gout is a common and very painful inflammatory arthritis caused by hyperuricaemia. This Review provides an update on the genetics of hyperuricaemia and gout, including findings from genome-wide association studies. Most of the genes that associated with serum uric acid levels or gout are involved in the renal urate-transport system. For example, the urate transporter genes SLC2A9, ABCG2 and SLC22A12 modulate serum uric acid levels and gout risk. The net balance between renal urate absorption and secretion is a major determinant of serum uric acid concentration and loss-of-function mutations in SLC2A9 and SLC22A12 cause hereditary hypouricaemia due to reduced urate absorption and unopposed urate secretion. However, the variance in serum uric acid explained by genetic variants is small and their clinical utility for gout risk prediction seems limited because serum uric acid levels effectively predict gout risk. Urate-associated genes and genetically determined serum uric acid levels were largely unassociated with cardiovascular–metabolic outcomes, challenging the hypothesis of a causal role of serum uric acid in the development of cardiovascular disease. Strong pharmacogenetic associations between HLA-B*5801 alleles and severe allopurinol-hypersensitivity reactions were shown in Asian and European populations. Genetic testing for HLA-B*5801 alleles could be used to predict these potentially fatal adverse effects.
Introduction
Gout, the most common inflammatory arthritis, is caused by hyperuricaemia and leads to substantial morbidity associated with excruciating pain.1,2 The disease affects ~8.3 million people in the USA1 and is a substantial socioeconomic burden.3 Gout is strongly associated with metabolic syndrome,4 myocardial infarction,5–7 diabetes8 and premature death.7
Thomas Sydenham recognized the familial nature of hyperuricaemia and gout in the 17th century.9 However, until the past decade, knowledge of the roles of specific genes in the pathogenesis of gout was limited to those that are associated with rare monogenic metabolic and renal disorders (Table 1).10–22 Mutations in genes that encode enzymes involved in purine syn thesis and interconversion lead to overproduction of uric acid and are associated with familial disorders such as glycogen storage diseases, which are charac terized by excessive cell death and ATP degradation.12–14 By contrast, mutations that lead to reduced excretion of urate are associated with hereditary renal disorders, such as medul lary cystic kidney disease type 1 and 2.18–20 Monogenic dis orders of purine pathways usually present in childhood or early adulthood and are often associated with additional non gouty clinical features.23
Table 1.
Mendelian syndromes associated with hyperuricaemia and gout
| Syndrome | Gene | Chromosome | Inheritance | Phenotype |
|---|---|---|---|---|
| Congenital errors of purine metabolism | ||||
| Hypoxanthine guanine phosphoribosyl transferase-related disease9 | HPRT1 | Xq26.2–q26.3 | XD | Neurological dysfunction, hyperuricaemia, gout |
| Phosphoribosyl pyrophosphatase synthetase-related disease22 | PRPS1 | Xq22.3 | XD | Hyperuricaemia, gout, neurological impairment |
| Excessive cell death and urate generation | ||||
| Glycogen storage disease-Ia12 | G6PC | 17q21.31 | AR | Growth retardation, lactic acidosis, hypoglycaemia, hepatomegaly, hyperuricaemia, gout |
| Glycogen storage disease-Ib12,13 | SLC37A4 | 11q23.3 | AR | Growth retardation, lactic acidosis, hypoglycaemia, hepatomegaly, hyperuricaemia, gout |
| Glycogen storage disease-III12,13 | AGL | 1q21.2 | AR | Early-onset hyperuricaemia, gout |
| Glycogen storage disease-V12 | PYGM | 11q13.1 | AR | Early-onset hyperuricaemia, gout |
| Glycogen storage disease-VII12,14 | PFKM | 2q13.11 | AR | Early-onset hyperuricaemia, gout |
| Late-onset carnitine palmitoyltransferase II deficiency116 | CPT2 | 1p32.3 | AR | Rhabdomyolysis, myoglobinuria, hyperuricaemia, gout |
| Myoadenylate deaminase deficiency15 | AMPD1 | 1p13.2 | AR or AD | Myopathy, hyperuricaemia, gout |
| Short chain, acyl-CoA dehydrogenase deficiency16 | ACADS | 12q24.31 | AR | Metabolic acidosis, neurological impairment, myopathy, hyperuricaemia, gout |
| Fructose-1-phosphate aldolase deficiency17 | ALDOB | 9q31.1 | AR or AD | Fructose intolerance, liver failure, renal tubulopathy, growth retardation, hyperuricaemia, gout |
| Reduced renal excretion of uric acid | ||||
| Medullary cystic kidney disease, type 118 | Unknown | 1q21 | AD | Variable penetrance, renal dysfunction, hypertension, gout |
| Medullary cystic kidney disease, type 219,20 | UMOD | 16p12.3 | AD or AR | Progressive renal dysfunction, variable hyperuricaemia, early-onset gout |
| Familial juvenile hyperuricemic nephropathy21 | UMOD | 16p12.3 | AD | Progressive renal dysfunction, variable hyperuricaemia, early-onset gout |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; XD, X-linked dominant.
Disproving the early contention that familial gout might be due to monogenic influences, studies of twins and families have shown a polygenic mode of inheritance for both hyperuricaemia and the fractional excretion of urate (FEua) by the kidneys.24 In one study of twins, the heritability of the renal clearance of urate was ~60%, whereas the estimated heritability of FEua was ~87%.25 Other studies have shown that serum urate levels have a substantial heritable component (~40%).26 The overall pattern of inheritance is best explained by a complex model incorporating interactions between more than one major gene, several modifying genes and environmental factors.26 This conclusion is supported by findings from the Framingham Heart Study,27 population studies of Pacific Islanders,23 and genome-wide association studies (GWAS) that have identified multiple novel genetic variants that associate with serum uric acid levels.23,28
Over the past 7 years, GWAS, replication studies and meta-analyses have led to a remarkable increase in our knowledge of common genetic variants that predispose to hyperuricaemia and gout (Figure 1). The discovery of these novel genes has substantially increased our understanding of the role of renal urate transporters in the pathogenesis of these disorders. In this Review, we describe the associations between common genetic variants, serum uric acid levels and gout as well as the role of these variants in hyperuricaemia and gout patho genesis. We also briefly discuss the association between gout, cardiovascular disease and metabolic syndrome and the pharmacogenetic associations between HLA-B*5801 and severe allopurinol-hypersensitivity reactions.
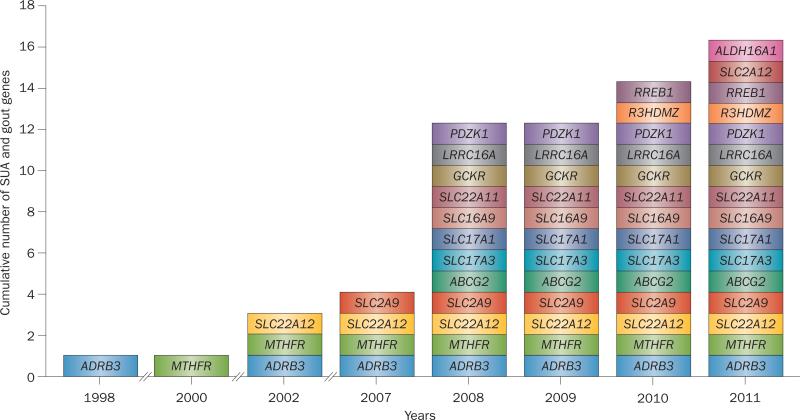
Figure 1.
Genetic variants implicated in the pathogenesis of hyperuricaemia or gout. Discovery timeline showing cumulative number of genes discovered from 2008–2011. Abbreviation: SUA, serum uric acid.
