Abstract
Twenty years after the proposal that pattern recognition receptors detect invasion by microbial pathogens, the field of immunology has witnessed several discoveries that have elucidated receptors and signaling pathways of microbial recognition systems and how they control the generation of T and B-lymphocyte-mediated immune responses. However, there are still many fundamental questions that remain poorly understood, even though sometimes the answers are assumed to be known. Here, we discuss some of these questions, including the mechanisms by which pathogen-specific innate immune recognition activates antigen-specific adaptive immune responses and the roles of different types of innate immune recognition in host defense from infection and injury.
Metazoans are transiently or constitutively colonized by a variety of microorganisms that can engage in mutualistic or antagonistic interactions with their hosts. The nature of these interactions is still poorly understood, although recent studies have begun elucidating the host receptors and signaling pathways involved in sensing both commensal and pathogenic microbes. These microbial sensing pathways are used by the immune system to maintain host-microbial homeostasis, and to induce anti-microbial defense mechanisms. In vertebrates, two types of immunity are used to protect the host from infections: innate and adaptive. The innate immune system is genetically programmed to detect invariant features of invading microbes. Innate immune cells include dendritic cells, macrophages, and neutrophils, among others. In contrast, the adaptive immune system, which is comprised on T and B lymphocytes, employs antigen receptors that are not encoded in the germ-line but generated de novo in each organism. Thus, adaptive immune responses are highly specific. The best characterized microbial sensors are the so-called pattern recognition receptors (PRRs) of the innate immune system, which detect relatively invariant molecular patterns found in most microorganisms of a given class (1). These structures are referred to as PAMPs (Pathogen-Associated Molecular Patterns), though they are not unique to microbes that can cause a disease. Several families of PRRs have been characterized over the past decade, thus elucidating the basic mechanisms of sensing microbial infections; however, several questions remain unresolved and new questions have arisen as a result of recent progress. Here we will discuss some of these emerging questions in the context of the current knowledge of innate immune recognition.
Are all PRRs created equal?
Microbial pathogens are recognized through multiple, distinct PRRs, which can be broadly categorized into secreted, transmembrane and cytosolic classes. Secreted PRRs, including collectins, ficolins and pentraxins, bind to microbial cell surfaces and activate classical and lectin pathways of the complement system, and opsonize pathogens for phagocytosis by macrophages and neutrophils.
The transmembrane PRRs include the Toll-like receptor (TLR) family and the C-type lectins. TLRs in mammals are either expressed on the plasma membrane or in endosomal/lysosomal organelles (2). Cell surface TLRs recognize conserved microbial patterns that are accessible on the cell surface, such as lipopolysaccharide (LPS) of gram negative bacteria (TLR4), lipoteichoic acids of gram positive bacteria and bacterial lipoproteins (TLR1/TLR2 and TLR2/TLR6) and flagellin (TLR5), whereas endosomal TLRs mainly detect microbial nucleic acids, such as dsRNA (TLR3), ssRNA (TLR7), and dsDNA (TLR9). Expression of TLRs is cell-type specific, allowing allocation of recognition responsibilities to various cell types (3). Dectin-1 and Dectin-2, are transmembrane receptors of the C-type lectin family that detect β-glucans and mannan, respectively, on fungal cell walls (4, 5).
The cytosolic PRRs include the retinoic acid inducible gene I (RIG-I) like receptors (RLRs) and the nucleotide-binding domain and leucine-rich repeat containing receptors (NLRs). RLRs detect viral pathogens (6). Unlike TLRs, most cell types express RLRs. RLR members RIG-I and melanoma differentiation factor 5 (MDA5) recognize viral RNA through their helicase domain and signal through their caspase recruitment domains (CARD) (7, 8). RLRs use a common adaptor molecule mitochondria antiviral signaling protein (MAVS) (9). Engagement of MAVS by RLRs leads to the activation of transcription factors nuclear factor κB (NF-κB) and interferon regulatory factor 3 (IRF3). RIG-I recognizes 5’ triphosphate-single stranded RNA with a double stranded RNA component – PAMPs associated with many single stranded RNA viruses (8). Similar RNA species are also generated by RNA polymerase III (Pol III) in the cytosol upon transcription of poly dA-dT rich double stranded DNA (10, 11). Thus, RIG-I is a sensor for both ssRNA viruses and some dsDNA viruses (via Pol III). MDA5 preferentially recognizes long dsRNA structures in the cytosol, a PAMP associated with positive single stranded RNA virus infections (9).
