Summary
Clearance or control of pathogens or tumors usually requires T-cell-mediated immunity. As such, understanding the mechanisms that govern the function, maintenance, and persistence of T cells will likely lead to new treatments for controlling disease. During an immune response, T-cell development is marked by striking changes in metabolism. There is a growing appreciation that these metabolic changes underlie the capacity of T cells to perform particular functions, and this has led to a recent focus on the idea that the manipulation of cellular metabolism can be used to shape adaptive immune responses. Although interest in this area has grown in the last few years, a full understanding of the metabolic control of T-cell functions, particularly during an immune response in vivo, is still lacking. In this review, we first provide a basic overview of metabolism in T cells, and then we focus on recent studies providing new or updated insights into the regulation of metabolic pathways and how they underpin T-cell differentiation and memory T-cell development.
Keywords: T cell, metabolism, immune response
Conventional view of T-cell metabolism
T cells develop in the thymus and then enter the periphery, where their role is to protect the body from invading pathogens and cancers. Circulating naive T cells are quiescent, but upon T-cell receptor (TCR)-mediated recognition of antigen and the receipt of costimulatory signals, these cells become activated, undergo extensive proliferation, and acquire effector functions, such as the ability to produce cytokines and cytolytic molecules. The differences in functional and phenotypic characteristics of quiescent T cells and activated T cells are supported by distinct metabolic requirements. Specifically, quiescent T cells (i.e. naive or memory T cells) have a catabolic metabolism where nutrients are broken down to fuel cell survival. In contrast, activated T cells adopt an anabolic metabolism, where nutrients will also be used to construct the molecular building blocks that are incorporated into cellular biomass to support proliferation (1–3). The balance of catabolic and anabolic pathways in a cell determines how much adenosine triphosphate (ATP) is generated versus consumed, the availability of biosynthetic precursors, and the redox status of the cell.
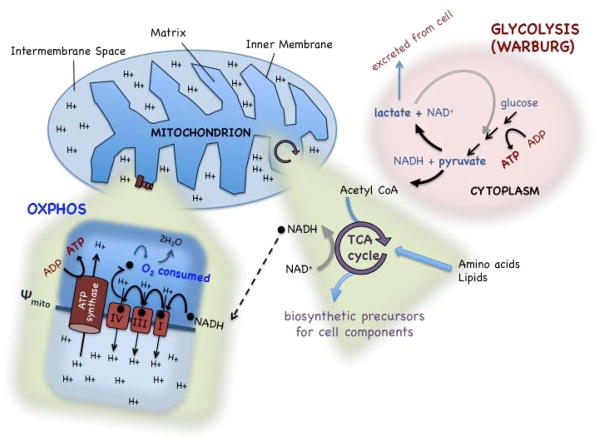
Upon activation, T cells increase in size and undergo a metabolic transition from oxidative phosphorylation (OXPHOS) to glycolysis (4–6) (Fig. 1). Signals from growth factor cytokines like interleukin-2 (IL-2) and the ligation of costimulatory CD28 promote this switch to glycolysis by inducing the phosphoinositol 3-kinase (PI3K)-dependent activation of AKT (7, 8). Mamalian target of rapamycin (mTOR) is a key regulator of translation (9) and cell size (10) that stimulates glycolysis and cellular metabolism (11) by increasing glycolytic enzyme activity and enhancing the expression of nutrient transporters. Active AKT promotes the mTOR pathway, thereby enabling increased utilization of glucose and amino acids (2, 12–16). Activated T cells aerobically ferment glucose into lactate, a process that occurs despite the presence of sufficient oxygen to support mitochondrial OXPHOS (5, 6) (Fig. 1). This phenomenon is referred to as the Warburg effect, after Otto Warburg who in the 1920s first described this process in cancer cells (17). The current view of metabolism in activated T cells is one where the induction and maintenance of Warburg metabolism, in addition to glutamine uptake and utilization, is essential for activation, proliferation, and the expression of effector functions (5, 7, 18–21) (Fig. 2).
Fig. 1. ATP-generating metabolic pathways.
Quiescent T cells mainly use oxidative phosphorylation (OXPHOS) to generate ATP, a process that takes place in mitochondria. A variety of substrates, such as glucose, amino acids, and lipids, are oxidized in the TCA cycle; these reactions generate biosynthetic precursors as well as the reducing agents NADH. The NADH coenzyme, which is used in many biochemical reactions, donates electrons to the electron transport chain for OXPHOS. Upon activation, T cells upregulate glycolysis, the process in which glucose is converted into pyruvate, thereby generating ATP and reducing NAD+ to NADH. To fuel OXPHOS, pyruvate can be shuttled into the TCA cycle; however, during Warburg metabolism, the majority of pyruvate is converted into lactate, which is excreted from the cell. In the process of converting pyruvate to lactate, NADH is oxidized to NAD+, which in turn can support continued glycolysis.
Fig. 2. Metabolic pathways in activated T cells.
Activated T cells use glucose and glutamine as their main nutrients to support rapid proliferation and effector functions. Glucose uptake is dependent on glucose transporter expression, and once inside the cell, glucose is converted into glucose-6-phosphate. This intermediate is the starting point for glycolysis, which ultimately leads to the production of pyruvate. In Warburg metabolism, the majority of pyruvate is excreted as lactate. Glutamine uptake and metabolism is also critical in activated T cells. As proliferation requires rapid synthesis of macromolecules for the generation of daughter cells, biosynthetic intermediates provided by the TCA cycle are necessary to support this process. Citrate from the TCA cycle can be excreted into the cytosol and used for lipid synthesis. Lipid synthesis also requires reductive power in the form of NADPH. NADPH is generated by the conversion of malate, excreted from the mitochondria, into pyruvate. Alternatively, using glucose-6-phosphate in the pentose phosphate pathway also generates NADPH. This latter pathway is important for nucleotide synthesis. Dashed arrows indicate multiple steps. Red, green, and purple arrows indicate glycolysis, glutaminolysis, and pentose phosphate pathways, respectively.
