Abstract
Enterohemorrhagic Escherichia coli O157:H7 is a major foodborne pathogen causing severe disease in humans worldwide. Healthy cattle are a reservoir of E. coli O157:H7, and bovine food products and fresh produce contaminated with bovine waste are the most common sources for disease outbreaks in the United States. E. coli O157:H7 also survives well in the environment. The abilities to cause human disease, colonize the bovine gastrointestinal tract, and survive in the environment require that E. coli O157:H7 adapt to a wide variety of conditions. Three major virulence factors of E. coli O157:H7 have been identified including Shiga toxins, products of the pathogenicity island called the locus of enterocyte effacement, and products of the F-like plasmid pO157. Among these virulence factors, the role of pO157 is least understood. This review provides a board overview of E. coli O157:H7 with an emphasis on pO157.
Keywords: E. coli O157:H7, pO157
Escherichia coli O157:H7
Escherichia coli
Escherichia coli (E. coli) is a Gram-negative, rod-shaped, facultative anaerobic bacterium. This microorganism was first described by Theodor Escherich in 1885. Most E. coli strains harmlessly colonize the gastrointestinal tract of humans and animals as a normal flora. However, there are some strains that have evolved into pathogenic E. coli by acquiring virulence factors through plasmids, transposons, bacteriophages, and/or pathogenicity islands. These pathogenic E. coli can be categorized based on serogroups, pathogenicity mechanisms, clinical symptoms, or virulence factors [33, 47]. Among them, enterohemorrhagic E. coli (EHEC) is defined as pathogenic E. coli strains that produce Shiga toxins (Stxs) and cause hemorrhagic colitis (HC) and the life-threatening sequelae hemolytic uremic syndrome (HUS) in humans. Several serotypes in EHEC are frequently associated with human diseases such as O26:H11, O91:H21, O111:H8, O157:NM, and O157:H7 [44, 51]. E. coli O157:H7 is the most frequently isolated serotype of EHEC from ill persons in the United States, Japan, and the United Kingdom and it the focus of this review.
History
EHEC serotype O157:H7 was first recognized in 1982 as a human pathogen associated with outbreaks of bloody diarrhea in Oregon and Michigan, U.S.A. [57, 71] and is also linked to sporadic cases of HUS in 1983 [34]. Since then, many outbreaks associated with EHEC have been reported in the United States and E. coli O157:H7 has become one of the most important foodborne pathogens.
Prevalence and Economic Cost
The Centers for Disease Control and Prevention (CDC) has estimated that E. coli O157:H7 infections cause 73,000 illnesses, 2,200 hospitalizations, and 60 deaths annually in the United States [43]. The outbreak surveillance data from CDC reported that E. coli O157:H7 infections are decreasing after the peak in 1999. However, large outbreaks and sporadic cases continue to occur. The annual cost of illness due to E. coli O157:H7 infections was 405 million dollars, including lost productivity, medical care, and premature deaths [21]. The high cost of illness requires additional efforts to control this pathogen.
Isolation and Identification
E. coli O157:H7 expresses somatic (O) antigen 157 and flagella (H) antigen 7. E. coli O157:H7 has unique features of delayed D-sorbitol fermentation (>24 h) and inability of producing β-glucuronidase, which can hydrolyze a synthetic molecule, 4-methyl-umbelliferyl-D-glucuronide (MUG) [68]. Thus, Sorbitol MacConkey (SMAC) agar supplemented with MUG has been used for detection of E. coli O157:H7. To increase the selectivity for E. coli O157:H7, cefixime, potassium tellurite, and vancomycin have been added to SMAC agar plates to inhibit other Gram-negative flora. The serotypes O157 and H7 can be further confirmed by a commercially available latex agglutination assay.
Genomic Organization
The chromosomal size of E. coli O157:H7 is 5.5 Mb. This genome includes a 4. 1 Mb backbone sequence conserved in all E. coli strains. The remaining are specific to E. coli O157:H7 [53]. Additionally, genome comparison of E. coli O157:H7 with nonpathogenic E. coli K12 shows that 0.53 Mb of DNA is missing for E. coli O157:H7, suggesting genomic reduction has also played a role in E. coli O157:H7 evolution [17, 53]. The majority of E. coli O157:H7-specific DNA sequences (1.4 Mb) are horizontally transferred foreign DNAs such as prophage and prophage-like elements. E. coli O157:H7 contains 463 phage-associated genes compared with only 29 in E. coli K-12 [72]. A change in G+C contents is one of the indications that a genomic region has been acquired by horizontal transfer, and Putonti et al. [55] estimated that at least 53 different species have contributed to these unique sequences in E. coli O157:H7. Virulence-associated genes between two sequenced E. coli O157:H7 strains are nearly identical (99%). Clearly, both the acquisition and loss of DNA have played an important role in the evolution of pathogenesis of E. coli O157:H7.
Evolution
Several comparative and epidemiological studies indicate that E. coli O157:H7 may have descended from the non-toxigenic and less virulent strain E. coli O55:H7 [72]. E. coli O15:H7 has emerged through four sequential events; (i) acquisition of an stx2-containing bacteriophage, (ii) acquisition of pO157 and the rfb region, (iii) acquisition of the stx1-containing bacteriophage, and (iv) loss of the ability to ferment D-sorbitol and loss of beta-glucuronidase (GUD) activity.
Animal Reservoir
Cattle are the major reservoir of E. coli O157:H7 and this reservoir host is generally asymptomatic when carrying this microorganism. There are rare cases of diarrheal disease in young calves with this serotype. The proportion of cattle shedding at any one time varies. Sheep, goats, pigs, and turkeys have also been found to shed E. coli O157:H7 in their feces.