NLRs comprise a large family of intracellular sensors that can detect pathogens and stress signals (12). NLRs are multidomain proteins that contain a C-terminal leucine-rich repeat (LRR) domain, a central nucleotide-binding oligomerization (NOD) domain and a N-terminal effector domain (13). They can be divided into three subfamilies depending on their N-terminal domains. NLR family members detect (in most cases indirectly) degradation products of peptidoglycans, various forms of stress (e.g., UV-irradiation), microbial products and non-infectious crystal particles (12).
Interestingly, most, if not all PRRs that activate the transcription factors NF-κB, IRF or nuclear factor of activated T cells (NFAT) are sufficient to induce both T- and B-cell responses, whereas secreted PRRs and some endocytic PRRs (scavenger receptors and mannose receptor) cannot induce adaptive immunity by themselves (14). TLRs are the best characterized receptors that can trigger activation of adaptive immune responses of several effector classes, including immunoglobulin (Ig)M, IgG and IgA antibody responses, T helper (Th)1 and Th17 CD4+ T cell responses, and CD8+ T cell responses (3). In a pathological setting, TLR4 can also induce Th2 and IgE responses, although the functional significance of this pathway in protective immunity is currently unknown. Engagement of Dectin-1 and Dectin-2 can drive Th17 responses, which are required to clear fungal infections (4). Several recent studies have demonstrated that cytosolic PRRs, including RLRs and some NLRs, can also activate adaptive immunity (15–19). A cytosolic DNA sensor pathway is also sufficient for activation of Th1, cytotoxic CD8+ T cell and antibody responses through TANK-binding kinase-1 (TBK-1) (20).
The relative contribution of different PRRs to activation of specific arms of adaptive immune response during microbial infections is not fully understood. Somewhat surprisingly, inflammasomes, rather than signaling through the viral sensors RLRs, are required for adaptive immunity against influenza infection (21, 22). Similarly, infection with respiratory syncytial virus (RSV) activates MAVS-dependent anti-viral innate host defenses and yet, mice deficient in both MAVS and MyD88 (and adaptor used by most TLRs) can mount adaptive immune responses and clear RSV infection (23). Protective immunity to RSV was instead shown to depend on the NLR, NOD2 (24). For many microbial infections, including the well-studied pathogens Mycobacterium tuberculosis and Listeria monocytogenes, the relevant innate immune recognition pathways are unknown, though some obvious candidates have been excluded. Activation of protective CD8+ T cell responses to L. monocytogenes is MyD88-independent (25) and the intracellular stage of infection activates the transcription factor IRF3 (26), but the sensor responsible for the induction of the adaptive immune response is unknown. In the case of Mycobacteria infection, immune protection is MyD88 dependent, but this is likely due to the requirement for the interleukin (IL)-1 receptor signaling rather than TLR signaling (27). The sensors responsible for activation of adaptive immunity to Mycobacteria infection remain to be elucidated. Finally, the innate recognition events that trigger activation of adaptive immunity in response to retroviral and lentiviral infections, including HIV-1, are not fully understood. HIV-1 can activate TLR7 and TLR9 in plasmocytoid dendritic cells (28) and the antibody response to Friend murine leukemia virus infection is MyD88 dependent, whereas CD8+ T cells responses only partially depend on MyD88 (29). Thus, it is likely that retroviruses may also activate one of the intracellular nucleic acid sensing pathways, but the receptors and ligands involved still need to be determined.
Are cell intrinsic and cell extrinsic innate immune recognition equivalent?
Innate immune recognition can be cell intrinsic or cell extrinsic, depending on whether it is mediated by infected or non-infected cells (30). Cell extrinsic innate immune recognition is mediated by transmembrane receptors (including TLRs and Dectins); their activation does not require the cells expressing these receptors to be infected. In contrast, cell intrinsic innate immune recognition is mediated by intracellular sensors, including NLRs and RLRs. Activation of these receptors generally requires that the cell is infected. Accordingly, these PRRs are broadly expressed because most cells can potentially be infected by pathogens, especially by viruses. In contrast, cell extrinsic recognition is mainly mediated by specialized cells of the immune system, such as macrophages and dendritic cells (DC). Although both types of recognition can induce antimicrobial effectors upon activation they may trigger adaptive immunity by different mechanisms, as discussed in more detail below.