Glucose and proliferating T cells
Glucose is an essential nutrient for T cells, and its uptake is regulated by glucose transporter expression (22). TCR stimulation leads to trafficking of the glucose transporter Glut1 from intracellular vesicles to the cell surface (8), while signals via CD28 and growth factor cytokines further increase its expression and cell surface localization (1, 5, 7). Naive T cells isolated from Glut1 overexpressing mice are larger, and these mice have increased numbers of CD44hi T cells, suggesting that the earliest steps in T-cell activation, the increase in cell size and activation marker expression, are dependent on glucose utilization (18). It has also been shown that in the absence of glucose, T-cell survival and proliferation is impaired, even in the presence of an alternative carbon source such as glutamine (18, 23). In addition, T-cell cytokine production is also thought to be dependent on glucose. For example, glucose deprivation has been shown to strongly inhibit IFN-γ gene expression (18, 24), while transgenic expression of Glut1 leads to enhanced IL-2 and IFN-γ production (18). Also, in the absence of glucose, CD8+ T cells have a defect in cytolytic activity marked by reduced granzyme and perforin production (19). Collectively, many studies illustrate that glucose is an essential nutrient for T cells.
Once inside the cell, glucose is degraded and used to (i) generate energy, either through Warburg metabolism or the TCA cycle and OXPHOS, and (ii) to provide the building blocks for cellular maintenance and proliferation (Fig. 1). To produce ATP from OXPHOS, glucose is converted to pyruvate via the glycolytic pathway and then to acetyl CoA, where it can enter the TCA cycle by condensing with oxaloacetate (Fig. 2). TCA cycle reactions generate the reduced coenzyme NADH, which acts as an electron donor for the electron transport chain (ETC), fueling ATP production through OXPHOS. Glycolysis can also generate ATP in the cytoplasm via substrate level phosphorylation. Here, glucose is not fully oxidized in the mitochondria, even though there is sufficient oxygen present to do so (Warburg effect), but rather glucose-derived pyruvate is converted to lactate in the cytoplasm and excreted from the cell. Lactate excretion supports continued Warburg metabolism by regenerating NADH to NAD+, a coenzyme that facilitates earlier steps of glucose breakdown (2) (Fig. 1). As an alternative to glycolysis, glucose-derived glucose-6-phosphate can be directed into the pentose phosphate pathway (Fig. 2). The pentose phosphate pathway is important for proliferating cells as it provides precursors for the synthesis of nucleotides and aromatic amino acids, as well as reducing equivalents in the form of NADPH. NADPH is required to maintain the pool of reduced glutathione, as well as to support lipid and cholesterol synthesis (Fig. 2).
Glutamine and proliferating T cells
In addition to the switch to glycolysis, several other metabolic pathways are altered upon T-cell activation. Changes include decreased fatty acid oxidation, decreased pyruvate flux into the TCA cycle, increased glucose flux into the pentose phosphate pathway, and increased glutamine metabolism (21, 25, 26) (Fig. 2). Similar to glucose uptake, growth factor signaling controls the import of glutamine into the cell. It was recently shown that in an IL-3-dependent cell line glutamine uptake is dependent on glucose flux through the hexosamine pathway (27). The IL-3 receptor, like receptors for other growth factor cytokines, is a glycoprotein whose surface expression is dependent on glucose availability for glycosylation, thus providing an explanation as to why in certain settings glucose is essential for mammalian cells even when there is an abundant supply of amino acids available (28). Glutamine uptake and metabolism has been shown to be critical for T-cell function (7, 20). Metabolism of glutamine, termed glutaminolysis in analogy to glycolysis, is an energy-producing pathway. In contrast to glycolytic energy production, glutaminolytic energy production requires mitochondrial OXPHOS (3). In addition to energy, proliferating T cells, much like tumor cells, require the rapid synthesis of macromolecules that can be incorporated into cellular machinery and membrane components to form daughter cells. Synthesis of macromolecules requires biosynthetic intermediates, which are provided by the TCA cycle through the conversion of metabolites such as pyruvate and glutamine.
As described earlier, pyruvate derived from glycolysis is converted into acetyl CoA, which condenses with oxaloacetate to generate citrate. Glutamine can enter the TCA via conversion to α-ketoglutarate, which can also be processed to oxaloacetate and subsequently citrate. To generate new lipids, citrate has to be excreted into the cytosol, where it can be converted to acetyl CoA, the backbone for lipid synthesis (29) (Fig. 2). Thus, both glucose and glutamine can be used as carbon sources for lipid synthesis. Cells must be capable of rapidly replenishing TCA cycle intermediates if any are drawn off for biosynthesis. Under normoxic conditions, most of the carbon for fatty acid synthesis is derived from glucose; however, glutamine is also important for lipid synthesis, as it replenishes oxaloacetate for continued TCA cycle function (23, 30–32). In addition to carbon, reductive power in the form of NADPH is required for lipid synthesis (29). Along with NADPH production from the pentose phosphate pathway, NADPH is also produced during glutaminolysis when glutamine is metabolized in the TCA cycle to pyruvate and then converted into lactate (31) (Fig. 2). The need for NADPH to support lipid and nucleotide biosynthesis as well as for maintenance of reduced glutathione may explain why the majority of glutamine-derived carbon is excreted as lactate in rapidly proliferating cells (31). In addition, high rates of glutamine metabolism in proliferating cells results in excess intracellular nitrogen that must be secreted as alanine or ammonia to avoid toxicity (29). Glutamine also provides a nitrogen source for non-essential amino acids and nucleotides (31, 33).
Much more is known about the metabolic pathways that support T-cell activation and proliferation than the metabolic pathways that support T-cell quiescence. Naive and memory T cells are smaller than activated T cells, have decreased metabolic activity that is primarily catabolic in nature, and are more resistant to apoptosis than activated cells (34). The conventional view suggests that quiescent T cells interchangeably break down glucose, amino acids, and fatty acids to fuel the TCA cycle and generate ATP in their mitochondria via OXPHOS (1, 4). Although TCR stimulation and CD28 costimulation are required for the switch from a quiescent metabolism to the glycolytic metabolism of an activated cell (7, 18), it has been shown that maintaining quiescence in naive T cells is not simply due to a lack of these signals but is an active process that involves metabolic regulation (34–36).
Regulation of metabolic pathways in T cells
The factors that regulate metabolism in cells are highly interconnected. Given the fundamental role of metabolism in virtually every cellular process, there has been accumulating interest in how T-cell metabolism is controlled during different phases of T-cell differentiation. For organizational purposes, we have chosen to discuss some of the latest findings in this field as they pertain to naive T cells, effector T cells, and memory T cells independently.