Molecular Subtyping
A variety of molecular subtyping methods have been developed to improve the understanding of the epidemiology of E. coli O157:H7 outbreaks. These methods include pulse-field gel electrophoresis (PFGE), restriction fragment length polymorphisms (RFLP), amplified fragment-length polymorphisms (AFLP), and phage typing [65, 73]. Among them, the PFGE method was standardized by CDC and has been applied successfully to discriminate outbreak-associated, sporadic, or unrelated infections since 1993 [3].
Infection
E. coli O157:H7 infection is a major public health concern in North America, Europe, and other areas of the world. Although the total case numbers of E. coli O157:H7 infections are lower than those of other enteric pathogens such as Salmonella or Campylobacter spp., the diseases caused by E. coli O157:H7 showed much higher hospitalization and fatality rates [43]. Human infection caused by E. coli O157:H7 can present a broad clinical spectrum ranging from asymptomatic cases to death. Most cases initiate with non-bloody diarrhea and self-resolve without further complication. However, some patients progress to bloody diarrhea or HC in 1–3 days. In 5–10% of HC patients, the disease can progress to the life-threatening sequelae, HUS or thrombocytopenic purpura (TTP) [1]. E. coli O157:H7 is the most common cause of HUS in the United States. Children and the elderly are at increased risk of severe clinical symptoms such as HUS.
Several strategies for therapy have been studied including the use of antibiotics and vaccination. However, there is no specific treatment for E. coli O157:H7 infection and the use of antibiotics may be contraindicated. Therefore, treatment is mainly supportive to limit the duration of symptoms and prevent systemic complications. Given this status, highly effective measures for prevention and control of E. coli O157:H7 infections are essential.
Transmission
In the United States, the most frequent route of transmission for E. coli O157:H7 infections is via consumption of contaminated food and water [56]. However, it can also be spread directly from person to person, particularly in child day-care facilities, and from animal to person. Infections have been documented from people visiting petting zoos, dairy farms, or camp grounds where cattle have previously grazed [28, 31]. Recently, potential airborne transmission has been reported in a contaminated building having an animal exhibit [70]. Of the 350 outbreaks reported to the CDC from 1982 to 2002, the determined transmission routes were foodborne (52%), unknown (21%), person-to-person (14%), waterborne (9%), and animal contact (3%) [56]. The model of transmission of E. coli O157:H7, which is updated from the diagram by Gansheroff and O’Brien [23] is shown in Fig. 1. These various transmission routes can be explained by the very low infectious dose (~50 CFU) of E. coli O157:H7.
Fig. 1.
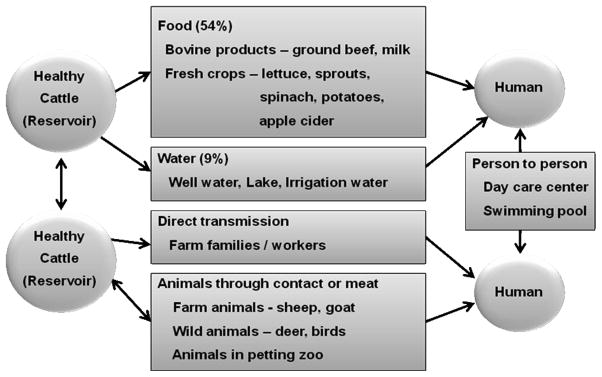
Transmission of E. coli O157:H7.
Healthy cattle are the major reservoirs of E. coli O157:H7 and carry this microorganism transiently without symptoms. Contaminated bovine products and crops are predominant sources for human infections.
Cattle are the natural reservoir of E. coli O157:H7. Between 1% and 50% of healthy cattle carry and shed E. coli O157:H7 in their feces at any given time [13, 18, 27]. Contaminated ground beef is the most common vehicle for E. coli O157:H7 outbreaks. Beef products may become contaminated during slaughter, and the process of grinding beef may transfer pathogens from the surface of the meat to the interior. Therefore, if ground beef is incompletely cooked, the bacteria can survive. In addition, there are a variety of contaminated food vehicles other than ground beef that have been linked to E. coli O157:H7 incidences, including unpasteurized milk, drinking water, salami, beef jerky, and fresh produce such as lettuce, radish sprouts, fresh spinach, and apple cider. The largest outbreak was traced to radish sprout contamination (1996) in Osaka, Japan, where 7,966 individuals were diagnosed with confirmed infections [45]. Epidemiological studies indicate that these food products seem to have been contaminated through bovine fecal materials. Therefore, prevention of E. coli O157:H7 in cattle can be one of the most important control methods. To control E. coli O157:H7 on the farm, the improvement of cattle management practices, the identification of inhibitory feeds, immunization, the utilization of feeding additives, and the use of probiotic cultures have been proposed.
Acid Resistance of E. coli O157:H7
Acid resistance (AR) is the ability of bacteria to protect themselves from extremely low pH (<pH 3.0). The low pH in the stomach (pH 1.5 to 3.0) is one of the first host defenses against foodborne enteric pathogens [54]. The ability to survive in the acidic environment of the stomach increases the chance of bacteria to colonize the intestines and cause infection. Acid resistance is associated with a lowering of the infectious dose of enteric pathogens [60]. The low infectious dose is one of the best known characteristics of E. coli O157:H7, making this bacterium highly infectious.