The principal distinction is the way the origin of antigens is established by cell extrinsic and cell intrinsic innate immune recognition. In the former case, the detection of a microbial cell or a viral particle by, for example a TLR expressed on DC, is followed by endocytosis or phagocytosis of the pathogen and subsequent processing and presentation of microbial antigens to T cells by major histocompatability complex (MHC) molecules. This presentation occurs in the context of several signals that are induced by the TLR and that are required for naïve T cell activation, including co-stimulatory signals and cytokines (Fig. 1). The microbial origin of the antigens is established through the physical association between an antigen and a PAMP that triggered the TLR. The physical association is primarily due to the co-occurrence of the antigen and a PAMP within the same particle (bacterial, yeast or protozoan cell or a viral particle). In cell biological terms, the association is interpreted, in part, through co-delivery of an antigen and a TLR ligand to the same phagosome or endosome, where the antigens are preferentially selected for presentation by MHC class II (31). During immunization, an antigen and a PAMP are generally mixed together and thus would not be perceived as having a common (microbial) origin unless both end up in the same endosome. This normally would require a large excess of PAMP over what is minimally required for activation of DCs. The co-recognition, however, is strongly facilitated by some adjuvants, such as mineral oil and alum, which promote antigen persistence and the co-recognition of the antigen and a PAMP. This effect of adjuvants can be substituted for by a physical association of an antigen and a PAMP, either by direct conjugation, or by co-absorption on the same particles, because in both cases they will localize to the same endosomes and the immune system will interpret this as an indication that the antigen is of microbial origin. Such associative recognition also explains why immunodominant antigens generally have both PAMP activity and antigenicity embedded within the same molecule. Examples include Toxoplasma profilin (32), several bacterial lipidated outer membrane proteins and flagellin; in each case these antigens are also PAMPs that can activate various TLRs. Similarly, in the case of auto-antigens that trigger TLR7- and TLR9-mediated autoimmunity, self antigens – ribonucleoproteins and chromatin complexes, contain both self antigens and TLR agonists.
Fig. 1.
Cell extrinsic recognition of pathogens. Bacteria detected by dendritic cells through Toll like receptors (TLRs) are internalized into phagosome where bacterial antigens are processed for presentation on MHC class II. Bacterial antigens (red) and pathogen-associated molecular patterns (PAMPs) (blue) are present in the same phagosome, which indicates to the decdritic cell their common origin. TLR-mediated recognition of bacterial PAMPs promotes the selection of bacterial antigens for optimal presentation on MHC class II. TLR signaling also leads to the induction of costimulatory molecules and cytokines necessary for activation and differentiation of T lymphocytes.
Associative recognition also plays an important role in cell extrinsic recognition by B cells. The co-engagement of the B cell receptor (BCR) with one of several innate immune signaling pathways, such as the C3dg complement component, results in a profound enhancement of antibody responses (33). In this case, C3dg ‘flags’ the antigen as foreign, thus instructing B cells about the origin of the antigen. Similarly, co-engagement of BCR and TLRs enhances antibody responses, as exemplified by the strong immunogenicity of flagellin and other antigens that have TLR agonist activity (14).
The situation is less clear for cell intrinsic immune recognition (Fig. 2). Indeed, the intracellular (cytosolic) sensors, such as RIG-I and MDA-5, detect viral nucleic acids in infected cells, which in most cases are not professional antigen presenting cells (APCs). In contrast to TLR-mediated recognition, where microbial antigens are ‘marked’ by physical association with microbial PAMPs, cell intrinsic sensing of viral nucleic acid is not known to be coupled to the viral antigens. It is possible that such an association does exist, and that RLR-mediated recognition of viral nucleic acids somehow promotes the selection of viral antigens for presentation to T cells. It is not obvious how this might work, especially if RLR-mediated recognition and antigen presentation occur in different cells – for example, in virally infected cells and in DC, respectively. Even when intracellular sensors are activated within an APC, it is unclear whether and how the microbial antigens are preferentially targeted for presentation to T cells. One possibility is that the innate recognition event somehow directs the microbial antigens for autophagic degradation followed by MHC presentation (Fig. 2). Indeed autophagy has been linked to MHC class II antigen presentation (34). Alternatively, cell intrinsic innate immune recognition may be coupled with induction of adaptive immunity by a mechanism that is independent of physical association but rather depends on another type of coincidence detection. Finally, a trivial, but likely incomplete explanation of selective activation of pathogen-specific T cells following cell intrinsic innate immune recognition is that all self antigen-specific T cells are deleted during negative selection, which would only allow for pathogen-specific T cells to become activated during an infection. This possibility, however, is inconsistent with the presence of mature self reactive T cells in peripheral tissues that are activated when mechanisms of peripheral tolerance are compromised. Moreover, cell intrinsic activation of a cytosolic DNA sensing pathway by endogenous DNA was recently shown to result in an autoimmune disease (35).