Naive T cells
KLF and Fox transcription factors
Naive T cells maintain a catabolic metabolism, which is actively enforced (34–36). The Forkhead box (Fox) and Kruppel-like family of transcription factors (KLF) are thought to be responsible for T-cell quiescence by inducing inhibitors of cellular activation (34). Several recent studies have suggested that Fox and KLF2 transcription factors indirectly maintain quiescence by regulating the survival and migration of T cells (37–40) (Fig. 3). A study by Takada et al. (40) showed that KLF2-deficient CD8 T cells had only a minimal defect in cell cycle control and displayed normal proliferative capacity. However, expression of the homing receptors S1P1 and CD62L were decreased and thus trafficking of T cells in vivo was impaired (40). In another study (41), T cells deficient in the transcription factor ELF4 showed reduced KLF2 and KLF4 expression, which resulted in diminished CD62L expression and increased homeostatic and antigen-driven proliferation, respectively (41). It was later shown by the same group that mTOR signaling, which is induced during T-cell activation (Fig. 3), inhibits ELF4 expression in activated CD8= T cells, releasing them from their quiescent state (42). Altered homing receptor expression changes trafficking patterns of naive T cells and thereby affects exposure to growth factors and subsequently the metabolic pathways in these cells.
Fig. 3. Factors regulating metabolism in T cells.
Metabolism has a fundamental role in virtually every cellular process, and there has been a growing interest in how metabolic pathways regulate T-cell function and fate. This diagram highlights some of the factors recently described in the literature to regulate metabolism in quiescent and activated T cells. Factors promoting anabolic pathways support the metabolic needs of T cells during activation and proliferation (red). Factors that regulate quiescence maintain catabolic mitochondrial metabolism and promote cell survival and persistence (blue). Depending on the context, signaling through growth factor receptors (red) can drive proliferation and effector T-cell differentiation (e.g. IL-2 signals), or promote survival (blue) through enhanced mitochondrial function (e.g. IL-15 and IL-7 signals).
Foxo1 has been found to be essential for IL-7Rα expression in peripheral T cells (38), and IL-7 signaling is critical for naive T-cell survival (43, 44) (Fig. 3). In line with these observations, it was recently shown that inactivation of Foxo1 was essential for effector differentiation of CD8+ T cells, a process regulated through the transcription factor T-bet and the mTOR kinase (45). In fasted muscle cells, Foxo1 is activated and will suppress glucose oxidation (46) while enhancing surface levels of the fatty acid translocase FAT/CD36 (47) and thus promoting fatty acid uptake and oxidation. These data suggest that Foxo1 prepares the muscle cell for the increased reliance on fatty acid metabolism that is characteristic of fasting and is consistent with the idea that Foxo1 is contributing to the maintenance of metabolic quiescence in naive T cells. Interestingly, a different Fox factor, Foxp1, has been demonstrated to compete with Foxo1 for the control of IL-7Rα expression, and loss of Foxp1 induced naive T cells to gain an effector phenotype (48). Whereas normal naive T cells do not proliferate in response to IL-7 (49), when Foxp1 was lost, IL-7Rα expression increased and proliferation was induced. In addition to promoting IL-7Rα expression, Foxp1 deficiency induced other changes, including enhanced Erk activation, and inhibition of Erk and MEK severely attenuated the proliferative response of Foxp1 deficient T cells. Thus, Foxp1 appears to affect more than one pathway to maintain naive T-cell quiescence (48).
MST1 is a kinase that directly activates FOXO transcription factors. The MST/FOXO pathway has been shown to mediate oxidative stress responses and extend lifespan in both mammalian neurons and nematodes (50). A recent study reported a primary immunodeficiency phenotype in humans that was associated with the loss of MST1 function and primarily characterized by a progressive decline in naive T cells (51). In T cells, MST-1 was necessary for Foxo1 protein stability and expression, and indeed IL-7Rα and CD62L expression was impaired on T cells from these patients (51). In line with impaired IL-7R signaling, Bcl-2 expression levels were low and cell survival was diminished in MST-deficient T cells (51). Collectively, these observations indicate that effective maintenance of T-cell quiescence and survival is likely a balanced interplay between regulating metabolism and apoptosis, activation, and migration of naive T cells.
Tsc and mTOR
Several recent studies investigated the role of the tumor suppressor Tsc1, a regulator of the mTOR pathway, in maintaining naive T-cell quiescence (Fig. 3). It was found that Tsc1 established a quiescent gene-expression program linking cell cycle and metabolism with the expression of immune response genes in T cells (35). T cells that lacked Tsc1 were larger and proliferated more than control T cells, indicating a loss of quiescence and a more activated state, which resulted from increased mTORC1 activation (35, 52). Loss of Tcs1 also led to more ROS production and thus enhanced apoptosis and lower numbers of peripheral T cells (35, 36, 52, 53). Unexpectedly, although Tsc1 deficiency caused hyperactive responses to TCR stimulation in vitro, expansion of Tsc1-deficient CD8+ T cells upon bacterial infection in vivo was impaired, demonstrating that maintenance of T-cell quiescence is important for a productive immune response (35). Failure to mount an efficient T-cell response in vivo was also observed when the excessive apoptosis evident in Tsc1-deficient T cells was blocked by transgenic expression of Bcl2, suggesting that Tsc1 deficiency impairs T-cell responses through a mechanism that is largely independent of cell survival. It was previously shown that hematopoietic stem cells (HSCs) deficient for Tsc1 have increased expression of many mitochondrial genes and higher rates of OXPHOS that leads to increased levels of ROS (54). Loss of Tsc1 function resulted in reduced hematopoiesis and self-renewal of HSCs. Treatment with a ROS antagonist in vivo restored HSC numbers and function, suggesting that the Tsc1-mTOR pathway serves to suppress mitochondrial respiration and ROS production, and thereby maintains quiescence in hematopoietic stem cells (54). In a separate study, it was also shown that scavenging ROS decreased cell death of Tsc1-deficient peripheral CD4+ T cells (36). Together, these studies illustrate how the Tsc1-mTOR pathway maintains quiescence by regulating cellular metabolism.