A variety of studies have reported the AR of E. coli O157:H7 strains [5, 12]. These studies have determined three efficient AR systems. The first AR system requires the alternative sigma factor RpoS and glucose repression. The rpoS mutant of E. coli O157:H7 was shed in lower numbers in experimentally infected mice and calves. The second AR system requires the addition of arginine during exposure of acidic condition. The arginine decarboxylase (adiA) and the regulator of adiA (cysB) were reported in this second AR system. The third AR system requires glutamate for protection at low pH condition. Essential components of this AR system include two isozymes of glutamate decarboxylase (gadA or gadB), and a putative glutamate, γ-amino butyric acid antiporter (gadC). Whereas only one of the two glutamate decarboxylases is neededfor protection at pH 2.5, bothglutamate decarboxylase isozymes are required at pH 2.0. Previous results revealed that glutamate-dependent AR is the most effective protection at pH 2.0 in complex medium. E. coli O157:H7 possesses three overlapping AR systems, but the control and requirements for AR activity are different in each AR system.
Other than these three AR systems, several proteins involved in AR of E. coli O157:H7 have been identified. These proteins include chaperone HdeA, RNA polymerase-associated protein SspA, and DNA-binding protein Dps. Moreover, it was shown that alterations in the cell wall membrane or colonic acid production are associated with successful AR. Thus, E. coli O157:H7 utilizes different AR systems based on the type of acidic environment naturally encountered.
Colonization of Cattle
E. coli O157:H7 naturally colonizes the gastrointestinal tracts of cattle, and the lymphoid follicle-dense mucosa at the terminal rectum, called the rectoanal junction (RAJ) mucosa, is known as a principal site of colonization in cattle [39, 48].
Three distinct patterns of E. coli O157:H7 carriage in cattle have been described previously [14, 39, 58]. First, animals can be transiently culture positive for short durations of a few days and are considered passive shedders and are likely not colonized at the RAJ mucosa. Second, cattle can be colonized and shed the bacteria for an average of 1 month and typically not longer than 2 months. Third, a few rare animals are colonized for a long duration and shed the bacteria from 3 to 12 months or longer. This unique situation, in which a few animals develop long-duration colonization (>2 months) with E. coli O157:H7, is likely due to bacterial association at the RAJ mucosa; however, it may be due to unique colonization by the bacteria at a site(s) in addition to the RAJ mucosa.
Age, diet, and immunity of individual cattle could also potentially affect bacterial colonization. Cray and Moon [14] reported that calves shed E. coli O157:H7 longer than adult cattle given the same level of E. coli O157:H7 inoculums.
Reducing the level of carriage of E. coli O157:H7 in cattle, as a major source of E. coli O157:H7 infection, would play a key role in decreasing the risk of human infection. The understanding of colonization factors of E. coli O157:H7 will be necessary to develop effective strategies for reducing or preventing bovine carriage of E. coli O157:H7.
Environmental Survival
E. coli O157:H7 can survive and persist in numerous environments such as soil, water, and food as well as in animal reservoirs (Fig. 2). E. coli O157:H7 has been shown to survive for a year in manure-treated soil and for 21 months in raw manure that had not been composted [30]. Composting manure is effective in destroying E. coli O157:H7, if the temperature is maintained above 50°C for 6 days. E. coli O157:H7 can survive for a long time in water, especially at cold temperatures. Water trough sediments contaminated with bovine feces can serve as a long-term (>8 months) reservoir of E. coli O157:H7, and the surviving bacteria in contaminated troughs is a source of infection [38]. Barker et al. [2] showed that E. coli O157:H7 survives and replicates in Acanthamoeba polyphaga. A. polyphaga is a common environmental protozoan that is widely distributed in soil, water, and fecal slurry. Thus, it can be an efficient transmission vehicle of E. coli O157:H7 in these environments.
Fig. 2.
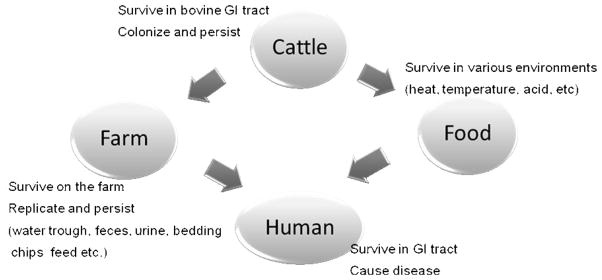
Ecological scheme of E. coli O157:H7.
GI tract, gastrointestinal tract.
To survive in varied environments, E. coli O157:H7 requires the ability to adapt to variations or extreme changes in temperature, pH, and osmolarity conditions commonly encountered in nature. For example, the exopolysaccharide (EPS) production of E. coli O157:H7 is associated with heat and acid tolerance, and the alteration of lipid composition in membranes is induced by heat stress [77].
These environmental adaptations of E. coli O157:H7 play an important role in the persistence and dissemination of this microorganism on farms and the increasing transfer from cattle to cattle. In addition, the ability to survive outside the host reservoir increases the risk that the pathogen may contaminate crops and produce via bovine manure contamination, irrigation with contaminated water, or direct contact with infected animals [42].
Major Virulence Factors
Defining the virulence factors and mechanisms of E. coli O157:H7 pathogenesis has been a focus of numerous studies (Fig. 3). The production of Stxs is considered essential but not solely responsible for disease. In addition, E. coli O157:H7 associated with severe human disease must colonize the intestinal mucosa-and the possessing of pO157 also correlates with the ability to cause disease. Each of these aspects is described below.
Fig. 3.