Fig. 2.
Cell intrinsic recognition. Dendritic cells directly infected by viruses recognize pathogen-associated molecular patterns(PAMPs) (blue) within the cytosol via RIG-I like receptors (RLRs). Cytosolic viral proteins (red) are processed and presented on MHC class I (via the conventional ER pathway) or major histocompatability complex (MHC) class II (via autophagy). RLR signaling leads to the induction of costimulatory molecules and cytokines necessary for activation and differentiation of T lymphocytes. How the origin of antigen is established in this case is unclear. In the case of MHC class I pathway, this may depend on the abundance of viral antigens. In the case of MHC class II pathway, it may depend on targeting of viral antigens by the autophagy machinery.
Whether the pathogen detection occurs through cell intrinsic or cell extrinsic mechanisms, DCs presumably always have to be directly activated by a PRR in order to activate T cell responses (36). But can DCs use cell extrinsic and cell intrinsic innate immune recognition pathways equally? Activation of the cell intrinsic pathway generally implies that the cell where recognition occurs is infected; however, infected DCs succumb to various pathogen-encoded strategies that can interfere with their function. Furthermore, most pathogens do not infect and replicate in DCs (37). When they do, such pathogens often utilize the biology of DCs to gain access to the target tissue within which they can replicate and disseminate. Examples of these include HIV-1 (38) and Venezuelan equine encephalitis virus (39). Indeed, in many cases, non-infected DCs present antigen derived from infected cells. The most extreme case of this is the antigen transfer model, in which DCs that have migrated from the infected tissue transfer antigen to the lymph node-resident DCs, thus amplifying T cell activation (40). How the origin of the antigen could be established in these models is not clear, although recent studies have suggested possible mechanisms. In one pathway, TLR3 in DC phagosomes detects viral nucleic acids (double stranded RNA) from infected cells and triggers DC activation and presentation of viral antigens on MHC class I (41) (Fig. 3). This finding is particularly interesting given the lack of evidence for the requirement of TLR3 in direct viral recognition by DCs (6). Thus, TLR3 might recognize viral double stranded RNA from infected cells but not the viruses themselves. Infected apoptotic cells also induce the production of transforming growth factor (TGF)-β and IL-6 by DCs, thus driving the differentiation of Th17 cells (42). Thus, cell extrinsic recognition of pathogens through the uptake of infected cells provides an additional layer of control of T cell responses generated by DCs.
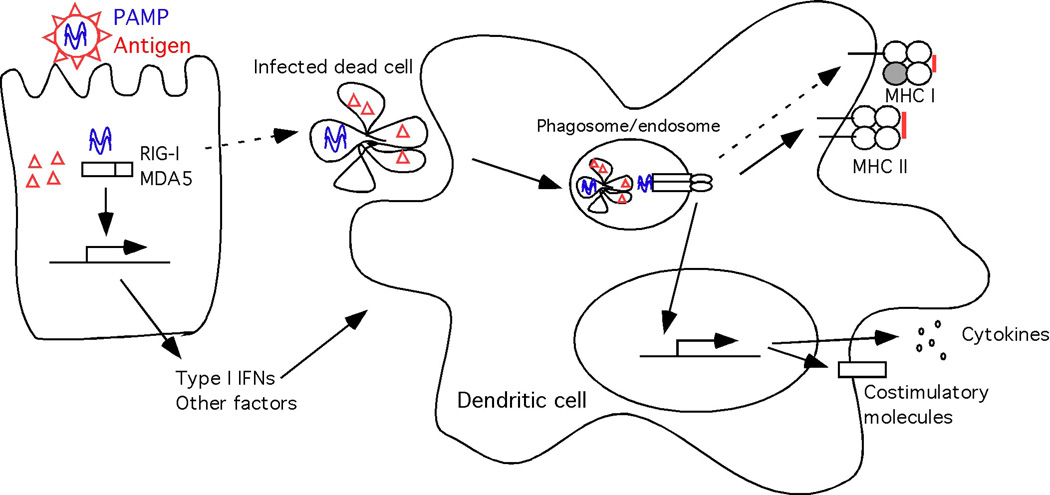
Fig. 3.