Lkb1 and AMPK
Evidence is emerging that the liver kinase B1 (LKB1) is also important in controlling T-cell metabolism and quiescence (Fig. 3). LKB1 is a serine/threonine kinase and is part of an evolutionarily conserved energy-sensing pathway (55, 56). It was first identified as the tumor repressor responsible for Peutz-Jeghers syndrome (57) and was recently described to regulate quiescence and metabolism in hematopoietic stem cells (58–60). LKB1 can exert its effects on cellular metabolism via the phosphorylation of the α subunit of AMP-activated protein kinase (AMPK), and AMPK activation by LKB1 is essential under conditions of bioenergetic stress (61, 62). AMPK has a central role in the regulation of energy homeostasis by mediating glucose, protein, and fatty acid metabolism (63). In contrast to the P13K/AKT/mTOR pathway, which promotes anabolism, AMPK supports catabolism and inactivates growth-inducing pathways. As quiescent cells have a catabolic metabolism and mostly use OXPHOS for their energy, the regulatory effects of AMPK in quiescent cells are likely to merge with the oxidation of fatty acids to supply energy needs. Activation of AMPK can promote the oxidation of fatty acids in two ways (64, 65): through suppression of mTOR activity (66) and by phosphorylating and thereby inactivating acetyl-CoA carboxylase (ACC) (67, 68) (Fig. 3). Although it was shown that AMPK was not the critical effector of LKB1 in the maintenance of hematopoiesis (58–60), LKB1 regulated mitochondrial function is at least partly AMPK dependent (58, 60). Recently, it was found that LKB1 is essential for the survival of thymocytes and development of T-cell progenitors (69, 70) and is required for CD4+ and CD8+ T-cell development and peripheral homeostasis (71). LKB1-deficient peripheral T cells were shown to have enhanced glucose uptake and a higher glycolytic rate (71), while LKB1-deficient thymocytes were shown to have reduced expression of the CD98 subunit of the amino L-acid transporter (70). These results suggest that LKB1 is critical for matching metabolism to energy demand and in this way acts as an important regulator of cellular quiescence. Future studies are required to further elucidate the role of mTOR and AMPK signaling in maintaining quiescence in T cells and its precursors.
Effector T cells
AKT, LKB1, and AMPK
T cells increase their metabolism in response to immune stimulation via the serine-threonine kinase AKT (7, 8) (Fig. 3). AKT is rapidly activated in response to TCR triggering or common γ-chain cytokines such as IL-2 and IL-15 (72–75). However, it was recently reported that AKT-independent pathways also control T-cell metabolism, survival, and proliferation (76). AKT was shown to be dispensable for the regulation of glucose uptake in effector CD8+ T cells (77) but rather maintained a critical role in T-cell effector functions by regulating the expression of cytolytic molecules and the repertoire of cytokine/chemokine-receptors and adhesion molecules (77). In addition to describing the role of LKB1 in controlling cellular quiescence, a recent study by MacIver et al. (71) investigated the role of LKB1 in T-cell development and peripheral function. In response to activation, LKB1-deficient T cells displayed defects in proliferation, viability, and altered metabolism. Loss of LKB1 resulted in enhanced glycolytic gene expression and impaired oxidation of lipids in response to metabolic stress induced by IL-2 withdrawal. In line with enhanced glucose metabolism, LKB1-deficient T cells had greater expression of surface activation markers and increased production of IFN-γ. Furthermore, T cells deficient for AMPK displayed similar defects in activation, metabolism, and cytokine production. As expected, both LKB1 and AMPK-1α-deficient T cells showed increased phosphorylation of mTOR targets, and rapamycin blocked the enhanced IFN-γ production in these cells (71). This study demonstrates that the metabolic effects of LKB1 and AMPK are mTOR dependent (71) and highlights the multifaceted role of these proteins in regulating T-cell development and function through metabolism.
HIF-1 and Myc
Recently, the metabolism of various CD4+ T-cell subsets has been explored. It was shown that upon activation, Th1, Th2, Th17, and Treg cells all increased their glycolytic rates but that this occurred to a much greater extent in Th1, Th2, and Th17 cells than in Treg cells (78, 79). Hypoxia inducible factor 1α (HIF-1α) is a transcription factor that regulates the expression of genes that encode for glycolytic enzymes (80) as well as downregulates mitochondrial oxygen consumption by blocking the entrance of pyruvate in the TCA cycle (81) (Fig. 3). A recent study showed that HIF-1α was strongly induced in Th17 cells but not in Th1 or Th2 cells and that Th17 cell differentiation in vitro and in vivo was impaired in the absence of this transcription factor (79). These results demonstrated that HIF-1α controls the glycolytic activity that is essential for Th17 cell development. Interestingly, the addition of fatty acids to Th1, Th2, and Th17 cells suppressed lineage-specific cytokine production (78). While this effect appeared to be at least in part due to enhanced cell death, precisely how fatty acids impair viability and cytokine production in these cells remains to be determined. In addition to the idea that the presence of a lipid fuel source alters cellular metabolism from glycolysis to lipid oxidation, it is also possible that the presence of fatty acids results in signaling that is not compatible with the glycolytic program of these cells. Alternatively, lipids could be promoting lipotoxicity, as non-adipose tissues have a limited capacity to store lipids, that when exceeded, cellular dysfunction and death may result (82). In contrast to CD4+ effector T cells, Treg cells demonstrated high rates of fatty acid oxidation, and blocking this type of metabolism impaired Treg development (78). Inhibiting mTOR with rapamycin also resulted in an increased frequency of Treg cells, an effect that was dependent on fatty acid oxidation (78). In line with this finding, Treg cells demonstrated elevated AMPK activation when compared to effector and naive CD4+ T cells, and administration of the AMPK activator metformin reduced Glut1 expression, promoted fatty acid oxidation, and increased the percentage and number Treg cells (78). Together these results demonstrate that HIF-1α is essential for Th17 cell development and associated glycolytic metabolism, while the absence of HIF-1α drives Treg cell differentiation and fatty acid oxidation.