Genetic associations
The majority of the novel genes that have associated with hyperuricaemia or gout in GWAS encode proteins that are involved in the renal urate-transport system (Figure 2). This finding is unsurprising because reduced renal excretion of urate is the cause of the disorders in up to 90% of cases.2 The group of related urate transporters that are expressed on the apical border of renal proximal tubules have been termed the ‘uric acid transportasome’ because of their interactive nature in regulating urate homeostasis (Figure 2).29,30 Components of the transporta some include glucose transporter type 9 (GLUT-9 also known as SLC2A9);31 urate anion transporter 1 (URAT1, also known as SLC22A12); the organic anion transporters, solute carrier family 22 member 6, member 8, member 11, and member 13 (SLC22A6, SLC22A8, SLC22A11 and SLC22A13, also known as OAT1, OAT3, OAT4, and ORCTL-3); multi-drug resistance-associated protein 4 (MRP4); sodium-coupled monocarboxylate transporter 1 and 2 (SLC5A8 and SLC5A12) and ATP-binding cassette subfamily G member 2 (ABCG2, also known as breast cancer resistance protein).23 Almost all of the apical absorptive and secretory transporters end with recognition motifs for binding to PDZ domain proteins, such as PDZK1. These scaffolding proteins tether the transporters in an apical complex.30 Notably, PDZK1 is also a genetic determinant of serum uric acid levels.32,33
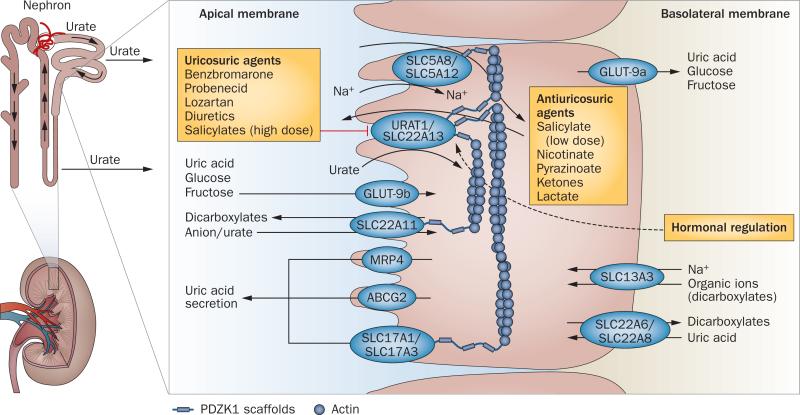
Figure 2.
The uric acid transportasome. Urate transporters in renal proximal tubules are involved in the secretion and reabsorption of urate. The balance between these processes determines the net proximal renal excretion. Urate secretion involves SLC22A6 and SLC22A8, which transport uric acid into the epithelial cell across the basolateral membrane, and URAT1, SLC22A13, SLC17A1, SLC17A3, MRP4 and ABCG2, which transport uric acid out of the epithelial cell across the apical membrane. Reabsorption of urate across the apical membrane involves the urate-anion exchangers URAT1 and SLC22A13, which facilitate the entry of urate into the cell in exchange for monocarboxylates (transported into the cell by the sodium-dependent transporters SCL5A8 and SCL5A12) as well as SLC22A11, which exchanges urate and dicarboxylates (transported into the cell by SLC13A3). Antiuricosuric drugs can serve as the exchanging anion for URAT1 and, therefore, enhance urate transport. URAT1 is inhibited by uricosuric agents and might be regulated by hormones. The glucose transporter GLUT-9 also has an important role in reabsorption of urate; GLUT-9b transports uric acid across the apical membrane and GLUT-9a transports uric acid out of the epithelial cell across the basolateral membrane. The scaffolding protein, PDZK1, is involved in the assembly of a transport complex in the apical membrane.
SLC2A9
GLUT-9 is encoded by the SLC2A9 gene. Several GWAS identified genetic variants of SLC2A9 that were robustly associated with serum uric acid levels.34–38 In these studies, variation in SLC2A9 was the most statistically significant genetic determinant of serum urate; accounting for 3.4–8.8% of the variance in women and 0.5–2.0% of the variance in men.34–39 Loss-of-function mutations in the SLC2A9 gene cause renal hypouricaemia40,41 and SLC2A9 variants have been associated with case definitions of gout in white, Chinese and Polynesian populations,26,34,37,39,42 as well as with low FEua in German, British and Croatian cohorts (Table 2).31,34,35,43,44 These finding are consistent with the critical role of GLUT-9 in the reabsorption of filtered urate by proximal tubules.
Table 2.
SLC2A9, ABCG2 and SLC22A12 variants associated with serum uric acid levels, FeUA and gout
| Variant | Location | Phenotype | Populations |
|---|---|---|---|
| SLC2A9 (chromosome 4) | |||
| rs1014290 | Intron 3 | SUA, FeUA, gout | European ancestry34 |
| rs6449213 | Intron 4 | SUA, FeUA, gout | White,34–37,43 African American51,72 |
| rs16890979 | Exon 6 | SUA, gout | White,37,42,44 African American,72 Amish55 |
| rs734553 | Intron 6 | SUA, gout | White,32,39,44 Icelandic,13 African American72 |
| rs7442295 | Intron 6 | SUA, gout | White 35,38,39,44 |
| rs737267 | Intron 7 | SUA, FeUA, gout | European ancestry34,44 |
| rs6855911 | Intron 7 | SUA, gout | White,35,38,39,44 African American72 |
| rs13129697 | Intron 7 | SUA, gout | White,33,44 African American72 |
| rs2241480 | Intron 8 | SUA, gout | European ancestry72 |
| rs7663032 | Intron 9 | SUA, gout | African American,72 Croatian44 |
| rs3775948 | Intron 9 | SUA | Croatian,44 African American51 |
| rs16890979 | Intergenic | SUA, gout | White,37 Amish,55 Croatian,44 Pacific Islander,42 New Zealander42 |
| rs717615 | Intergenic | SUA | Croatian44 |
| rs6856396 | Intergenic | SUA | African American51 |
| rs10489070 | Intergenic | Gout | Amish55 |
| ABCG2 (chromosome 4) | |||
| rs2231137 | Exon 2 | SUA | Japanese63 |
| rs72552713 (Q126X) | Exon 4 | SUA, gout | Japanese63 |
| rs2231142 (Q141K) | Exon 5 | FeUA, SUA, gout | White,32,37,39,44,62 African,37,72 Chinese,60 Icelandic,13 Japanese,59,63 Pacific Islander,61 New Zealander31,61 |
| rs2199936 | Intergenic | SUA | White 32,33,44 |
| SLC22A12 (chromosome 11) | |||
| rs11231825 | Exon 1 | FeUA, SUA | Chinese,70 White,32,68 African American72 |
| rs3825016 | Exon 2 | FeUA | German68 |
| rs12800450 | Exon 2 | SUA | African American72 |
| rs161109885 | Intron 3 | SUA | Chinese73 |
| rs893006 | Intron 4 | SUA | Japanese,67 Chinese71 |
| rs1529909 | Intron 4 | FeUA, SUA | Korean74 |
| rs475688 | Intron 4 | Gout | Chinese,70 Solomon Islander70 |
| rs17300741 | Intron 4 | SUA | European32,75 |
| rs7932775 | Exon 8 | SUA, FeUA, gout | German,68 Chinese,70,73 Solomon Islander70 |
| rs505802 | Intergenic | SUA | European,32,44 African American72 |
| rs11602903 | Intergenic | FeUA, SUA | German,68 Chinese73 |
Abbreviations: FeUA, fractional excretion of urate; SUA, serum uric acid.
Studies using Xenopus oocytes have shown that GLUT-9 is a robust urate transporter34,31,40,41 that can be partially inhibited by uricosuric agents, such as probenecid, losartan, benzbromarone31,32,41 and tranilast,45 as well as by the glucose transporter inhibitor phloretin.46 The protein was reported to be a fructose transporter,47 and potentially a fructose–urate exchanger,31,34 but some studies have not confirmed that fructose is a substrate.41 GLUT-9 does not undergo cis-inhibition or trans-stimulation in response to pyrazinoate and related anions so does not seem to function as a urate–anion exchanger (D. B. Mount, unpublished data). More over, GLUT-9 is activated by membrane depolarization41 and, therefore, might function as a urate uniporter or electrogenic exchanger.
Two distinct N-terminal isoforms of human GLUT-9 have been identified: GLUT-9a (540 residues) and GLUT-9b (511 residues, also known as GLUT9ΔN).48 These isoforms are generated by alternative splicing of 5' ends and differ in membrane trafficking.48 In Madin–Darby canine kidney cells that were transfected with human GLUT-9, GLUT-9a trafficked to the basolateral membrane and GLUT-9b to the apical membrane.48 Leukocyte expression of GLUT-9b mRNA correlates more closely with serum uric acid levels than GLUT-9a mRNA expression.35 This finding suggests that GLUT-9b has a more substantial role in urate homeostasis than GLUT-9a. Protein and mRNA from both isoforms have been detected in human kidney,48 but detailed localization studies have not yet been carried out. However, expression at the basolateral membrane suggests that GLUT-9a is likely to function as the exit site for urate from proximal tubule cells, whereas GLUT-9b might transport urate into the proximal tubule cells across the apical membrane (Figure 2).
GLUT-9 is expressed in hepatocytes (both isoforms), chondrocytes (GLUT-9a),34,49 intestinal cells (GLUT-9a) and leukocytes (both isoforms), as well as in renal epithelial cells (both isoforms).48,50 Expression of GLUT-9 in articular cartilage chondrocytes suggests a possible role of SLC2A9 variants in the development of mono-sodium urate crystal-induced arthritis.34,49 Further understanding of the regulation of urate transporters at the tissue level might provide insight into the mechanisms of tophi formation, and perhaps crystal-induced joint inflammation.