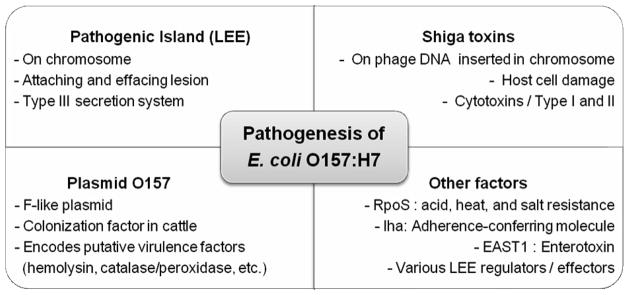
Virulence factors of E. coli O157:H7.
Shiga Toxins (Stxs)
Stx is a potent cytotoxin and is bacteriophage-encoded. Stx is expanded from a single transcriptional unit and causes damage to a variety of cell types [29]. Stxs can be divided into two groups called Stx1 and Stx2 but do not generate cross-reactive antibodies that are 56% homologous in amino acid sequences. Stx1 is identical to Stx from Shigella dysenteriae I, but for a single amino acid difference. Virulent isolates of E. coli O157:H7 can express Stx1 only, Stx2 only, or both toxins. Stx2 is known to be more toxic and is more often associated with HC or HUS in human infections than are Stx1 strains [6, 50].
Stx has a conserved structure consisting of one enzymatically active A subunit (A1) and five identical receptor-binding B subunits (B5). The B5 subunit binds to the specific host receptors globotriaosylceramide (Gb3) or globotetraosylceramide (Gb4) [47]. After binding of Stx (A1B5) to the host cell, the A subunit is internalized to the cytoplasm. Al inhibits protein synthesis by the specific removal of a single adenine residue from the 28S rRNA of the 60S ribosomal subunit [59]. The detailed mechanisms of Stx translocation to various tissues are not fully understood.
The Locus of Enterocyte Effacement
E. coli O157:H7 colonizes the intestinal mucosa and induces a characteristic histopathological lesion referred to as attaching and effacing (A/E) lesions. The A/E lesion is characterized by effacement of microvilli and bacterial adherence to the epithelial cell membrane. Attached bacteria stimulate host cell actin polymerization accumulation, resulting in a raised attachment pedestal [11]. Genetic studies have shown that the genes responsible for A/E lesions map to a 13 region, which has been designated the locus of enterocyte effacement (LEE). The LEE of E. coli O157:H7 is conserved in EPEC as well, and it is well known that the presence of the LEE is strongly associated with disease [24]. The LEE of E. coli O157:H7 is 43 kb in size and contains an additional 7.5 kb prophage sequence compared with EPEC strains. The role of this additional sequence is not clearly defined. The LEE is composed of at least 41 different genes organized into three major regions; (i) a type III secretion system (TTSS) that exports effecter molecules; (ii) an adhesion called intimin and its translocated receptor, Tir, which is translocated into the host cell membrane by the TTSS; and (iii) several secreted proteins (Esp) as a part of TTSS, which are important in modification of host cell signal transduction during the formation of A/E lesions [15, 52]. Recently, non-LEE encoded effectors have also been identified, and the elucidation of their roles will further increase the understanding of the pathological phenomena in E. coli O157:H7 infections [16].
Plasmid O157 (pO157)
In addition to Stxs and the LEE, which both are chromosomally encoded, all clinical isolates of E. coli O157:H7 possess a putative virulence plasmid called pO157.
Plasmid O157 (pO157)
Plasmids
A plasmid is an extrachromosomal DNA that is capable of replicating independently of the chromosomal DNA. Plasmids are mobile elements that provide various host beneficial traits, such as resistance to antibiotics and heavy metals, production of toxins and other virulence factors, biotransformations of hydrocarbons, and symbiotic nitrogen fixation [22]. Plasmid-encoded genes are required for full pathogenesis in many enteropathogenic bacteria including Shigella, Yersinia, Salmonella, and E. coli species.
pO157
E. coli O157:H7 contains a highly conserved plasmid, named pO157. The pO157 is a nonconjugative F-like plasmid with a range size from 92 to 104 kb. The complete sequence of pO157 in two different outbreak isolates has been published [10, 41]. The pO157 shows a dynamic structure and includes different mobile genetic elements such as transposons, prophages, insertion sequences (IS), and parts of other plasmids. The heterogeneous composition of pO157 can delimit the co-responses to functional regions of pO157. Among them, IS or remnants of IS are frequently associated with the virulence-related segments, which are similar to compositions of the large virulence plasmid in Shigella spp. [10, 41]. These results indicate that the actual pO157 is formed by integration of fragments from evolutionally different species origins into an F-like plasmid, and thus virulence factors or putative virulence factors on the different segments of pO157 may be from different origins. The complete sequence of pO157 reveals 100 open reading frames (ORFs) [10]. Among them, 43 ORFs showed sufficient similarities to known proteins, suggesting functions, and 22 ORFs had no convincing similarity with any known proteins. Thirty-five proteins are presumably involved in the pathogenesis of E. coli O157:H7 infections, but of which only 19 genes have been previously characterized including a hemolysin (ehxA) [63], a catalase-peroxidase (katP) [9], a type II secretion system apparatus (etp) [62], a serine protease (espP) [8], a putative adhesin (toxB) [67], a zinc metalloprotease (stcE) [37] and an eae conserved fragment (ecf) [75]. However, the biological significance of pO157 in pathogenesis is not fully understood.