Cell extrinsic recognition of infected cells. A virally-infected non-antigen presenting cells recognizes pathogen-associated molecular patterns (PAMP) (blue lines indicate viral nucleic acids) within the cytosol via the RIG-I-like receptors (RLRs), leading to secretion of type I interferons (IFNs) and other factors that activate dendritic cells. Infected dead cells are taken up by non-infected dendritic cells and viral PAMPs (blue) are recognized through endosomal Toll like receptors (TLRs). Viral antigens (red) are processed and presented on major histocompatability (MHC) class II (via the conventional endosomal pathway) or MHC class I (via cross-presentation). TLR signaling leads to the induction of costimulatory molecules and cytokines necessary for activation and differentiation of T lymphocytes.
What is the role of pathogen recognition by APCs versus infected cells?
Besides being a source of microbial antigens, infected cells can engage DCs in other ways to provide critical signals for T cell activation. TLR-dependent signals in infected epithelial cells are required for DC-mediated induction of Th1 responses in response to herspes simplex virus (HSV)-2 or Toxoplasma gondii (43, 44). Moreover, epithelial cell-specific inactivation of NF-κB directs CD4+ T cell differentiation towards unprotective Th1/Th17 responses against Trichuris muris infection in the gut (45). Furthermore, the TLR3 ligand polyinosinic:polycytidylic acid (Poly I:C)-induced Th1 response to an HIV gag protein vaccine depends on MDA5-mediated recognition in both hematopoieitic and stromal cell compartments (16). These studies indicate that at least in some cases, PRR signaling in DCs alone cannot generate robust protective immunity, and that DCs must receive additional cues from the infected cells. Recognition of pathogens by the infected epithelial cells alone, however, cannot induce CD4+ T cell responses (43, 44). Rather, direct recognition of PAMPs by TLRs in DCs is required for CD4+ T cell activation (36). Collectively, these studies indicate that at least in some cases, DCs require two signals for CD4+ T cell activation: direct sensing of the PAMPs associated with the invading pathogen and detection of a PRR-induced signal from the infected cells. The observation that tissue-migrant DCs exposed to both of these signals are the primary APC for CD4+ T cell activation (46) supports this idea. The nature of the second signal likely depends on the pathogen and the tissue microenvironment.
Whether similar requirements also apply to the generation of CD8+ T cell responses is unclear. Infected cells present antigens on MHC class I, so that they can be killed by activated CD8+ T cells. To become activated, however, CD8+ T cells must recognize antigen presented by DCs, which in most cases are not infected by the same pathogen. Thus, to activate a CD8+ T cell response, uninfected DCs must present microbial antigens on MHC class I, in a process known as cross-presentation. In the antigen transfer model, migrant DCs from peripheral tissues, upon arrival in the draining lymph node, transfer antigens to blood-derived lymph node resident DCs (40). Blood-derived CD8α+ DCs in the lymph node are the predominant APCs for CD8+ T cells upon infection with influenza, HSV-1 or Listeria, or encounter with apoptotic cells (40). How do these DCs receive the proper cues to become competent to prime CD8+ T cells? PAMPs and antigens may be preserved within the migrant DCs, allowing CD8α+ DCs to acquire such information and drive CD8+ T cell activation. Alternatively, cross priming by DCs may not require signals from pathogens or infected cells, but instead may require a signal from antigen-specific CD4+ T cells. For example, CD4+ T cell help is necessary for DCs to activate the CD8+ T cell response during HSV-1 infection (47). Here, the information regarding the pathogen and the microenvironment already has been processed by the CD4+ T cells, thus alleviating the need for CD8+ T cells to do the same. Future studies will help resolve this issue.
Are endogenous and microbial TLR ligands equivalent?
In addition to recognition of microbial structures, several studies have demonstrated that some TLRs are also involved in sensing endogenous signals generated during tissue injury. Although some initial reports were likely artifacts caused by contamination of recombinant protein preparations, more recent analyses revealed a number of cases of bona fide endogenous TLR stimulators. One class of endogenous TLR ligands is chromatin fragments and ribonucleoprotein complexes released from dead cells. When clearance of apoptotic cells is insufficient, these complexes can activate TLR7 and TLR9 on DC and B cells, which can result in the development of systemic autoimmune diseases (48). In these cases TLR activation by self nucleic acids is clearly unintended. Self nucleic acids are simply mistaken for microbial.