The causal variant or variants within SLC2A9 for determination of serum uric acid concentration have not yet been identified. However, progress has been made in fine-mapping the GWAS signal.51 Considerable variation in coding sequence exists in SLC2A9 and at least 24 annotated nonsynonymous variants have been identified.52 With the exception of loss-of-function mutations associated with familial hypouricaemia,40,41,53,54 there has been no systematic characterization of the transport phenotypes of SLC2A9 coding variants. The overall contribution of coding sequence variation in SLC2A9 to urate homeostasis is not clear because of population heterogeneity and linkage disequilibrium (nonrandom association of alleles at two or more loci) within the gene. For example, the biochemically conservative Val253Ile variant associated strongly with serum uric acid levels in some,37,55 but not all, studies.34,38 However, the presence of both the Val253Ile allele and the common allele on the protective and susceptibility haplotypes in Pacific Islanders suggests that there is not a substantial role for the Val253Ile variant in gout susceptibility in this population.42 Similarly, the Arg265His variant was associated with hyperuricaemia56,57 and severe gout57,58 in some, but not all, populations.55
ABCG2
Associations between ABCG2 polymorphisms and uric acid levels have been consistently demonstrated in GWAS and replicated in gout cases from several populations (Table 2).37,39,59–62ABCG2 encodes ABCG2, a multifunctional transporter that belongs to the ATP-binding cassette family and mediates the efflux of various compounds in an ATP-dependent manner.63 ABCG2 is expressed in the brush border membrane of the proximal tubules of the kidney and has a role in the apical secretion of urate.62 The transporter is also abundantly expressed in the apical membrane of epithelial cells in the small intestine and in the liver, suggesting a possible role in the extrarenal excretion of uric acid.64 Interestingly, ABCG2 mRNA levels are upregulated by statins in the human hepatoma cell line, HepG2.65 Functional studies of the mechanism of this upregulation might provide data that could potentially lead to the development of improved therapies for gout.
In white, African and Asian populations, the strongest association between ABCG2 and gout involved the single nucleotide polymorphism (SNP) rs2231142 in exon 5,37 which causes a Glu141Lys amino acid substitution. A meta-analysis of GWAS data showed that the Glu141Lys polymorphism accounted for 0.57% of the variation in serum urate.32 Interestingly, the poly morphism had a larger effect on serum urate levels in men than in women.32 Functional studies of the Glu141Lys substitution have shown that it causes a 53% reduction in the rate of ABCG2-mediated urate transport compared with wild-type ABCG2.62 The associated reduction in urate secretion might lead to an imbalance between net secretion and absorption, favouring absorption and thus leading to hyperuricaemia.
An analysis of the Atherosclerosis Risk in Communities study showed that at least 10% of all gout cases in white individuals were attributable to the Gln141Lys causal variant.62 However, the frequency of the Gln141Lys risk allele has been reported to be as high as 32% in the Asian population.32,60 Although both Maoris and Pacific Islanders are known to have an increased risk of gout, the Gln141Lys variant is strongly associated with gout only in the western Polynesian population.61 This distribution might be the result of chance depletion of the allele during eastward migration, as the frequency of the Gln141Lys variant in white populations has been reported to be <12%.37,61
SLC22A12
The SLC22A12 gene encodes URAT1, an essential urate transporter in proximal tubules that has a role in the apical absorption of urate66 and is a target of both urico suric and antiuricosuric agents (Figure 2). Loss-of-function mutations in SLC22A12 have been identified in patients with familial renal hypouricaemia2,66 and several associations between SLC22A12 variants, serum uric acid concentrations and gout have been reported (Table 2).67–72 A Japanese study showed that an intron SNP of SLC22A12 (rs893006) was associated with serum uric acid levels in Japanese men (n = 326).67 Another study showed statistically significant associations between three different polymorphisms in the N-terminus of SLC22A12 (–788T>A in the promoter region, C258T in exon 1 and C426T in exon 2) and reduced FEua levels in a German population.68 The strongest association was found for the C426T mutation (P = 0.0002).68 Variations in SLC22A12 were also associated with gout in Mexican69 and Asian populations.70,71,73,74 The Wellcome Trust Case-Control Consortium (WTCCC) genome-wide scans did not show associations between known urate transporter genes, including SLC22A12, and serum urate levels.36 However, this lack of association might be attributable to poor tagging of the transporter genes by the gene array chip used by the consortium.28,36 A subsequent meta-analysis of 14 GWAS (including the WTCCC studies) did show a significant association between SLC22A12 and serum urate levels (P = 2 × 10–9).32
In populations with European ancestry, SCLA22A12 might only account for 0.13% of serum uric acid variance, and the observed association might be the result of close linkage disequilibrium with the SLC22A11 gene.33 However, a novel rare nonsynonymous variant of SLC22A12 with a large serum uric acid effect size (71.38 μmol/L) was identified as being associated with serum urate (P = 2.7 × 10–16), and the identification replicated, in a meta-analysis of data from GWAS of African American populations.72 The variant SNP, rs12800450, caused a glycine-to-tryptophan substitution at position 65 (Gly65Trp) and had a minor allele frequency of 1%.7214C-urate transport assays showed that the Gly65Trp mutant protein had reduced urate transport function compared with wild-type URAT1, suggesting that the mutation might lead to a reduction in urate absorption.72
Other reproducibly associated variants
Two large meta-analyses, each involving >28,000 participants, identified several other genome-wide genetic variants that are reproducibly associated with serum uric acid levels or gout (Table 3).32,33 The first, carried out in 2009, showed associations between nine common genetic variant loci and serum uric acid concentrations—the previously discussed variants SLC2A9 (P = 5.2 × 10–201), ABCG2 (P = 3.1 × 10–26) and SLC22A12 (P = 2.0 × 10–9), and six other variants: SLC17A1 (P = 3.0 × 10–14), SLC22A11 (P = 6.7 × 10–14), SLC16A9 (P = 1.1 × 10–8), GCKR (P = 1.4 × 10–9), LRRC16A (P = 8.5 × 10–9) and near PDZK1 (P = 2.7 × 10–9).32 In 2010, six of these loci (SLC2A9, ABCG2, SLC17A1, SLC22A11, CGKR and PDZK1) were confirmed in a meta-analysis of five population-based cohorts of the CHARGE consortium.33 Of the remaining three loci, SLC22A12 was not considered to be an independent locus because of its proximity to SLC2A11 and the genome-wide significance of SLC16A9 and LRRC16 was border line (P = 1.7 × 10–7 and 1.3 × 10–7 respectively).33 How ever, a UK population-based study (n = 7,795) later con firmed the association between SLC16A9, but not LRRC16, and serum uric acid concentrations.75 The CHARGE meta-analysis also identified the R3HDM2–INHBC region and RREB1 loci as having genome-wide significance with serum urate levels.33 Subsequently, a meta-analysis72 and a GWAS51 showed that 10 of the 11 loci that have been shown to influence serum urate concentration in individuals with European ancestry were significantly associated with serum uric acid or gout in African-American populations.
Table 3.
Other* genetic variants associated with serum uric acid levels and gout
| Variant | Location | Phenotype | Populations |
|---|---|---|---|
| SLC16A9 (chromosome 10) | |||
| rs12356193 | Intron 1 | SUA | European,32 Icelandic13 |
| SLC17A1 (chromosome 6) | |||
| rs1165196 | Exon 7 | SUA, gout | White,33 Icelandic,13 Japanese37,117 |
| rs1183201 | Intron 10 | SUA | European32 |
| rs1179086 | Intron 12 | Gout | Japanese117 |
| rs3757131 | Intron 12 | Gout | Japanese117 |
| rs11751616 | Intergenic | SUA | African American72 |
| rs2051541 | Intergenic | SUA, gout | European ancestry72 |
| SLC17A3 (chromosome 6) | |||
| rs1165205 | Intron 1 | SUA | White37 |
| SLC22A11 (chromosome 11) | |||
| rs17300741 | Intron 4 | SUA | European32 |
| rs10792443 | Intron 4 | SUA, gout | European ancestry72 |
| rs2078267 | Intron 6 | SUA, gout | White,33 Icelandic13 |
| GCKR (chromosome 2) | |||
| rs780094 | Intron 16 | SUA | European32,75 |
| rs780093 | Intron 17 | SUA, gout | White,33 Icelandic13 |
| rs814295 | Intron 17 | SUA, gout | African American72 |
| LRRC16A (chromosome 6) | |||
| rs9467527 | Intron 12 | SUA | African American72 |
| rs742132 | Intron 30 | SUA | European32,75 |
| PDZK1 (chromosome 1) | |||
| rs882211 | Intron 1 | SUA, gout | African American72 |
| rs12129861 | Intergenic | SUA | European,32,38 Icelandic13 |
| rs1967017 | Intergenic | SUA, gout | White33 |
| R3HDM2-INHBC region (chromosome 12) | |||
| rs1106766 | Intergenic | SUA | White,33 Icelandic13 |
| RREB1 (chromosome 6) | |||
| rs675209 | Intergenic | SUA | White,33 Icelandic,13 Croatian44 |
In addition to SLC2A9, ABCG2 and SLC22A12 variants (Table 2).