Hemolysin (ehx)
Hemolysin was the first described virulence factor of pO157 [4, 61]. The hemolysin operon (ehxCABD) may be foreign in origin because it has a different G+C% and codon usage than the surrounding genetic contents. A 3.4-kb fragment encodes genes required for hemolysin synthesis and transport, and this region has been used as a diagnostic probe for E. coli O157:H7 and often EHEC isolates. Several studies showed that hemolysin is highly conserved among different serotypes of EHEC such as O157:H7, O111:H8, and O8:H19, but it is not known if these have identical biological activities [7].
Catalase-Peroxidase (katP)
A gene for a catalase-peroxidase activity (katP) was identified from pO157 [9]. This gene is 2.2 kb in size and is highly homologous to the bacterial bifunctional catalase-peroxidase. The KatP enzyme activity of E. coli O157:H7 was shown in both cytoplasm and periplasm fractions. The N-terminal signal sequence suggests that this enzyme is transported through the cytoplasmic membrane. The katP gene was found in all E. coli O157:H7 strains but is not found in EPEC, ETEC, EIEC, and EAggEC strains. This enzyme may help E. coli O157:H7 colonize host intestines by reducing oxidative stress and using the by-product oxygen in diminished or deprived oxygen conditions of the host intestine.
Type II Secretion System (T2SS) (etp)
The pO157 encodes 13 ORFs named etpC to etpO, which show high similarities to T2SS of Gram-negative bacteria [62]. These genes are located adjacent to the hemolysin locus. An IS911-like insertion element was found to be located far from the etp and ehx genes. Similar to the katP gene, etp genes were also found in all E. coli O157:H7 strains, some in non-O157 EHEC strains, and not found in EPEC, ETEC, EIEC, and EAggEC strains. This T2SS is similar to the pullulanase secretion pathway (pulO) of Klebsiella oxytoca, but its function has not been identified.
Serine Protease (espP)
EspP is the pO157-encoded type V secreted serine protease and is known to cleave pepsin A and human coagulation factor V [8]. This extracellular enzyme is similar to several secreted or surface-bound proteins, including PssA in EHEC O26:H-, EspC in EPEC, and IgA1 protease in Neisseria species [69]. Recently, Dziva et al. [19] reported that EspP influences the intestinal colonization of calves and adherence to bovine primary intestinal epithelial cells. Moreover, degradation of human coagulation factor V via EspP could contribute to the mucosal hemorrhage observed in HC patients.
Metalloprotease (stcE)
A metalloprotease, StcE, is encoded on pO157 and specifically cleaves the C1 esterase inhibitor [37]. The C1 esterase inhibitor is a host regulator of multiple proteolytic cascades related to inflammation pathways, such as the classical complement, intrinsic coagulation, and contact activation. StcE is secreted through T2SS encoded on pO157 and is regulated by the LEE-encoded regulator (ler) [20, 37]. Grys et al. [25] demonstrated that StcE can contribute to intimate adherence of E. coli O157:H7 to Hep2 cells in vitro. The stcE gene was found all in E. coli O157:H7, some in EPEC serotype O55:H7, and it is not found in other diarrheagenic E. coli.
Putative Adhesion (toxB)
The toxB gene is encoded on a sequence 9.5 kb in size, and its predicted product shows 20% similarity with toxin B of Clostridium difficile [41]. Recent studies showed that ToxB contributes to the adherence of E. coli O157:H7 to Caco-2 cells through the increased secretion of TTSS [67]. Moreover, a sequence comparison revealed that ToxB shares 28% of amino acids identity and 47% of similarity to the predicted product of efa-1/lifA, another virulence gene frequently found on the chromosome of EPEC and non-O157 EHEC isolates [46]. The presence of the efa-1/ lifA gene is known to inhibit the activation of human and murine gastrointestinal lymphocytes, and therefore ToxB might be involved in inhibiting host lymphocytes [36]. However, a mutation of the toxB and efa-1 genes did not influence intestinal colonization in calves or sheep [66].
Eae Gene-Positive Conserved Fragments (ecf)
Recently, we reported that pO157 encoded the ecf operon (ecf1–4) that is temperature regulated by an intrinsically curved DNA [76]. ecf1 and ecf2 encode a putative polysaccharide deacetylase and an LPS α-1, 7-N-acetylglucosamine transferase, respectively, and both are unique to pO157 [32]. ecf3 shows similarity to the putative outermembrane protein in E. coli K1, associated with bacterial invasion [49]. ecf4, also named msbB2, encodes the second copy of a lipid A myristoyl transferase [35, 76]. The double mutant carrying deletions in the ecf4 and its chromosomal copy lpxM of E. coli O157:H7 had an altered lipid A structure and membrane fatty acid composition, and showed decreased persistence in bovine gastrointestinal tracts [76]. However, a single mutant of ecf4 did not show significant difference compared with wild-type E. coli O157:H7.
Pathogenesis of pO157
After the first report that pO157 was required for the expression of fimbriae and adhesion to epithelial cells, several studies reported conflicting results on the role of pO157 in adherence to epithelial cells (Table 1) [74]. In vivo studies of pO157, using animal models including the mouse, rabbit, and gnobiotic piglet, also showed conflicting results. However, the in vivo studies have limitations because there is no suitable animal model reproducing all aspects of the disease. Therefore, the precise role of pO157 in the pathogenesis of E. coli O157:H7 still needs to be defined.
Table 1.