Unlike the accidental TLR recognition of endogenous nucleic acids, detection of other endogenous ligands might serve a physiological purpose. The two common sources of endogenous TLR ligands are components of extracellular matrix (ECM), and intracellular proteins. Inflammation and injury cause degradation and accumulation of several ECM components. Small molecular weight fragments of hyaluronic acid (HA) (49), biglycan (50) and versican produced by tumor cells (51) can trigger TLR2 and/or TLR4 activation. Fragments of heparan sulfate, an acidic polysaccharide found in cell membranes and extracellular matrices, activates DCs through TLR4 (52). Furthermore, several intracellular proteins have been suggested to activate TLRs, including the high-mobility group box 1 (HMGB1) protein, which normally resides in the nucleus but is thought to be secreted or released from damaged or necrotic cells. Extracellular HMGB1 has pro-inflammatory effects, which are mediated by TLRs 2, 4 and 9 and the receptor for advanced glycation end products (RAGE) (53).
Both biglycan and HA fragments accumulate during tissue injury and activate macrophages to produce inflammatory chemokines and cytokines via TLR2 and TLR4 (49, 50). Biglycan-deficient mice were less susceptible to death caused by TLR2 or TLR4-dependent sepsis due to lower amounts of circulating tumor necrosis factor (TNF)-α and reduced leukocyte infiltration in the lung (50). Similarly, TLR4-mutant mice secrete less inflammatory cytokines after ischemic reperfusion, and this effect was mimicked by neutralization of HMGB1 in control but not TLR4-mutant mice (54). In contrast, in a non-infectious lung injury model, mice deficient in both TLR2 and TLR4 show impaired leukocyte recruitment, increased tissue injury and decreased survival (49). These studies indicate that several endogenous ligands provide signals through TLR2 and TLR4 to initiate inflammatory responses and promote tissue protection and repair.
These studies raise an important issue: do microbial and endogenous agonists of TLR2 or TLR4 trigger identical responses? Microbial stimulators of TLRs activate inflammatory, tissue repair and adaptive immune responses. Endogenous stimulators of TLRs 2 and 4 are only known to induce the inflammatory and tissue reparative responses. Activation of TLRs in the absence of infection can lead to autoimmune responses, as illustrated by the effects of accidental stimulation of TLR7 and TLR9 by self nucleic acids (48). It is thus reasonable to assume that the endogenous TLR2/4 agonists, unlike their microbial counterparts, do not induce activation of adaptive immune responses (Fig. 4). Indeed, TLR2 activation by necrotic cells was shown to induce expression of inflammatory and tissue repair genes, but not genes associated with adaptive immunity (55). Likewise, HA triggers signals distinct from LPS, by engagement of TLR4, MD2, and CD44 (56).
Fig. 4.
Proposed consequences of Toll like receptor (TLR) recognition of exogenous versus endogenous ligands. TLR engagement by exogenous (A) or endogenous (B) agonists leads to signaling from distinct subcellular compartments (indicated by blue and green, respectively) and/or engagement of coreceptors. Consequently, exogenous ligands induce transcription of genes leading to inflammation, tissue repair and the initiation of adaptive immunity (A). In contrast, endogenous ligands induce TLR signaling for activation of inflammation and tissue repair but not the initiation of adaptive immunity (B).
Thus, the physiological, endogenous ligands of TLR2 and TLR4 likely trigger signals distinct from their microbial counterparts and promote specifically genes involved in tissue homeostasis and repair. The differential signaling by microbial versus endogenous TLR ligands may be induced due to the engagement of different co-receptors (Figure Fig. 4). Indeed, HA, but not LPS, signals through both CD44 and TLR4, while HMGB1 signals through TLRs and RAGE (53, 56). Thus, differential co-receptors usage may potentially influence the signaling pathways induced by microbial and endogenous TLR ligands. Moreover, there are examples of differential TLR signaling from distinct sub-cellular compartments (57). It will be important to address these and other possibilities in the future studies because the prevailing view of the role of endogenous ligands as ‘danger signals’ that activate the adaptive immune responses is likely incorrect. Physiological endogenous activators of TLRs, or any other PRRs, have not yet been shown to be sufficient to activate adaptive immune responses, whereas ‘unintended’ stimulation of TLR7 and TLR9 results in autoimmune responses. This also applies to the endogenous activators of inflammasomes. This is illustrated by the lack of autoimmunity in patients with gout, a condition caused by inflammasome activation by endogenous uric acid crystals (12). Furthermore, genetic mutations in the inflammasome components lead to autoinflammatory diseases that differ from autoimmune disorders in that they do not involve activation of autoreactive T and B cell responses (58).