Abbreviation: SUA, serum uric acid.
A reproducible genome-wide association between the SLC17A3 loci, serum uric acid levels and gout has been shown in a white population.37SLC17A1 and SLC17A3 encode SLC17A1 (also known as NPT1) and SLC17A3 (also known as NPT4), respectively. These proteins are voltage-dependent, multispecific anion transporters that are expressed at the apical membrane of the proximal tubule and are thought to be involved in the apical secretion of urate.76,77,78 Other identified substrates for these transporters include p-amminohippuric acid, bumetanide and estrone sulphate.79 Coding polymorphisms in the SLC17A1 (Arg138Ala) and SLC17A3 genes (Val257Phe, Gly279Arg and Phe378Leu) that are associated with hyperuricaemia generate transport proteins with reduced function.76,77,80 The scaffolding protein, PDZK1-interacting protein 1, interacts with the protein products of SLC22A12 (which encodes URAT1), SLC22A13, SLC5A8, SLC5A12, SLC22A11, SLC17A1 andSLC17A3.81 Thus, PDZK1 might link the transporter proteins URAT1 (responsible for urate reabsorption) and SLC17A1 (responsible for secretion) into a complex that could potentially regulate urinary urate excretion (Figure 2).41,81 PDZK1 variants that are associated with gout and serum uric acid levels might affect the function of this putative regulatory complex.
MCT9, a monocarboxylic acid, sodium-dependent transporter protein that is expressed in the kidney, is encoded by SLC16A9. The substrate or substrates of this transporter are unknown. However, a meta-analysis showed that the rs12356193 SNP of the SLC16A9 gene was associated with dl-carnitine and propionyl-l-carnitine concentrations as well as with serum uric acid levels.32
The LRRC16A gene codes for LRRC16A (also known as CARMIL), which is present in kidney and epithelial cells. This protein is a key inhibitor of actin capping protein and, therefore, a regulator of actin polymerization.82 However, the underlying mechanism behind the association between LRRC16A and serum uric acid levels is not known. The biochemical mechanism underlying the association between SNPs of the RREB1 and INHBC genes and serum uric acid levels is also unknown. RREB1 encodes a zinc finger transcription factor (ras-responsive element-binding protein 1) that has been reported to regulate the androgen receptor and calcitonin gene,83 whereas INHBC encodes a member of the transforming-β growth factor family, Inhibin β C chain.84
The GCKR gene encodes GCKR, which is predominantly expressed in the liver and does not seem to function in the direct handling of renal urate. In the liver, GCKR regulates glucokinase75,85 by mediating the phosphorylation of glucose to glucose-6-phosphate, a precursor of liver glycogen synthesis and of de novo purine synthesis.75,85 A deficiency in the activity of glucose-6-phosphatase causes glycogen storage disease type 1 (also known as von Gierke disease), which is characterized by hypertriglyceridaemia and hyperuricaemia.75,85 The GCKR SNP rs780094 might affect both serum uric acid and triglyceride levels (which are associated with insulin resistance) via a common mediator.75 Interestingly, GCKR is associated with insulin resistance,32 glucose level, triglyceride level, and C-reactive protein,86 which are components of metabolic syndrome.75
Potentially associated variants
Several other genes that might be associated with serum uric acid levels or gout have been identified (Table 4). An Icelandic study that involved whole-genome sequencing of 16 million SNPs for low frequency variants, showed that a risk allele of ALDH16A1 was associated with an OR of 3.1 for gout (P = 1.5 × 10–16), which probably represents the largest single gene effect among recently discovered variants.13 However, the ALDH16A1 variant is rare (~1% in Icelandic and <1% in white populations) and is expected to have a limited population attributable risk compared with common variants such as those of SLC2A9. The results of the Icelandic study further confirmed an association between the previously identified genetic variants of SLC2A9, ABCG2, SLC17A, SLC16A9, SLC22A11, GCKR, and INHBC, and serum uric acid levels.13 In addition, a novel variant (rs12129861) that associated with gout was identified in chromosome 1.13 Other reported genetic polymorphisms that could potentially be associated with serum uric acid levels and gout include variants of SGK1-SLC2A12,72ADRB3,87–89MTHFR,90–95PRKG296 and TGFB197 (Figure 3).32 Most of these genes can be considered candidate genes as they have not yet been validated in large-scale studies in separate populations.
Table 4.
Genetic variants potentially associated with serum uric acid levels and gout
| Variant | Phenotype | Potential functional mechanism | Populations |
|---|---|---|---|
| ADRB3 (chromosome 8) | |||
| Trp64Arg | SUA | Might cause reduced lipolysis and increased adipose tissue that could result in insulin resistance | Korean,88 Spanish,87 Italian,95 Japanese,89 Chinese95 |
| rs4994 | Gout | Unknown | Chinese94 |
| MTHFR (chromosome 1) | |||
| Cys677Thr | SUA | Might cause increased de novo synthesis of purines | Japanese,90,92 Korean,91 Iranian,118 Brazilian93 |
| PRKG2 (chromosome 4) | |||
| rs10033237 | Gout | Might cause increased renin activity | Taiwanese96 |
| rs7688672 | Gout | Might cause increased renin activity | Taiwanese96 |
| rs6837293 | Gout | Might cause increased renin activity | Taiwanese96 |
| SGK1-SLC2A12 Locus (chromosome 6) | |||
| rs9321453 | SUA | Unknown | European ancestry, African American72 |
| ALDH16A1 (chromosome 19) | |||
| Cg1580C>G | SUA, gout | Might be involved in purine metabolism | Icelandic13 |
| Chr 1-centromere (chromosome 1) | |||
| Chr1_142697422 | SUA, gout | Unknown | Icelandic13 |
| TGFB1 (chromosome 19) | |||
| 869T/C | Tophi formation | Might regulate local inflammatory responses and/or tophi formation | Taiwanese97 |
Abbreviation: SUA, serum uric acid.
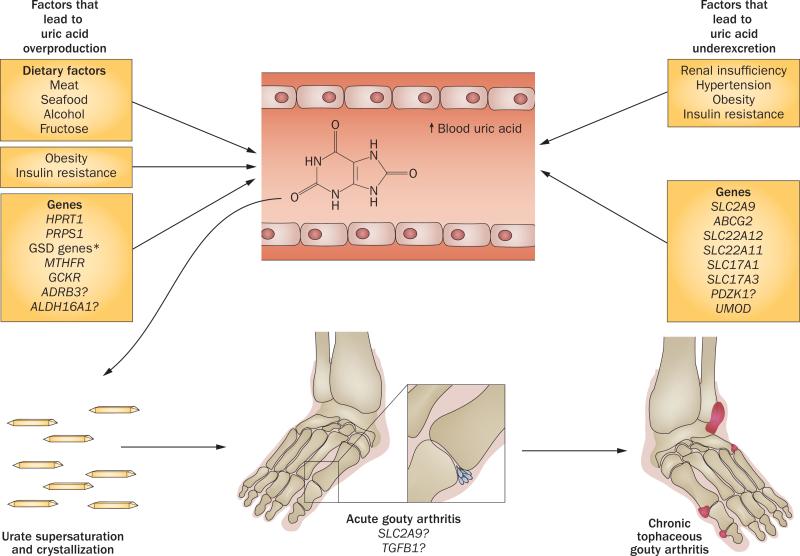
Figure 3.
The pathogenesis of hyperuricaemia and gout. Genetic and environmental factors are juxtaposed with the two major mechanisms that lead to hyperuricaemia—exogenous and endogenous overproduction of uric acid and underexcretion of urate. Hyperuricaemia results in the formation of monosodium urate crystals in oversaturated tissue fluids. As well as urate concentration (serum uric acid levels >404.5 μmol/L), crystallization is dependent on pH, temperature and other factors.2 Stimulation of the NALP3 inflammasome and other humoral and cellular inflammatory mediators by monosodium urate crystals results in acute gouty arthritis.115 Chronic cumulative urate crystal formation in tissue fluids leads to deposition of monosodium urate crystals in the synovium, cartilage, tendons and soft tissues, resulting in tophi formation and chronic tophaceous gouty arthritis. The vast majority of newly identified common genetic variants that are associated with hyperuricaemia and/or gout are involved in urate renal excretion. However, some of these variants are also expressed in extrarenal tissues and might be involved in the regulation of serum urate homeostasis (ABCG2) or the development of monosodium-urate-crystal-induced inflammation and arthritis (SCL2A9, TGF-β).*Including CPT2, AMP1, ACDS and ALDOB. Abbreviation: GSD, glycogen storage disease.