Summary of studies on pO157 pathogenesis in vitro and in vivo.
| Year | Target | Pathogenesis | Effect | |
|---|---|---|---|---|
| In vitro | 1987 | Whole plasmid | Expression of fimbriae | Yes |
| Adherence to epithelial cells | ||||
| 1990 | Whole plasmid | Adherence to epithelial cells | Yes | |
| 1993 | Whole plasmid | Production of pilli | No | |
| Adherence to epithelial cells | ||||
| 2001 | toxB gene on pO157 | Adherence to epithelial cells | Yes | |
| 2005 | stcE gene on pO157 | Adherence to epithelial cells | Yes | |
| 2007 | espP gene on pO157 | Adherence to bovine primary intestinal epithelial cells | Yes | |
| In vivo | 1987 | Whole plasmid | Attaching and effacing lesion in gnotobiotic piglets | No |
| 1990 | Whole plasmid | Colonization of mouse | No | |
| 1993 | Whole plasmid | Clinical symptoms in rabbit | No | |
| 2006 | Whole plasmid | Colonization of cattle | Yes | |
| 2007 | Whole plasmid | Colonization of cattle | Yes | |
| 2007 | espP gene on pO157 | Colonization of calves | Yes |
Recently, we showed that the pO157 affects the efficiency of E. coli O157:H7 colonization of healthy cattle and survival in acidic conditions [40, 64]. An isogenic ΔpO157 E. coli O157:H7 mutant is more resistant to acidic synthetic bovine gastric fluid and bile than wild type [40]. This enhanced acid resistance in the ΔpO157 mutant is due to increased glutamate decarboxylase (GAD) expression. The mechanism of gad regulation by the pO157 is not known, but is likely due to pO157 regulation of chromosomal genes. In vivo, the ΔpO157 mutant survives passage through the bovine gastrointestinal tracts better than wild type, but does not colonize the bovine RAJ mucosa well as wild type [40, 64].
pO157-Like Plasmids in EHEC
Large plasmids resembling pO157 are found in most non-O157 EHEC isolates, but not all isolates from humans, and the size is varied from 70 to 200 kb [26]. These plasmids usually carry the hemolysin operon (ehx), but the etpC-O, katP, and espP genes are found in less than 50% of the isolates [11]. Some of these EHEC–hemolysin plasmids are reported to be involved in adhesion, but some are not. Epidemiological evidence suggests a stronger correlation of the presence of this EHEC–hemolysin plasmid with the development of HUS rather than diarrhea. In addition to pO157 or EHEC–hemolysin plasmids, a number of other plasmids ranging in size from 2 to 87 kb have been reported in E. coli O157:H7 isolates. However, no correlation has been seen with possession of any of these plasmids and clinical disease.
Concluding Remarks
This review focuses on the serotype E. coli O157:H7 and its 92-kb plasmid. E. coli O157:H7 causes severe human disease worldwide. Three major virulence factors include Shiga toxins, products of the pathogenicity island called the locus of enterocyte effacement, and products of the F-like plasmid pO157. This pathogen survives well in diverse environments, from its silent reservoir in healthy cattle to the farm environment. Genes encoded on the pO157 influence bacterial adherence to eukaryotic cells, colonization of cattle, and acid resistance. Further study to understand the mechanisms of E. coli O157:H7 pathogenesis and persistence in the environment will lead to effective interventions to prevent human disease.
References
- 1.Banatvala N, Griffin PM, Greene KD, Barrett TJ, Bibb WF, Green JH, Wells JG. The United States National Prospective Hemolytic Uremic Syndrome Study: Microbiologic, serologic, clinical, and epidemiologic findings. J Infect Dis. 2001;183:1063–1070. doi: 10.1086/319269. [DOI] [PubMed] [Google Scholar]
- 2.Barker J, Humphrey TJ, Brown MW. Survival of Escherichia coli O157 in a soil protozoan: Implications for disease. FEMS Microbiol Lett. 1999;173:291–295. doi: 10.1111/j.1574-6968.1999.tb13516.x. [DOI] [PubMed] [Google Scholar]
- 3.Barrett TJ, Lior H, Green JH, Khakhria R, Wells JG, Bell BP, Greene KD, Lewis J, Griffin PM. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer ME, Welch RA. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1996;64:167–175. doi: 10.1128/iai.64.1.167-175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brashears MM, Galyean ML, Loneragan GH, Mann JE, Killinger-Mann K. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J Food Prot. 2003;66:748–754. doi: 10.4315/0362-028x-66.5.748. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 9.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli 0157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 10.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucl Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caprioli A, Morabito S, Brugère H, Oswald E. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 12.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho S, Bender JB, Diez-Gonzalez F, Fossler CP, Hedberg CW, Kaneene JB, Ruegg PL, Warnick LD, Wells SJ. Prevalence and characterization of Escherichia coli O157 isolates from Minnesota dairy farms and county fairs. J Food Prot. 2006;69:252–259. doi: 10.4315/0362-028x-69.2.252. [DOI] [PubMed] [Google Scholar]
- 14.Cray WC, Jr, Moon HW. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delahay RM, Frankel G, Knutton S. Intimate interactions of enteropathogenic Escherichia coli at the host cell surface. Curr Opin Infect Dis. 2001;14:559–565. doi: 10.1097/00001432-200110000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vázquez A, et al. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, et al. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003;185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn JR, Keen JE, Thompson RA. Prevalence of Shiga-toxigenic Escherichia coli O157:H7 in adult dairy cattle. J Am Vet Med Assoc. 2004;224:1151–1158. doi: 10.2460/javma.2004.224.1151. [DOI] [PubMed] [Google Scholar]