In contrast to the findings by Shi et al. (79) showing that HIF-1α was strongly and selectively induced only in the Th17 cells subset, a recent study by Wang et al. (21) showed that HIF-1α expression was highly induced upon activation in a mixed culture of CD4+ and CD8+ T cells. Despite its quick induction, HIF-1α appeared dispensable for the initiation of metabolic changes and activation-associated proliferation in these T cells (21). This study went on to show that Myc, a transcription factor that regulates glycolytic metabolism in transformed cells (83, 84), was highly induced upon T-cell activation, and its expression was required for activation-induced metabolic programming (21) (Fig. 3). Acute Myc deletion inhibited proliferation and cell growth upon T-cell activation, while leaving AKT and ERK signaling pathways intact. Consistent with the important role for glycolysis and glutaminolysis in T-cell activation, Myc-deficient T cells showed impaired activation-induced glycolysis and glutaminolysis and decreased activation of the downstream targets of mTOR. Furthermore, Myc-driven glutaminolysis was tightly coupled to the biosynthesis of polyamines that are essential for T-cell proliferation (21). In agreement with these findings, Carr et al. (20) recently showed that T cells grown without glutamine did not increase in size to the same extent as in cells cultured in media with glutamine, implicating that this amino acid is essential for optimal cell growth. Interestingly, in renal carcinoma cells and fibroblasts, other effects of Myc and HIF-1α signaling have been described. It has been shown that Myc stimulates nuclear-encoded mitochondrial genes and mitochondrial biogenesis (85), and HIF-1α inhibits these changes (86). Although metabolic processes in cancer cells can closely mimic those in activated T cells, future studies in T cells are required to determine the relative expression and dominance of Myc and HIF-1α upon activation and how this may affect mitochondrial changes.
mTOR
The conserved kinase mTOR has a central role in the regulation of metabolism by sensing and integrating signals in response to nutrients, growth factors, energy, and stress (87) (Fig. 3). mTOR is present in two distinct complexes: mTOR complex 1 (mTORC1) and mTORC2 (88). It was previously shown that mTOR signaling is required for CD4+ effector T-cell differentiation, as mTOR-deficient T cells fail to develop into Th1, Th2, and Th17 cells both in vitro and in vivo. However, mTOR-deficient T cells do differentiate into Treg cells (89). More recently it was shown by this same group that mTORC1 signals specifically regulate Th1 and Th17 lineage differentiation, whereas mTORC2 signaling drives Th2 development (90). In line with their previous observation, inhibition of both mTORC1 and mTORC2 was required for spontaneous Treg induction (90). These studies provide a strong link between metabolic control and CD4+ T-cell differentiation. Future studies are needed to further elucidate the pathways by which mTORC1 and mTORC2 differentially affect helper T-cell development and metabolism.
ERRα
Another important regulator of transcription of metabolic genes is the nuclear hormone receptor, estrogen-related receptor-α (ERR-α) (91, 92) (Fig. 3). Recently, it was shown that this protein acts as a metabolic regulator of effector CD4+ T-cell homeostasis and function by broadly affecting metabolic gene expression and glucose metabolism (93). ERR-α-deficient CD4+ T cells exhibited reduced activation and proliferation, which was accompanied by impaired glucose uptake and mitochondrial metabolism. Consistent with these observations in T cells, it was recently shown that the expression of the Drosophila ortholog of ERR mid-embryogenesis induced a switch in gene expression that promoted a metabolic program associated with proliferating cells, supporting the dramatic growth that occurs during larval development (94). Interestingly, the failure of T cells to become activated in the presence of an ERR-α inhibitor was a result of impaired glucose metabolism, as this defect was rescued by the addition of exogenous fatty acids, but not pyruvate, indicating that ERR-α positively regulates pyruvate flux into the TCA cycle. This illustrates that mitochondrial metabolism, and not only glycolysis, is important for the early phase of T-cell activation. This is consistent with another study showing that mitochondrial biogenesis is induced in the early phase after TCR activation (95). The inability to direct pyruvate into the TCA cycle may lead to decreased lipid synthesis and cell growth and thus impaired activation and proliferation. Although fatty acid oxidation could rescue CD4+ T-cell proliferation during initial activation, lipids were not sufficient to optimally restore cell division and effector function of Th1, Th2, and Th17 cells upon ERR-α inhibition. However, as would be expected from cells that mainly use lipid metabolism, Treg cells were efficiently generated in the presence of an ERR-α inhibitor. Since ERRs have been shown to associate with HIF-1α and regulate its function in tumors (96), future studies will need to determine how nuclear hormone receptors and transcription factors like HIF-1α and Myc interact in CD4+ T cells.
Targeting effector T-cell metabolism for immunotherapy
Implicit in the fact that metabolism underlies the functional capacity of T cells is the idea that manipulating T-cell metabolism in vivo will alter the outcome of adaptive immune responses. In a recent study, the bioenergetics of alloreactive T cells was examined (97). It was found that bone marrow cells that were transplanted into irradiated syngeneic hosts proliferated and adopted a glycolytic metabolism, as evidenced by increased Glut1 expression and lactate production when compared to naive bone marrow cells. Oxygen consumption by these cells, however, was unaffected, suggesting that glycolysis was the predominant form of metabolism rather than OXPHOS in bone marrow cells transplanted into irradiated syngeneic hosts. In contrast, lymphocytes that proliferated in response to alloantigens in a model of graft versus host disease (GVHD) also showed increased Glut1 expression and lactate production but had elevated oxygen consumption levels as well. Although large increases of intracellular acylcarnitines suggested changes in fatty acid metabolism, it was not directly determined if fatty acid oxidation was responsible for the increase in oxygen consumption. It was also observed that when compared to the transplanted bone marrow cells, alloreactive T cells proliferated faster, had decreased amounts of glutathione, had hyperpolarized mitochondrial membrane potential, and increased superoxide levels. Use of Bz-423, a compound that promotes superoxide production by inhibiting the mitochondrial F1F0-ATPase, selectively induced apoptosis in alloreactive T cells but not resting T cells or proliferating bone marrow cells. The authors speculate that this was most likely a result of the lack of sufficient anti-oxidant reserves in the alloreactive cells to cope with increased superoxide. Importantly, Bz-423 has therapeutic potential, as its use arrested ongoing GVHD by selectively inducing apoptosis in the alloreactive T cells. This study highlights the value of targeting T-cell metabolism for therapeutic benefit. To what extent the bioenergetics of alloreactive T cells differs from T cells activated by an infection or cancer needs further exploration.