Clinical utility of genetic variants
The common genetic variants identified in GWAS confer a modest risk of gout. However, an additive composite genetic urate score of high-risk alleles can confer up to a 41-fold increased risk for gout compared with individuals without any such alleles.33 A similarly constructed nongenetic risk factor score (based on BMI, alcohol intake, diuretic use and history of hypertension) conferred up to a 79-fold increased risk of gout in the US general population.98 The serum uric acid variance explained by common genetic variants is small (~6% in the CHARGE meta-analysis),33 particularly when compared with the 67% of serum uric acid variance that can be explained by nongenetic factors, such as serum creatinine level, metabolic syndrome components (including insulin resistance, waist circumference and systolic blood pressure) and FEua.75
The clinical utility of genetic variants or their composite genetic scores for gout remains unclear. Their utility for predicting gout seems limited because serum urate levels, which can be easily measured, effectively predict gout risk.2 Furthermore, genetic scores would have limited utility in justifying initiation of urate-lowering therapy in patients with asymptomatic hyperuricaemia37 because even onset of gout does not necessarily justify initiation of such therapy given the associated potential adverse effects.99,100 Further studies are required to determine if genetic scores could be used to predict worse outcomes, such as tophaceous erosive gout, complications of gout, or responses to certain classes of antigout drugs, particularly uricosuric drugs that might target the urate transportasome.29
Urate and cardiovascular events
Serum uric acid levels and gout are both strongly associated with metabolic syndrome and risk of cardiovascular disease. However, whether these relations are causal is unclear.2,33,101 In an attempt to answer this long-standing question, Mendelian randomization studies were included in some GWAS102,103 and in the CHARGE meta-analysis.33 In these studies the genetic urate score (rather than serum uric acid concentration) was used as the exposure variable for a potential causal role of serum uric acid in the development of cardiovascular–metabolic events.2,33,101,104 The Mendelian randomization studies could be thought of as natural randomized trials of the effects of genetically determined variation in serum uric acid concentration on cardiovascular–metabolic outcomes because the genes are randomly assigned during gamete formation and this assignment is unlikely to be affected by nongenetic or environmental factors.
The CHARGE Mendelian randomization analyses showed that genetically-determined serum uric acid levels (assessed using the urate genetic score) were not associated with blood pressure, glucose levels, renal function, chronic kidney disease or coronary heart disease.33 These findings did not support a causal role for serum uric acid in the development of cardiovascular disease or metabolic syndrome. However, the urate genetic scores were strongly associated with gout, confirming a causal relationship.33 Similarly, a Mendelian randomization analysis of SLC2A9, which used a Bayesian approach, found no evidence for a casual role of serum uric acid in metabolic syndrome.102 A similar lack of association between urate genetic variants and cardiovascular and metabolic outcomes has been shown in other GWAS.31,34,39
By contrast, a Mendelian randomization study published in 2012, showed that serum uric acid levels determined by SLC2A9 alleles were associated with systolic blood pressure in 526 Amish adults who were not receiving diuretic or antihypersensitive drugs.103 The difference in findings might be related to differences in study design. The participants in the Amish study received standardized diets and 24-h ambulatory blood-pressure monitoring,103 whereas the CHARGE study participants received a liberal diet and single-visit blood-pressure measurement.33 The contrasting findings of the two studies suggest the potential importance of a controlled setting and homogenous population when examining secondarily mediating gene effects in Mendelian randomization analyses.103
Pharmacogenetics of allopurinol
The most commonly used urate-lowering antigout drug, allopurinol, is associated with rare (affecting ~0.2% of patients), but severe, hypersensitivity reactions that are fatal in 25% of cases.105 Pharmacogenetic studies have shown a strong association between HLA-B*5801 and allopurinol-induced Steven–Johnson Syndrome (SJS) and toxic epidermal necrolysis (TEN) in Korean, Han Chinese, Japanese, Thai106–110 and European populations.111 A meta-analysis based on these studies showed that patients who expressed the HLA-B*5801 allele had an 80–97-fold increased risk of developing SJS or TEN in response to allopurinol compared with patients who did not express the allele.112
A GWAS that included 14 Japanese patients who had allopurinol-related SJS or TEN and 991 ethnically matched healthy controls, confirmed the strong association between the HLA-B*5801 locus and allopurinol-related hypersensitivity reactions (OR 62.8; P = 5.388 × 10–12).113 The study also identified several other SNPs in BAT1, HCP5, MICC and PSORS1C1 genes, which were significantly associated with allopurinol-related SJS or TEN (OR >61; P <4.62 × 10–8 for all associations).113 The PSORS1C1 SNP, rs9263726, was in absolute linkage disequilibrium with HLA-B*5801.113 If these findings are confirmed in other populations, rs9263726, which can easily be typed, could be a useful biomarker for prediction of allopurinol-related SJS or TEN.113
Conclusions
GWAS have led to a remarkable increase in replicable genetic association data for serum uric acid and gout. Reduced renal excretion of urate is the major cause of hyperuricaemia and gout and most of the common genes discovered in GWAS are involved in the renal urate-transport system. The genes for the urate transporters, GLUT-9 and ABCG2, which are important modulators of uric acid levels, consistently associate with serum uric acid levels and gout. Although the GWAS association data for SLC22A12 (which encodes URAT1) have been less impressive, many layers of evidence indicate that URAT1 is an essential component of renal urate handing. Loss-of-function mutations in the absorptive transporter genes SLC22A12 or SLC2A9 lead to a dominance of urate secretion and hypouricaemia, whereas loss-in-function or reduction-in-function mutations in the secretory urate transporter genes, ABCG2, SLC17A1 or SLC17A3, cause hyperuricaemia. These findings indicate that serum uric acid levels are largely determined by the relative balance between urate absorption and secretion across the proximal tubule. Most of the other replicated genetic variants discussed in this Review require further functional characterization. However, these genetic data add considerably to our understanding of the pathogenesis of hyperuricaemia and gout. Future research directions might include replication in other ethnic cohorts, fine mapping and resequencing, functional studies of the identified genetic variants and evaluation of potential interactions between genes and key environmental factors.2,28,100,114 Genetic markers that are associated with severe gout (such as tophaceous erosive gout), other complications of gout, or efficacy and adverse reactions to antigout drugs should also be evaluated.
Key points.
■ The majority of the genes that associate with hyperuricaemia and gout in genome-wide association studies have been implicated in the renal urate-transport system
■ Genetic variation explains only a modest level of variance in serum uric acid levels (~6%)
■ Serum uric acid levels are determined by the net balance between urate absorption and secretion, which is mediated by separate sets of transporters in the renal proximal tubule
■ The clinical utility of testing for urate-associated genes seems limited because serum urate levels themselves can effectively predict gout risk at a low cost
■ Urate-associated genes and genetically determined urate levels have been largely unassociated with cardiovascular or metabolic outcomes, suggesting that serum uric acid does not have a causal role in these outcomes
■ Strong pharmacogenetic associations between HLA-B*5801 alleles and severe allopurinol-hypersensitivity reactions have been shown in Asian and European populations, suggesting clinical utility of testing for these alleles
Review criteria.
A search for original articles published between 1965 and 2012 and focusing on the genetics of hyperuricaemia and gout was performed in MEDLINE and PubMed. The search terms used were “uric acid”, “urate”, “urate transporters”, “hyperuricaemia”, “gout”, “gene”, “genetic”, and “loci”, alone and in combination. All articles identified were English-language, full-text papers. We also searched the reference lists of identified articles for further relevant papers.
Acknowledgements
This work was partly supported by grants from the NIH (R01AR056291 and P60AR047785).
Footnotes
Competing interests
A. M. Reginato declares associations with the following companies: Savient, Takeda, URL. D. B. Mount declares associations with the following companies: Nuon, URL. H. K. Choi declares associations with the following companies: Takeda, URL. See the article online for full details of the relationships. I. Yang declares no competing interests.
Author contributions
All authors contributed equally to researching the data for the article, discussions of the content, writing the article and editing of the manuscript before submission.