- 19.Dziva F, Mahajan A, Cameron P, Currie C, McKendrick IJ, Wallis TS, Smith DGE, Stevens MP. EspP, a type V-secreted serine protease of enterohaemorrhagic Escherichia coli O157:H7, influences intestinal colonization of calves and adherence to bovine primary intestinal epithelial cells. FEMS Microbiol Lett. 2007;271:258–264. doi: 10.1111/j.1574-6968.2007.00724.x. [DOI] [PubMed] [Google Scholar]
- 20.Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frenzen PD, Drake A, Angulo FJ. Economic cost of illness due to Escherichia coli O157 infections in the United States. J Food Prot. 2005;68:2623–2630. doi: 10.4315/0362-028x-68.12.2623. [DOI] [PubMed] [Google Scholar]
- 22.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 23.Gansheroff LJ, O’Brien AD. Escherichia coli O157:H7 in beef cattle presented for slaughter in the US: Higher prevalence rates than previously estimated. Proc Natl Acad Sci USA. 2000;97:2959–2961. doi: 10.1073/pnas.97.7.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 25.Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect Immun. 2005;73:1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hales BA, Hart CA, Batt RM, Saunders JR. The large plasmids found in enterohemorrhagic and enteropathogenic Escherichia coli constitute a related series of transfer-defective Inc F-IIA replicons. Plasmid. 1992;28:183–193. doi: 10.1016/0147-619x(92)90050-k. [DOI] [PubMed] [Google Scholar]
- 27.Hancock DD, Besser TE, Rice DH, Herriott DE, Tarr PI. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuvelink AE, van Heerwaarden C, Zwartkruis-Nahuis JT, van Oosterom R, Edink K, van Duynhoven YT, de Boer E. Escherichia coli O157 infection associated with a petting zoo. Epidemiol Infect. 2002;129:295–302. doi: 10.1017/s095026880200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacewicz MS, Acheson DW, Binion DG, West GA, Lincicome LL, Fiocchi C, Keusch GT. Responses of human intestinal microvascular endothelial cells to Shiga toxins 1 and 2 and pathogenesis of hemorrhagic colitis. Infect Immun. 1999;67:1439–1444. doi: 10.1128/iai.67.3.1439-1444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X, Morgan J, Doyle MP. Fate of Escherichia coli O157:H7 in manure-amended soil. Appl Environ Microbiol. 2002;68:2605–2609. doi: 10.1128/AEM.68.5.2605-2609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RP, Wilson JB, Michel P, Rahn K, Renwick SA, Gyles CL, Spika JS. Human infection with verotoxigenic Escherichia coli associated with exposure to farms and rural environments. In: Stewart CS, Flints HJ, editors. Escherichia coli O157 in Farm Animals. CABI Publishing; Wallingford, U.K: 1999. pp. 147–168. [Google Scholar]
- 32.Kaniuk NA, Vinogradov E, Li J, Monteiro MA, Whitfield C. Chromosomal and plasmid-encoded enzymes are required for assembly of the R 3-type core oligosaccharide in the lipopolysaccharide of Escherichia coli O157:H7. J Biol Chem. 2004;279:31237–31250. doi: 10.1074/jbc.M401879200. [DOI] [PubMed] [Google Scholar]
- 33.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 34.Karmali MA, Steele BT, Petric M, Lim C. Sporadic cases of haemolytic–uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;1:619–620. doi: 10.1016/s0140-6736(83)91795-6. [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Jia W, Bishop RE, Gyles C. An msbB homologue carried in plasmid pO157 encodes an acyltransferase involved in lipid A biosynthesis in Escherichia coli O157:H7. Infect Immun. 2004;72:1174–1180. doi: 10.1128/IAI.72.2.1174-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klapproth JMA, I, Scaletsky CA, McNamara BP, Lai LC, Malstrom C, James SP, Donnenberg MS. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect Immun. 2000;68:2148–2155. doi: 10.1128/iai.68.4.2148-2155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol. 2002;45:277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- 38.LeJeune JT, Besser TE, Hancock DD. Cattle water troughs as reservoirs of Escherichia coli O157. Appl Environ Microbiol. 2001;67:3053–3057. doi: 10.1128/AEM.67.7.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim JY, Li J, Sheng H, Besser TE, Potter K, Hovde CJ. Escherichia coli O157:H7 colonization at the rectoanal junction of long-duration culture-positive cattle. Appl Environ Microbiol. 2007;73:1380–1382. doi: 10.1128/AEM.02242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JY, Sheng H, Seo KS, Park YH, Hovde CJ. Characterization of an Escherichia coli O157:H7 Plasmid O157 deletion mutant and its survival and persistence in cattle. Appl Environ Microbiol. 2007;73:2037–2047. doi: 10.1128/AEM.02643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, et al. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Maule A. Survival of verocytotoxigenic Escherichia coli O157 in soil, water and on surfaces. Symp Ser Soc Appl Microbiol. 2000;29:71S–78S. doi: 10.1111/j.1365-2672.2000.tb05334.x. [DOI] [PubMed] [Google Scholar]