Memory T cells
Fatty acid oxidation and memory T cells
The metabolic demands of proliferating T cells in culture have been studied extensively, and the importance of glucose is well documented. Much less is known about metabolic requirements of memory T cells. Several years ago we published a study showing that CD8+ T cells that lacked TRAF6 were impaired in their ability to form a stable population of antigen-specific memory T cells after bacterial infection, and microarray analysis suggested that this was due to defects in several catabolic pathways of metabolism, including fatty acid oxidation (98). We demonstrated that augmenting fatty acid oxidation in CD8+ T cells, by treating mice with either the AMPK activator metformin or the mTOR inhibitor rapamycin, two drugs that are known to induce fatty acid oxidation, promoted the development of memory CD8+ T cells after immunization (64–68). This study was the first to suggest that targeting catabolic pathways of metabolism, such as fatty acid oxidation, after vaccination could enhance the development of stable CD8+ T-cell memory. Our findings showing that rapamycin promoted CD8+ T-cell memory development were supported by another study published at the same time demonstrating that this drug also promoted immunity to viral infection (99). Importantly, this study by Araki et al. (99) established mTOR as a cell-intrinsic regulator of CD8+ T-cell memory development. Since rapamycin is traditionally used clinically for its immuosuppressive properties, these findings generated considerable interest.
Focusing on this ability of rapamycin to enhance immunity, it was more recently shown by Rao et al. (100) that CD8+ T cells activated and cultured in the presence of rapamycin for 3 days and subsequently transferred into recipient mice formed better memory T cells in terms of quantity and quality in comparison to control T cells. Furthermore, it was found that mTOR regulated the expression of the transcription factors T-bet and Eomesodermin, which are known to direct effector and memory CD8+ T-cell fates, respectively (100–103), further illustrating how this central metabolic regulator specifies T-cell fate. Interestingly, a recent paper from this same laboratory (45) showed that the ability of rapamycin to augment CD8+ memory T-cell development was ablated when Foxo1 was silenced. Inactivation of Foxo1 appeared to be important for T-bet mediated effector functions, whereas active Foxo1 resulted in a switch to more Eomesodermin transcription factor activation, and more CD8+ memory precursor T cells. Given these findings, it will be interesting to determine whether Eomesodermin regulates the expression of metabolism genes that dictate a quiescent phenotype in CD8+ T cells.
Mitochondrial spare respiratory capacity
Research in our laboratory continues to focus on the role of metabolism in the development of memory CD8+ T cells after infection. In a recent study (104), we showed that memory CD8+ T cells maintain significant mitochondrial spare respiratory capacity (SRC) in comparison to naive or effector T cells. SRC is the extra mitochondrial capacity available in a cell to produce energy under conditions of increased work or stress and is thought to be important for long-term cellular survival and function (105–109). We found that greater SRC was underscored by enhanced mitochondrial biogenesis in memory CD8+ T cells, and that this process was modulated by IL-15. In addition, we demonstrated that SRC was abrogated when carnitine palmitoyltransferase 1a (CPT1a), the mitochondrial protein that is the rate-limiting step for β-oxidation of fatty acids in the mitochondria (110–112) was inhibited. Our findings led us to a model where bioenergetics regulates memory T-cell development after infection (Fig. 4). As effector CD8+ T cells rapidly proliferate during an immune response, they primarily use glycolysis to support bioenergetic needs. We speculate that during this process, the majority of CD8+ effector T cells fail to maintain and/or induce mitochondrial biogenesis resulting in a reduction in the ratio between mitochondrial mass and overall cell mass, and thereby lose reserve energy-generating capacity, i.e. SRC. Reduced mitochondrial mass increases dependence on glycolysis and renders effector T cells bioenergetically unstable due to a decline in their ability to create energy from diverse substrates (e.g. fatty acids) via OXPHOS, and thus are unable to maintain viability when infection-associated signals decline and elements such as IL-2 that support glycolysis dissipate. Some antigen-specific T cells from the primary infection survive as long-lived memory T cells, because these cells have maintained mitochondrial function via exposure to IL-15 or other growth factor cytokines. More mitochondrial mass allows greater use of fatty acids for energy, thereby facilitating cell survival in the absence of pro-glycolysis signals. Ongoing experiments in our laboratory will determine whether, in addition to simply more mitochondrial mass, there are qualitative differences between mitochondria in effector T cells as compared to memory T cells. For example, increased expression of ETC complexes or CPT1a on a per mitochondrion basis could facilitate the use of fatty acids for energy via OXPHOS. Given that mitochondria undergo significant changes in T cells during various phases of the immune response we think that it is very likely that there will also be qualitative differences in these organelles between these cell types.
Fig. 4. Bioenergetic model of memory CD8+ T-cell survival after infection.

As effector CD8+ T cells rapidly proliferate during an immune response, they primarily use glycolysis to support bioenergetic needs. We speculate that during this process, the majority of CD8+ effector T cells fail to maintain and/or induce mitochondrial biogenesis resulting in a reduction in the ratio between mitochondrial mass and overall cell mass and thereby lose reserve energy-generating capacity, i.e., spare respiratory capacity. Reduced mitochondrial mass increases dependence on glycolysis and renders effector T cells bioenergetically unstable due to a decline in their ability to create energy from diverse substrates via OXPHOS, and thus are unable to maintain viability when infection-associated signals decline and elements such as IL-2 that support glycolysis dissipate. Some antigen-specific T cells from the primary infection survive as long-lived memory T cells, because these cells have maintained mitochondrial mass via exposure to IL-15 or other growth factor cytokines. More mitochondrial mass allows greater use of fatty acids for energy via OXPHOS, thereby facilitating cell survival in the absence of pro-glycolysis signals. Moreover, the enhanced spare respiratory capacity provided by increased mitochondrial mass might allow memory T cells to respond quickly if pathogen is reencountered.
Our studies revealed that maintaining/increasing mitochondrial mass is critical for CD8+ T-cell memory development. Unlike the case for effector T cells which rely predominantly on glycolytic metabolism, memory T cells maintain substantial mitochondrial mass, which provides the flexibility to use diverse substrates for energy generation, such as the oxidation of fatty acids. Our earlier microarray analyses comparing ‘memory-impaired’ TRAF6-deficient CD8+ T cells versus ‘memory-competent’ control CD8+ T cells revealed that in addition to fatty acid oxidation, amino acid catabolism may also be involved in CD8+ T-cell memory development (98). Previous studies have shown that antigen-presenting cells can regulate amino acid availability to T cells (113–115), which may influence T-cell development during an immune response. Furthermore, autophagy is an important survival pathway when cells are faced with metabolic stress (116). If T cells experience metabolic stress during an immune response, autophagy could be induced, and this presumably would include the breakdown of amino acids for fuel in addition to lipid oxidation. However, since fats are an efficient energy store, yielding more than twice the energy per gram than either carbohydrate or protein, it is logical that lipids would be the preferential fuel source for long-lived quiescent T cells. More work will need to be done in the area of substrate utilization in T cells after infection to reveal whether there is any role for amino acid catabolism fueling the development of memory T cells.