References
- 1.Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricaemia in the US general population: The National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011;63:3136–3141. doi: 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- 2.Choi HK, Mount DB, Reginato AM, American College of Physicians & American Physiological Society Pathogenesis of gout. Ann. Intern. Med. 2005;143:499–516. doi: 10.7326/0003-4819-143-7-200510040-00009. [DOI] [PubMed] [Google Scholar]
- 3.Wu EQ, et al. Disease-related and all-cause health care costs of elderly patients with gout. J. Manag. Care Pharm. 2008;14:164–175. doi: 10.18553/jmcp.2008.14.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi HK, Ford ES, Li C, Curhan G. Prevalence of the metabolic syndrome in patients with gout: the Third National Health and Nutrition Examination Survey. Arthritis Rheum. 2007;57:109–115. doi: 10.1002/art.22466. [DOI] [PubMed] [Google Scholar]
- 5.Abbott RD, Brand FN, Kannel WB, Castelli WP. Gout and coronary heart disease: the Framingham Study. J. Clin. Epidemiol. 1988;41:237–242. doi: 10.1016/0895-4356(88)90127-8. [DOI] [PubMed] [Google Scholar]
- 6.Krishnan E, Baker JF, Furst DE, Schumacher HR. Gout and the risk of acute myocardial infarction. Arthritis Rheum. 2006;54:2688–2696. doi: 10.1002/art.22014. [DOI] [PubMed] [Google Scholar]
- 7.Choi HK, Curhan G. Independent impact of gout on mortality and risk for coronary heart disease. Circulation. 2007;116:894–900. doi: 10.1161/CIRCULATIONAHA.107.703389. [DOI] [PubMed] [Google Scholar]
- 8.Choi HK, De Vera MA, Krishnan E. Gout and the risk of type 2 diabetes among men with a high cardiovascular risk profile. Rheumatology (Oxford) 2008;47:1567–1570. doi: 10.1093/rheumatology/ken305. [DOI] [PubMed] [Google Scholar]
- 9.Syndenham T. The Works of Thomas Sydndenham, MD on Acute and Chronic Diseases. II. G. J. & J. Robinson; London: 1853. A Treatise of the Gout and Dropsy. [Google Scholar]
- 10.Page T, Nyhan WL. The spectrum of HPRT deficiency: an update. Adv. Exp. Med. Biol. 1989;253A:129–133. doi: 10.1007/978-1-4684-5673-8_20. [DOI] [PubMed] [Google Scholar]
- 11.Mateos EA, Puig JG. Purine metabolism in Lesch–Nyhan syndrome versus Kelley–Seegmiller syndrome. J. Inherit. Metab. Dis. 1994;17:138–142. doi: 10.1007/BF00735419. [DOI] [PubMed] [Google Scholar]
- 12.Mineo I, et al. Myogenic hyperuricaemia. A common pathophysiologic feature of glycogenosis types III, V, and VII. N. Engl. J. Med. 1987;317:75–80. doi: 10.1056/NEJM198707093170203. [DOI] [PubMed] [Google Scholar]
- 13.Sulem P, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat. Genet. 2011;43:1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 14.Vora S, DiMauro S, Spear D, Harker D, Danon MJ. Characterization of the enzymatic defect in late-onset muscle phosphofructokinase deficiency. New subtype of glycogen storage disease type VII. J. Clin. Invest. 1987;80:1479–1485. doi: 10.1172/JCI113229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabina RL, et al. Myoadenylate deaminase deficiency. Functional and metabolic abnormalities associated with disruption of the purine nucleotide cycle. J. Clin. Invest. 1984;73:720–730. doi: 10.1172/JCI111265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson-Mundt A, Luder AS, Greene CL. Hyperuricaemia in medium-chain acylcoenzyme A dehydrogenase deficiency. J. Pediatr. 1992;120:444–446. doi: 10.1016/s0022-3476(05)80918-7. [DOI] [PubMed] [Google Scholar]
- 17.Perheentupa J, Raivio K. Fructose-induced hyperuricaemia. Lancet. 1967;2:528–531. doi: 10.1016/s0140-6736(67)90494-1. [DOI] [PubMed] [Google Scholar]
- 18.Vyletal P, et al. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 19.Vyletal P, Bleyer AJ, Kmoch S. Uromodulin biology and pathophysiology—an update. Kidney Blood Press. Res. 2010;33:456–475. doi: 10.1159/000321013. [DOI] [PubMed] [Google Scholar]
- 20.Hart TC, et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J. Med. Genet. 2002;39:882–892. doi: 10.1136/jmg.39.12.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamatani N, et al. Localization of a gene for familial juvenile hyperuricemic nephropathy causing underexcretion-type gout to 16p12 by genome-wide linkage analysis of a large family. Arthritis Rheum. 2000;43:925–929. doi: 10.1002/1529-0131(200004)43:4<925::AID-ANR26>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Becker MA, Smith PR, Taylor W, Mustafi R, Switzer RL. The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J. Clin. Invest. 1995;96:2133–2141. doi: 10.1172/JCI118267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merriman TR, Dalbeth N. The genetic basis of hyperuricaemia and gout. Joint Bone Spine. 2011;78:35–40. doi: 10.1016/j.jbspin.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 24.Reed DR, Price RA. X-linkage does not account for the absence of father–son similarity in plasma uric acid concentrations. Am. J. Med. Genet. 2000;92:142–146. [PubMed] [Google Scholar]
- 25.Emmerson BT, Nagel SL, Duffy DL, Martin NG. Genetic control of the renal clearance of urate: a study of twins. Ann. Rheum. Dis. 1992;51:375–377. doi: 10.1136/ard.51.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilk JB, et al. Segregation analysis of serum uric acid in the NHLBI Family Heart Study. Hum. Genet. 2000;106:355–359. doi: 10.1007/s004390000243. [DOI] [PubMed] [Google Scholar]
- 27.Yang Q, et al. Genome-wide search for genes affecting serum uric acid levels: the Framingham Heart Study. Metabolism. 2005;54:1435–1441. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Choi HK, Zhu Y, Mount DB. Genetics of gout. Curr. Opin. Rheumatol. 2010;22:144–151. doi: 10.1097/BOR.0b013e32833645e8. [DOI] [PubMed] [Google Scholar]
- 29.Dalbeth N, Merriman T. Crystal ball gazing: new therapeutic targets for hyperuricaemia and gout. Rheumatology (Oxford) 2009;48:222–226. doi: 10.1093/rheumatology/ken460. [DOI] [PubMed] [Google Scholar]
- 30.Endou H, Anzai N. Urate transport across the apical membrane of renal proximal tubules. Nucleosides Nucleotides Nucleic Acids. 2008;27:578–584. doi: 10.1080/15257770802136024. [DOI] [PubMed] [Google Scholar]
- 31.Caulfield MJ, et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:e197. doi: 10.1371/journal.pmed.0050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolz M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ. Cardiovasc. Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitart V, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 35.Doring A, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 36.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehghan A, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark K, et al. Association of common polymorphisms in GLUT9 gene with gout but not with coronary artery disease in a large case–control study. PLoS ONE. 2008;3:e1948. doi: 10.1371/journal.pone.0001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo H, et al. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricaemia. Am. J. Hum. Genet. 2008;83:744–751. doi: 10.1016/j.ajhg.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anzai N, et al. Plasma urate level is directly regulated by a voltage-driven urate efflux transporter URATv1 (SLC2A9) in humans. J. Biol. Chem. 2008;283:26834–26838. doi: 10.1074/jbc.C800156200. [DOI] [PubMed] [Google Scholar]
- 42.Hollis-Moffatt JE, et al. Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Maori, Pacific Island, and Caucasian case-control sample sets. Arthritis Rheum. 2009;60:3485–3492. doi: 10.1002/art.24938. [DOI] [PubMed] [Google Scholar]
- 43.Brandstätter A, et al. Sex-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care. 2008;31:1662–1667. doi: 10.2337/dc08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karns R, et al. Genome-wide association of serum uric acid concentration: replication of sequence variants in an island population of the Adriatic coast of Croatia. Ann. Hum. Genet. 2012;76:121–127. doi: 10.1111/j.1469-1809.2011.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal A, Emerling DE, Serafini TA, Mount DB. Tranilast inhibits urate transport mediated by URAT1 and GLUT9 [abstract]. Arthritis Rheum. 2010;62(Suppl. 10):164. [Google Scholar]