- 43.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melton-Celsa AR, Darnell SC, O’Brien AD. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. Massive outbreak of Escherichia coli O157:H7 infection in school children in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 46.Morabito S, Tozzoli R, Oswald E, Caprioli A. A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect Immun. 2003;71:3343–3348. doi: 10.1128/IAI.71.6.3343-3348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DGE, Gally DL. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orndorff PE, Wang Y, Huang SH, Wass CA, Stins MF, Kim KS. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect Immun. 1999;67:4751–4756. doi: 10.1128/iai.67.9.4751-4756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostroff SM, Tarr PI, Neill MA, Lewis JH, Hargrett-Bean N, Kobayashi JM. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 51.Paton AW, Paton JC. Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR. J Clin Microbiol. 1999;37:3362–3365. doi: 10.1128/jcm.37.10.3362-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perna NT, Mayhew GF, Posfai G, Elliott S, Donnenberg MS, Kaper JB, Blattner FR. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 54.Peterson WL, Mackowiak PA, Barnett CC, Marling-Cason M, Haley ML. The human gastric bactericidal barrier: Mechanisms of action, relative antibacterial activity, and dietary influences. J Infect Dis. 1989;159:979–983. doi: 10.1093/infdis/159.5.979. [DOI] [PubMed] [Google Scholar]
- 55.Putonti C, Luo Y, Katili C, Chumakov S, Fox GE, Graur D, Fofanov Y. A computational tool for the genomic identification of regions of unusual compositional properties and its utilization in the detection of horizontally transferred sequences. Mol Biol Evol. 2006;23:1863–1868. doi: 10.1093/molbev/msl053. [DOI] [PubMed] [Google Scholar]
- 56.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 58.Sanderson MW, Besser TE, Gay JM, Gay CC, Hancock DD. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet Microbiol. 1999;69:199–205. doi: 10.1016/s0378-1135(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 59.Saxena SK, O’Brien AD, Ackerman EJ. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28S RNA when microinjected into Xenopus oocytes. J Biol Chem. 1989;264:596–601. [PubMed] [Google Scholar]
- 60.Schlech WF, III, Chase DP, Badley A. A model of food-borne Listeria monocytogenes infection in the Sprague–Dawley rat using gastric inoculation: Development and effect of gastric acidity on infective dose. Int J Food Microbiol. 1993;18:15–24. doi: 10.1016/0168-1605(93)90003-y. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt H, Henkel B, Karch H. A gene cluster closely related to type II secretion pathway operons of Gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett. 1997;148:265–272. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt H, Karch H, Beutin L. The large-sized plasmids of enterohemorrhagic Escherichia coli O157 strains encode hemolysins which are presumably members of the E. coli alpha-hemolysin family. FEMS Microbiol Lett. 1994;117:189–196. doi: 10.1111/j.1574-6968.1994.tb06763.x. [DOI] [PubMed] [Google Scholar]
- 64.Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect Immun. 2006;74:4685–4693. doi: 10.1128/IAI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shima K, Yoshii N, Akiba M, Nishimura K, Nakazawa M, Yamasaki S. Comparison of PCR–RFLP and PFGE for determining the clonality of enterohemorrhagic Escherichia coli strains. FEMS Microbiol Lett. 2006;257:124–131. doi: 10.1111/j.1574-6968.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 66.Stevens MP, Roe AJ, Vlisidou I, Van Diemen PM, La Ragione RM, Best A, Woodward MJ, Gally DL, Wallis TS. Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect Immun. 2004;72:5402–5411. doi: 10.1128/IAI.72.9.5402-5411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, Taguchi H, Kamiya S, Hayashi T. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001;69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson JS, Hodge DS, Borczyk AA. Rapid biochemical test to identify verocytotoxin-positive strains of Escherichia coli serotype O157. J Clin Microbiol. 1990;28:2165–2168. doi: 10.1128/jcm.28.10.2165-2168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Diemen PM, Dziva F, Stevens MP, Wallis TS. Identification of enterohemorrhagic Escherichia coli O26:H-genes required for intestinal colonization in calves. Infect Immun. 2005;73:1735–1743. doi: 10.1128/IAI.73.3.1735-1743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varma JK, Greene KD, Reller ME, DeLong SM, Trottier J, Nowicki SF, et al. An outbreak of Escherichia coli O157 infection following exposure to a contaminated building. JAMA. 2003;290:2709–2712. doi: 10.1001/jama.290.20.2709. [DOI] [PubMed] [Google Scholar]
- 71.Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willshaw GA, Smith HR, Cheasty T, Wall PG, Rowe B. Vero cytotoxin-producing Escherichia coli O157 outbreaks in England and Wales, 1995: Phenotypic methods and genotypic subtyping. Emerg Infect Dis. 1997;3:561–565. doi: 10.3201/eid0304.970422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoon JW, Hovde CJ. All blood, no stool: Enterohemorrhagic Escherichia coli O157:H7 infection. J Vet Sci. 2008;9:219–231. doi: 10.4142/jvs.2008.9.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon JW, Lim JY, Park YH, Hovde CJ. Involvement of the Escherichia coli O157:H7(pO157) ecf operon and lipid A myristoyl transferase activity in bacterial survival in the bovine gastrointestinal tract and bacterial persistence in farm water troughs. Infect Immun. 2005;73:2367–2378. doi: 10.1128/IAI.73.4.2367-2378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoon JW, Minnich SA, Ahn JS, Park YH, Paszczynski A, Hovde CJ. Thermoregulation of the Escherichia coli O157:H7 pO157 ecf operon and lipid A myristoyl transferase activity involves intrinsically curved DNA. Mol Microbiol. 2004;51:419–435. doi: 10.1046/j.1365-2958.2003.03827.x. [DOI] [PubMed] [Google Scholar]
- 77.Yuk HG, Marshall DL. Adaptation of Escherichia coli O157:H7 to pH alters membrane lipid composition, verotoxin secretion, and resistance to simulated gastric fluid acid. Appl Environ Microbiol. 2004;70:3500–3505. doi: 10.1128/AEM.70.6.3500-3505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