The question of metabolic conversion, promotion, or maintenance
It is still unclear whether T cells that are destined to become memory cells fully commit to using Warburg metabolism in the effector phase and subsequently switch to a quiescent metabolism later, or whether memory cell precursors never fully switch to Warburg metabolism and promote or simply maintain a significant amount of mitochondrial respiratory capacity throughout activation. While our data as well as those from other laboratories indicate that the population of CD8+ effector T cells generated in vivo or after activation in vitro actively use both glycolysis and OXPHOS (21, 97, 104), it is not possible to determine whether these measurements are reflecting heterogeneity in the cell populations, where more terminally differentiated effectors T cells are mostly using glycolysis and less differentiated effectors T cells are using more oxidative metabolism, or whether individual cells actually use both metabolic programs simultaneously. There does seem to be a high degree in uniformity in the effector cell populations generated in vitro, to where activation marker expression and effector function are adopted evenly across the population, which might suggest that metabolic phenotypes are also evenly distributed across the population and that all cells are using both glycolysis and OXPHOS. However, the real answers to these questions will have to wait until it becomes possible to measure metabolism on a single cell level While it now has been shown that activated T cells use OXPHOS in addition to glycolysis (21, 97, 104), it has yet to be determined which one or if both of these energy-generating pathways are actually required for the survival and proliferation of effector T cells.
Our research has indicated that memory T cells have more mitochondrial mass than effector T cells and that IL-15 induces mitochondrial biogenesis (104), suggesting that exposure to IL-15 or perhaps other common-γ cytokines, such as IL-7 or IL-4, is critical to ensure that mitochondrial mass is maintained and long-lived memory T cells can develop/persist (Fig. 3). Whether effector T cells will receive these cytokine signals can be controlled at many levels, including cytokine and/or cytokine receptor expression, trans-presentation of the cytokine to T cells by other cells, or T-cell localization to niches where cytokine is expressed (117, 118) (Fig. 3). Most likely a combination of all these factors will determine whether a T cell receives a cytokine signal in vivo. Our unpublished preliminary data indicate that lymph node homing receptors are upregulated quickly after IL-2 withdrawal in vitro, suggesting that once infection-associated signals disappear, T cells become competent to traffic to the lymph nodes where growth factor cytokines like IL-15 are expressed. A recent study has shown that CD8+ DCs constitutively express higher levels of IL-15 when compared to other DC subsets (119). In addition to the role of CD8+ DC in cross-priming CD8+ T cells (120), perhaps these cells also support mitochondrial maintenance in CD8+ T cells that are destined to become long-lived memory T cells via IL-15 trans-presentation during priming.
Memory T-cell development in vivo
The question remains how memory cell formation is organized in vivo. One possibility is that effector T cells at the lowest end of the growth factor gradient will be the first to sense growth factor deprivation and in response will upregulate molecules like CD62L and migrate to the lymph nodes where IL-15 is presented, making these cells more likely to survive contraction. We speculate that the antigen-specific T cells that continue to rapidly proliferate in response to infection will eventually outpace their mitochondrial biogenesis, resulting in lower mitochondrial mass and respiratory capacity, and thus a higher dependency on glycolysis (Fig. 4). Alternatively, there may be heterogeneity in CD62L expression on antigen-stimulated T cells already in the early phase of the immune response as a result of variation in strength of T-cell activation. In response to TCR triggering or cytokines, AKT is activated and FOXO transcription factors are sequestered in the cytoplasm (121) (Fig. 3) and thus are unable to drive KLF2 expression (76). In the case of inadequate AKT activity, FOXO transcription factors are not retained in the cytoplasm and can still drive the expression of KLF2 (75) and thereby maintain the expression of several adhesion and chemokine receptors, such as CD62L, CCR7, and S1P1, which control T-cell migration (37, 74). Weak antigen-stimulation results in suboptimal activation of the AKT-mTOR pathway, that is sufficient to drive T-cell growth and proliferation but not to suppress KLF2 expression (73, 76). Thus, T cells that are activated by a weaker signal will proliferate but also maintain higher expression of CD62L and maintain the ability to migrate to lymph nodes and gain access to growth factor cytokines like IL-15.
Another possibility is that heterogeneity in the effector pool as a result of asymmetric cell division may contribute to the generation of T cells that are better equipped for memory development than others (122). Asymmetric division could result in unequal distribution of several critical factors, including cytokine receptors, lymph node homing receptors, transcription factors, and mitochondria. Accordingly, it has been shown that cells generated during the peak of infection that have not terminally differentiated, express IL-7R and persist as long-lived memory T cells (123). It is possible that mitochondria are also unequally distributed during the first divisions after activation, so that terminally differentiated effector T cells acquire fewer mitochondria, while memory precursor T cells acquire more.
Bioenergetics and recall responses
In addition to bioenergetic differences between memory CD8+ T cells and effector CD8+ T cells, we have also shown that naive CD8+ T cells have less SRC and mitochondrial mass than memory CD8+ T cells (104). We speculate that during an immune response, signals from IL-15, as well as other growth factor cytokines, support mitochondrial biogenesis and thus endow memory cells with greater mitochondrial mass as compared to naive T cells. In addition to simply enhanced mitochondrial content contributing to bioenergetic differences, qualitative differences in mitochondria may also exist between these cell types, i.e. differences in the expression of ETC complexes or other mitochondrial proteins could result in intrinsic differences in respiratory capacity. Regardless of the variation in the mitochondria between cell types, we suspect that greater mitochondrial mass would indicate that memory T cells have more readily available ATP-generating capacity than naive T cells. If this were the case, it may explain why antigen-specific CD8+ memory T cells respond faster to re-infection on a per cell basis when compared to their antigen-specific naive CD8+ T-cell counterparts. Investigating cell-intrinsic bioenergetics differences between naive and memory T cells and how this disparity impacts their functionality will be a focus of our future studies.