- 46.Bibert S, et al. Mouse GLUT9: evidences for a urate uniporter. Am. J. Physiol. Renal Physiol. 2009;297:F612–F619. doi: 10.1152/ajprenal.00139.2009. [DOI] [PubMed] [Google Scholar]
- 47.Manolescu AR, Augustin R, Moley K, Cheeseman C. A highly conserved hydrophobic motif in the exofacial vestibule of fructose transporting SLC2A proteins acts as a critical determinant of their substrate selectivity. Mol. Membr. Biol. 2007;24:455–463. doi: 10.1080/09687680701298143. [DOI] [PubMed] [Google Scholar]
- 48.Augustin R, et al. Identification and characterization of human glucose transporter-like protein-9 (GLUT9): alternative splicing alters trafficking. J. Biol. Chem. 2004;279:16229–16236. doi: 10.1074/jbc.M312226200. [DOI] [PubMed] [Google Scholar]
- 49.Richardson S, et al. Molecular characterization and partial cDNA cloning of facilitative glucose transporters expressed in human articular chondrocytes; stimulation of 2-deoxyglucose uptake by IGF-I and elevated MMP-2 secretion by glucose deprivation. Osteoarthritis Cartilage. 2003;11:92–101. doi: 10.1053/joca.2002.0858. [DOI] [PubMed] [Google Scholar]
- 50.Phay JE, Hussain HB, Moley JF. Cloning and expression analysis of a novel member of the facilitative glucose transporter family, SLC2A9 (GLUT9). Genomics. 2000;66:217–220. doi: 10.1006/geno.2000.6195. [DOI] [PubMed] [Google Scholar]
- 51.Charles BA, et al. A genome-wide association study of serum uric acid in African Americans. BMC Med. Genomics. 2011;4:17. doi: 10.1186/1755-8794-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Center for Biotechnology Information dbSNP Short Genetic Variations. 2012 NCBI [online] http://www.ncbi.nlm.nih.gov/projects/SNP/
- 53.Dinour D, et al. Two novel homozygous SLC2A9 mutations cause renal hypouricaemia type 2. Nephrol. Dial. Transplant. 2012;27:1035–1041. doi: 10.1093/ndt/gfr419. [DOI] [PubMed] [Google Scholar]
- 54.Dinour D, et al. Homozygous SLC2A9 mutations cause severe renal hypouricaemia. J. Am. Soc. Nephrol. 2010;21:64–72. doi: 10.1681/ASN.2009040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McArdle PF, et al. Association of a common nonsynonymous variant in GLUT9 with serum uric acid levels in old order Amish. Arthritis Rheum. 2008;58:2874–2881. doi: 10.1002/art.23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urano W, et al. Association between GLUT9 and gout in Japanese men. Ann. Rheum. Dis. 2010;69:932–933. doi: 10.1136/ard.2009.111096. [DOI] [PubMed] [Google Scholar]
- 57.Tu HP, et al. Associations of a non-synonymous variant in SLC2A9 with gouty arthritis and uric acid levels in Han Chinese subjects and Solomon Islanders. Ann. Rheum. Dis. 2010;69:887–890. doi: 10.1136/ard.2009.113357. [DOI] [PubMed] [Google Scholar]
- 58.Hollis-Moffatt JE, et al. The SLC2A9 nonsynonymous Arg265His variant and gout; evidence for a population-specific effect on severity. Arthritis Res. Ther. 2011;13:R85. doi: 10.1186/ar3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamagishi K, et al. The rs2231142 variant of the ABCG2 gene is associated with uric acid levels and gout among Japanese people. Rheumatology (Oxford) 2010;49:1461–1465. doi: 10.1093/rheumatology/keq096. [DOI] [PubMed] [Google Scholar]
- 60.Wang B, et al. Genetic analysis of ABCG2 gene C421A polymorphism with gout disease in Chinese Han male population. Hum. Genet. 2010;127:245–246. doi: 10.1007/s00439-009-0760-4. [DOI] [PubMed] [Google Scholar]
- 61.Phipps-Green AJ, et al. A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Maori, case and control sample sets. Hum. Mol. Genet. 2010;19:4813–4819. doi: 10.1093/hmg/ddq412. [DOI] [PubMed] [Google Scholar]
- 62.Woodward OM, et al. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl Acad. Sci. USA. 2009;106:10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuo H, et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci. Transl. Med. 2009;1:5ra11. doi: 10.1126/scitranslmed.3000237. [DOI] [PubMed] [Google Scholar]
- 64.Hosomi A, Nakanishi T, Fujita T, Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS ONE. 2012;7:e30456. doi: 10.1371/journal.pone.0030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodrigues AC, Curi R, Genvigir FD, Hirata MH, Hirata RD. The expression of efflux and uptake transporters are regulated by statins in Caco-2 and HepG2 cells. Acta Pharmacol. Sin. 2009;30:956–964. doi: 10.1038/aps.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enomoto A, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002;417:447–452. doi: 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- 67.Shima Y, Teruya K, Ohta H. Association between intronic SNP in urate-anion exchanger gene, SLC22A12, and serum uric acid levels in Japanese. Life Sci. 2006;79:2234–2237. doi: 10.1016/j.lfs.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 68.Graessler J, et al. Association of the human urate transporter 1 with reduced renal uric acid excretion and hyperuricaemia in a German Caucasian population. Arthritis Rheum. 2006;54:292–300. doi: 10.1002/art.21499. [DOI] [PubMed] [Google Scholar]
- 69.Vázquez-Mellado J, et al. Molecular analysis of the SLC22A12 (URAT1) gene in patients with primary gout. Rheumatology (Oxford) 2007;46:215–219. doi: 10.1093/rheumatology/kel205. [DOI] [PubMed] [Google Scholar]
- 70.Tu HP, et al. The SLC22A12 gene is associated with gout in Han Chinese and Solomon Islanders. Ann. Rheum. Dis. 2010;69:1252–1254. doi: 10.1136/ard.2009.114504. [DOI] [PubMed] [Google Scholar]
- 71.Guan M, et al. High-resolution melting analysis for the rapid detection of an intronic single nucleotide polymorphism in SLC22A12 in male patients with primary gout in China. Scand. J. Rheumatol. 2009;38:276–281. doi: 10.1080/03009740802572483. [DOI] [PubMed] [Google Scholar]
- 72.Tin A, et al. Genome-wide association study for serum urate concentrations and gout among African Americans identifies genomic risk loci and a novel URAT1 loss-of-function allele. Hum. Mol. Genet. 2011;20:4056–4068. doi: 10.1093/hmg/ddr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C, et al. Multiple single nucleotide polymorphisms in the human urate transporter 1 (hURAT1) gene are associated with hyperuricaemia in Han Chinese. J. Med. Genet. 2010;47:204–210. doi: 10.1136/jmg.2009.068619. [DOI] [PubMed] [Google Scholar]
- 74.Jang WC, et al. T6092C polymorphism of SLC22A12 gene is associated with serum uric acid concentrations in Korean male subjects. Clin. Chim. Acta. 2008;398:140–144. doi: 10.1016/j.cca.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 75.van der Harst P, et al. Replication of the five novel loci for uric acid concentrations and potential mediating mechanisms. Hum. Mol. Genet. 2010;19:387–395. doi: 10.1093/hmg/ddp489. [DOI] [PubMed] [Google Scholar]
- 76.Iharada M, et al. Type 1 sodium-dependent phosphate transporter (SLC17A1 protein) is a CI–-dependent urate exporter. J. Biol. Chem. 2010;285:26107–26113. doi: 10.1074/jbc.M110.122721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jutabha P, et al. Human sodium phosphate transporter 4 (hNPT4/SLC17A3) as a common renal secretory pathway for drugs and urate. J. Biol. Chem. 2010;285:35123–35132. doi: 10.1074/jbc.M110.121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright AF, Rudan I, Hastie ND, Campbell H. A ‘complexity’ of urate transporters. Kidney Int. 2010;78:446–452. doi: 10.1038/ki.2010.206. [DOI] [PubMed] [Google Scholar]
- 79.Jutabha P, et al. Functional analysis of human sodium-phosphate transporter 4 (NPT4/SLC17A3) polymorphisms. J. Pharmacol. Sci. 2011;115:249–253. doi: 10.1254/jphs.10228sc. [DOI] [PubMed] [Google Scholar]
- 80.Nabipour I, et al. Serum uric acid is associated with bone health in older men: a cross-sectional population-based study. J. Bone Miner. Res. 2011;26:955–964. doi: 10.1002/jbmr.286. [DOI] [PubMed] [Google Scholar]
- 81.Anzai N, Jutabha P, Amonpatumrat-Takahashi S, Sakurai H. Recent advances in renal urate transport: characterization of candidate transporters indicated by genome-wide association studies. Clin. Exp. Nephrol. 2012;16:89–95. doi: 10.1007/s10157-011-0532-z. [DOI] [PubMed] [Google Scholar]