Concluding remarks
As the basic functions of TLRs are becoming increasingly well defined, many new questions emerge. One fundamental issue is that microbial TLR ligands are not unique to pathogens, but instead are common to all microbes of a given class. This creates a problem of discrimination between commensals and pathogens. One possibility is that pathogens are distinguished from commensals due to their unique virulence activities, such as production of pore-forming toxins. The notion of pathogen-commensal discrimination is complicated, however, because distinctions between them are often arbitrary and conditional upon the host immune status. A widespread assumption is that the immune system has to discriminate between commensals and pathogens, such that immune responses are generated exclusively towards the latter. It could be argued, however, that the immune system handles all microbes in the same way. Immune responses are in fact generated against commensals, and moreover, commensals maintain their ‘innocuous’ status towards the host in part because they are actively suppressed by the immune system. Of course the immune response to microbes in highly colonized tissues, such as the intestine, is tightly regulated and has a distinct modality, so as to avoid immunopathology. Specific forms of immune responses to commensals do exist under normal conditions, however, as exemplified by commensal-specific IgA antibodies normally present in the intestinal lumen (59). This may be the reason why TLRs recognize structures present on all microbes, whether they are known to cause a disease under a particular condition or not.
Future studies will likely reveal additional mechanisms of immune recognition that may be superimposed on PRR-mediated recognition to ensure differential responses to commensals, pathogens, and endogenous TLR ligands. And perhaps the most interesting aspects of innate immune recognition are yet to be discovered. Though the field may be seen as approaching the beginning of the end, it is in fact just at the end of the beginning.
Acknowledgments
The work in the authors’ laboratories is supported by the grants from NIH: R01AI055502, R37AI046688, R01DK071754 (RM) and R01AI064705, R21AI083242, R01AI054359 (AI), by the Howard Hughes Medical Institute (RM), and by the Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (AI).
References and Notes
- 1.Janeway CA., Jr Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1. [PubMed] [Google Scholar]
- 2.Takeda K, Akira S. International Immunology. 2005;17:1. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki A, Medzhitov R. Nature immunology. 2004 Oct;5:987. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 4.Robinson MJ, et al. The Journal of experimental medicine. 2009 Aug 31;206:2037. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GD. Nature Rev. Immunol. 2006;6:33. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Reis e Sousa C. Immunity. 2007;27:370. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Saito T, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:582. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoneyama M, Fujita T. Immunity. 2008;29:178. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi O, Akira S. Immunol Rev. 2009 Jan;227:75. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu YH, Macmillan JB, Chen ZJ. Cell. 2009 Jul 22; doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ablasser A, et al. Nature immunology. 2009 Jul 16; [Google Scholar]
- 12.Martinon F, Mayor A, Tschopp J. Annu Rev Immunol. 2009;27:229. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 13.Kanneganti TD, Lamkanfi M, Nunez G. Immunity. 2007 Oct;27:549. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Palm NW, Medzhitov R. Immunol Rev. 2009 Jan;227:221. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 15.Kumar H, Koyama S, Ishii KJ, Kawai T, Akira S. J Immunol. 2008 Jan 15;180:683. doi: 10.4049/jimmunol.180.2.683. [DOI] [PubMed] [Google Scholar]
- 16.Longhi MP, et al. J Exp Med. 2009 Jul 6;206:1589. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngoi SM, Tovey MG, Vella AT. J Immunol. 2008 Dec 1;181:7670. doi: 10.4049/jimmunol.181.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz JH, et al. Immunity. 2007 Apr;26:445. doi: 10.1016/j.immuni.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi KS, et al. Science. 2005 Feb 4;307:731. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 20.Ishii KJ, et al. Nature. 2008 Feb 7;451:725. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 21.Koyama S, et al. J Immunol. 2007 Oct 1;179:4711. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 22.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. The Journal of experimental medicine. 2009 Jan 12; doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhoj VG, et al. Proc Natl Acad Sci U S A. 2008 Sep 16;105:14046. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabbah A, et al. Nature immunology. 2009 Oct;10:1073. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Way SS, Kollmann TR, Hajjar AM, Wilson CB. J Immunol. 2003 Jul 15;171:533. doi: 10.4049/jimmunol.171.2.533. [DOI] [PubMed] [Google Scholar]