- 82.Yang C, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thiagalingam A, et al. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol. Cell. Biol. 1996;16:5335–5345. doi: 10.1128/mcb.16.10.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scmitt J, et al. Structure, chromosomal localization, and expression analysis of the mouse inhibin/activin βC (Inhbc) gene. Genomics. 1996;32:358–366. doi: 10.1006/geno.1996.0130. [DOI] [PubMed] [Google Scholar]
- 85.Yang Chou J, Mansfield BC. Molecular genetics of type 1 glycogen storage diseases. Trends Endocrinol. Metab. 1999;10:104–113. doi: 10.1016/s1043-2760(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 86.Orho-Melander M, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morcillo S, et al. Trp64Arg polymorphism of the ADRB3 gene predicts hyperuricaemia risk in a population from southern Spain. J. Rheumatol. 2010;37:417–421. doi: 10.3899/jrheum.090637. [DOI] [PubMed] [Google Scholar]
- 88.Rho YH, Choi SJ, Lee YH, Ji JD, Song GG. The association between hyperuricaemia and the Trp64Arg polymorphism of the β-3 adrenergic receptor. Rheumatol. Int. 2007;27:835–839. doi: 10.1007/s00296-006-0300-7. [DOI] [PubMed] [Google Scholar]
- 89.Hayashi H, et al. Contribution of a missense mutation (Trp64Arg) in β3-adrenergic receptor gene to multiple risk factors in Japanese men with hyperuricaemia. Endocr. J. 1998;45:779–784. doi: 10.1507/endocrj.45.779. [DOI] [PubMed] [Google Scholar]
- 90.Zuo M, et al. The C677T mutation in the methylene tetrahydrofolate reductase gene increases serum uric acid in elderly men. J. Hum. Genet. 2000;45:257–262. doi: 10.1007/s100380070037. [DOI] [PubMed] [Google Scholar]
- 91.Hong YS, et al. The C677 mutation in methylene tetrahydrofolate reductase gene: correlation with uric acid and cardiovascular risk factors in elderly Korean men. J. Korean Med. Sci. 2004;19:209–213. doi: 10.3346/jkms.2004.19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Itou S, et al. Significant association between methylenetetrahydrofolate reductase 677T allele and hyperuricaemia among adult Japanese subjects. Nutr. Res. 2009;29:710–715. doi: 10.1016/j.nutres.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 93.Kimi Uehara S, Rosa G. Association of uricemia with biochemical and dietary factors in human adults with metabolic syndrome genotyped to C677T polymorphism in the methylenetetrahydrofolate reductase gene. Nutr. Hosp. 2011;26:298–303. doi: 10.1590/S0212-16112011000200009. [DOI] [PubMed] [Google Scholar]
- 94.Wang B, et al. Positive correlation between β-3-adrenergic receptor (ADRB3) gene and gout in a Chinese male population. J. Rheumatol. 2011;38:738–740. doi: 10.3899/jrheum.101037. [DOI] [PubMed] [Google Scholar]
- 95.Strazzullo P, et al. Relationship of the Trp64Arg polymorphism of the β3-adrenoceptor gene to central adiposity and high blood pressure: interaction with age. Cross-sectional and longitudinal findings of the Olivetti Prospective Heart Study. J. Hypertens. 2001;19:399–406. doi: 10.1097/00004872-200103000-00007. [DOI] [PubMed] [Google Scholar]
- 96.Chang SJ, et al. The cyclic GMP-dependent protein kinase II gene associates with gout disease: identified by genome-wide analysis and case–control study. Ann. Rheum. Dis. 2009;68:1213–1219. doi: 10.1136/ard.2008.093252. [DOI] [PubMed] [Google Scholar]
- 97.Chang SJ, et al. Associations between gout tophus and polymorphisms 869T/C and –509C/T in transforming growth factor β1 gene. Rheumatology (Oxford) 2008;47:617–621. doi: 10.1093/rheumatology/ken054. [DOI] [PubMed] [Google Scholar]
- 98.Choi HK, Curhan G. Beer, liquor, wine, and serum uric acid level—The Third National Health and Nutrition Examination Survey. Arthritis Rheum. 2004;51:1023–1029. doi: 10.1002/art.20821. [DOI] [PubMed] [Google Scholar]
- 99.Zhang W, et al. EULAR evidence based recommendations for gout. Part I: Diagnosis. Report of a task force of the Standing Committee for International Clinical Studies Including Therapeutics (ESCISIT). Ann. Rheum. Dis. 2006;65:1301–1311. doi: 10.1136/ard.2006.055251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neogi T. Clinical practice. Gout. N. Engl. J. Med. 2011;364:443–452. doi: 10.1056/NEJMcp1001124. [DOI] [PubMed] [Google Scholar]
- 101.Feig DI, Soletsky B, Johnson RJ. Effect of allopurinol on blood pressure of adolescents with newly diagnosed essential hypertension: a randomized trial. JAMA. 2008;300:924–932. doi: 10.1001/jama.300.8.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McKeigue PM, et al. Bayesian methods for instrumental variable analysis with genetic instruments (‘Mendelian randomization’): example with urate transporter SLC2A9 as an instrumental variable for effect of urate levels on metabolic syndrome. Int. J. Epidemiol. 2010;39:907–918. doi: 10.1093/ije/dyp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parsa A, et al. Genotype-based changes in serum uric acid affect blood pressure. Kidney Int. 2012;81:502–507. doi: 10.1038/ki.2011.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stark K, et al. Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS ONE. 2009;4:e7729. doi: 10.1371/journal.pone.0007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Terkeltaub RA. Clinical practice. Gout. N. Engl. J. Med. 2003;349:1647–1655. doi: 10.1056/NEJMcp030733. [DOI] [PubMed] [Google Scholar]
- 106.Kang HR, et al. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet. Genomics. 2011;21:303–307. doi: 10.1097/FPC.0b013e32834282b8. [DOI] [PubMed] [Google Scholar]
- 107.Jung JW, et al. HLA-B58 can help the clinical decision on starting allopurinol in patients with chronic renal insufficiency. Nephrol. Dial. Transplant. 2011;26:3567–3572. doi: 10.1093/ndt/gfr060. [DOI] [PubMed] [Google Scholar]
- 108.Hung SI, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc. Natl Acad. Sci. USA. 2005;102:4134–4139. doi: 10.1073/pnas.0409500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaniwa N, et al. HLA-B locus in Japanese patients with antiepileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008;9:1617–1622. doi: 10.2217/14622416.9.11.1617. [DOI] [PubMed] [Google Scholar]
- 110.Tassaneeyakul W, et al. Strong association between HLA-B*5801 and allopurinol-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in a Thai population. Pharmacogenet. Genomics. 2009;19:704–709. doi: 10.1097/FPC.0b013e328330a3b8. [DOI] [PubMed] [Google Scholar]
- 111.Lonjou C, et al. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet. Genomics. 2008;18:99–107. doi: 10.1097/FPC.0b013e3282f3ef9c. [DOI] [PubMed] [Google Scholar]
- 112.Somkrua R, Eickman EE, Saokaew S, Lohitnavy M, Chaiyakunapruk N. Association of HLA-B*5801 allele and allopurinol-induced Stevens Johnson syndrome and toxic epidermal necrolysis: a systematic review and meta-analysis. BMC Med. Genet. 2011;12:118. doi: 10.1186/1471-2350-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tohkin M, et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenomics J. doi: 10.1038/tpj.2011.41. http://dx.doi.org/10.1038/tpj.2011.41. [DOI] [PubMed]
- 114.Choi HK. A prescription for lifestyle change in patients with hyperuricaemia and gout. Curr. Opin. Rheumatol. 2010;22:165–172. doi: 10.1097/BOR.0b013e328335ef38. [DOI] [PubMed] [Google Scholar]
- 115.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 116.Bertorini TE, Shively V, Taylor B, Palmieri GM, Fox IH. ATP degradation products after ischemic exercise: hereditary lack of phosphorylase or carnitine palmityltransferase. Neurology. 1985;35:1355–1357. doi: 10.1212/wnl.35.9.1355. [DOI] [PubMed] [Google Scholar]
- 117.Urano W, et al. Sodium-dependent phosphate cotransporter type 1 sequence polymorphisms in male patients with gout. Ann. Rheum. Dis. 2010;69:1232–1234. doi: 10.1136/ard.2008.106856. [DOI] [PubMed] [Google Scholar]
- 118.Golbahar J, Aminzadeh MA, Al-Shboul QM, Kassab S, Rezaian GR. Association of methylenetetrahydrofolate reductase (C677T) polymorphism with hyperuricaemia. Nutr. Metab. Cardiovasc. Dis. 2007;17:462–467. doi: 10.1016/j.numecd.2006.02.002. [DOI] [PubMed] [Google Scholar]