- 26.O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Proc Natl Acad Sci U S A. 2002 Oct 15;99:13861. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiling N, Ehlers S, Holscher C. Immunol Lett. 2008 Feb 15;116:15. doi: 10.1016/j.imlet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Beignon AS, et al. J Clin Invest. 2005 Nov;115:3265. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browne EP, Littman DR. PLoS Pathog. 2009 Feb;5:e1000298. doi: 10.1371/journal.ppat.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stetson DB. Curr Opin Immunol. 2009 Jun;21:244. doi: 10.1016/j.coi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blander JM, Medzhitov R. Nature. 2006 Apr 6;440:808. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- 32.Yarovinsky F, Kanzler H, Hieny S, Coffman RL, Sher A. Immunity. 2006 Sep 22; doi: 10.1016/j.immuni.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. Science. 1996 Jan 19;271:348. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 34.Schmid D, Munz C. Immunity. 2007 Jul;27:11. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Cell. 2008 Aug 22;134:587. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sporri R, Reis e Sousa C. Nature immunology. 2005 Feb;6:163. doi: 10.1038/ni1162. [DOI] [PubMed] [Google Scholar]
- 37.Banchereau J, Steinman RM. Nature. 1998;392:245. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 38.de Jong MA, Geijtenbeek TB. J Intern Med. 2009 Jan;265:18. doi: 10.1111/j.1365-2796.2008.02046.x. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald GH, Johnston RE. Journal of virology. 2000 Jan;74:914. doi: 10.1128/jvi.74.2.914-922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbone FR, Belz GT, Heath WR. Trends Immunol. 2004 Dec;25:655. doi: 10.1016/j.it.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 41.Schulz O, et al. Nature. 2005 Feb 24;433:887. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 42.Torchinsky MB, Garaude J, Martin AP, Blander JM. Nature. 2009 Mar 5;458:78. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 43.Sato A, Iwasaki A. Proc Natl Acad Sci U S A. 2004 Nov 16;101:16274. doi: 10.1073/pnas.0406268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minns LA, et al. J Immunol. 2006 Jun 15;176:7589. doi: 10.4049/jimmunol.176.12.7589. [DOI] [PubMed] [Google Scholar]
- 45.Zaph C, et al. Nature. 2007 Mar 29;446:552. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 46.Germain RN, Jenkins MK. Curr Opin Immunol. 2004 Feb;16:120. doi: 10.1016/j.coi.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Smith CM, et al. Nature immunology. 2004 Nov;5:1143. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- 48.Marshak-Rothstein A, Rifkin IR. Annu Rev Immunol. 2007;25:419. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 49.Jiang D, et al. Nature medicine. 2005 Nov;11:1173. doi: 10.1038/nm1315. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer L, et al. J Clin Invest. 2005 Aug;115:2223. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, et al. Nature. 2009 Jan 1;457:102. [Google Scholar]
- 52.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. J Immunol. 2002 May 15;168:5233. doi: 10.4049/jimmunol.168.10.5233. [DOI] [PubMed] [Google Scholar]
- 53.van Beijnum JR, Buurman WA, Griffioen AW. Angiogenesis. 2008;11:91. doi: 10.1007/s10456-008-9093-5. [DOI] [PubMed] [Google Scholar]
- 54.Tsung A, et al. The Journal of experimental medicine. 2005 Apr 4;201:1135. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, et al. J Immunol. 2001 Jun 15;166:7128. doi: 10.4049/jimmunol.166.12.7128. [DOI] [PubMed] [Google Scholar]
- 56.Taylor KR, et al. J Biol Chem. 2007 Jun 22;282:18265. doi: 10.1074/jbc.M606352200. [DOI] [PubMed] [Google Scholar]
- 57.Barton GM, Kagan JC. Nat Rev Immunol. 2009 Aug;9:535. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masters SL, Simon A, Aksentijevich I, Kastner DL. Annu Rev Immunol. 2009;27:621. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. Mucosal Immunol. 2008 Jan;1:11. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]