STAT3
Two recent studies demonstrated that STAT3 has a critical role in the development and maintenance of memory T cells (124, 125). Siegel et al. (125) showed that patients with dominant-negative STAT3 mutations have a T-cell intrinsic defect resulting in decreased numbers of central memory T cells. Naive T cells isolated from these patients and cultured in IL-7 or IL-15 exhibited impaired differentiation into central memory T cells. The impaired capacity to form central memory T cells was associated with decreased expression of Bcl6 and SOCS3, two STAT3 responsive transcription factors important in the development of memory T cells (126–128). Cui et al. (124) demonstrated the intrinsic requirement of STAT3 in CD8+ T cells for memory differentiation and maturation in a mouse model of lymphocytic choriomeningitis virus (LCMV) infection. STAT3-deficient T cells displayed an effector phenotype, had a shortened lifespan, had reduced self-renewal capacity, and showed poor protective immunity upon secondary infection. In addition, Bcl6 and SOCS3-deficient CD8+ T cells also demonstrated impaired memory T-cell development. Interestingly, it was recently shown that STAT3 is not only present in the cytosol but also in mitochondria, where it regulates the function of ETC complexes I and II (129, 130). STAT3-deficient cells exhibited defects in ETC function and ATP generation but maintained normal mitochondrial gene expression and content (129). Importantly, it was suggested that STAT3 activity in mitochondria is unrelated to its actions as a transcription factor. Although Cui’s study suggests at least a partial role for STAT3-mediated transcriptional effects on memory T-cell formation, it is tempting to speculate that mitochondrial STAT3 plays a role in memory T-cell formation as well. As the importance of fatty acid oxidation and spare respiratory capacity for memory CD8+ T-cell development has recently become clear, the impaired ETC function and ATP-generating capacity evident in the absence of STAT3 suggests that mitochondrial STAT3 may have a key role in this process. Furthermore, in agreement with the idea that maintaining mitochondrial capacity to utilize fats for energy while not depending solely on glucose and IL-2 is a critical feature for CD8+ T cells to persist as long-lived memory T cells, it was suggested that the T-cell memory defect in STAT3-deficient patients was potentially resulting from increased reliance on IL-2 (125).
Diet and the immune response
Malnutrition and immunity
There is abundant epidemiological evidence which suggests that malnutrition impairs vaccine efficacy and increases susceptibility to infections; however, the underlying mechanism is poorly understood (131, 132). As the maintenance of long-lived memory T cells requires homeostatic proliferation and thus availability of building blocks to support the production of new daughter cells, dietary components can be critical to this process. A recent study from Iyer et al. (133) demonstrated that mice that were infected with LCMV on a normal diet but then switched to a low protein diet after memory T cells were established developed twofold lower numbers of antigen-specific T cells. Also, acute and basal homeostatic proliferation of CD44hi CD8+ T cells was impaired in mice on the low protein diet, which was not due to lower IL-7R or IL-15R expression. In addition, the memory CD8+ T cells in mice fed the low protein diet showed decreased proliferation upon challenge with LCMV clone 13, resulting in impaired viral clearance. Importantly, supplementation of appropriate protein levels in mice fed the low protein diet rescued the quantitative reduction of memory CD8+ T cells. This study demonstrates the importance of dietary amino acids for optimal maintenance of memory CD+8 T cells and an adequate secondary effector response.
Caloric restriction and T-cell responses
In contrast to malnutrition, caloric restriction, as defined as a reduction in nutrient availability without malnutrition, has long been known to increase lifespan in a variety of species ranging from yeast to mammals (134). In parallel with the proposed model linking T-cell longevity and maintaining the capacity to use a wide variety of fuels (Fig. 4), the capacity of animals to adapt to metabolic stress or harsh environmental conditions may have evolved along with genetic adaptation for longevity. Indeed, the stress resistance of vertebrate cells in culture positively correlates with the lifespan of the species from which they were isolated (135, 136). Most of the work exploring the molecular pathways that control longevity has been performed using worms, yeast, and fruit flies. Studies have shown that insulin/IGF signaling, DAF-16/FOXO transcription factors, and TOR all play crucial roles in the regulation of metabolism and lifespan (137–142). Inhibition of TOR by the AMPK-FOXO pathway has been shown to positively regulate longevity by caloric restriction in C. elegans (143). In agreement with these findings, administration of rapamycin or metformin, which inhibit mTOR and activate AMPK respectively, has been demonstrated to increase mammalian lifespan (144, 145), which corresponds with studies showing that these agents increase T-cell lifespan (98, 99, 146). Furthermore, consistent with our studies showing that increased fatty acid metabolism primes T cells for longevity (98, 104), a positive correlation between fat metabolism and increased lifespan has been observed in C. elegans (147).
Mitochondrial function and cellular longevity
As fatty acid oxidation occurs in the mitochondria, it is logical that maintenance of these organelles is critical for the survival of long-lived memory T cells (104). Support for this idea comes from studies that compared the transcriptional profiles of the aging process across humans, mice, flies, and worms, and identified the downregulation of nuclear-encoded genes of the mitochondrial ETC as the only age effect common to all four species (148). The decline in mitochondrial function with aging is opposed by caloric restriction by the induction of nuclear encoded genes that promote mitochondrial function (149). A key component in the transcriptional changes upon caloric restriction is increased expression of the transcriptional coactivator PGC-1α, a protein that is directly phosphorylated by AMPK (150). Activation of PGC1α results in induction of mitochondrial biogenesis, increased mitochondrial respiration and fatty acid oxidation, and regulation of ROS detoxification (151, 152). The latter process seems critical to prevent extensive oxidative damage as a result of increased respiration. It is likely that caloric restriction-induced PGC-1α expression results in a wide array of adaptations to provide a positive setting for increased OXPHOS. For instance, expression of uncoupling proteins, which have been associated with decreased ROS-induced damage, is increased in response to caloric restriction (153). In line with this, the Drosophila homolog of PGC-1 increases mitochondrial gene expression and activity and extends lifespan (154). Future studies are required to determine whether PGC-1 is also the critical regulator in T cells that links mitochondrial mass to cellular longevity.
It was shown that Tsc1-deficient T cells (discussed in section on naive T cells), which have hyperactive mTOR, have a survival defect and fewer mitochondria than cells with functional Tsc1, fitting with their exit from quiescence and decreased longevity and persistence (35). However, it was shown several years ago that mTOR inhibition decreased the gene expression of the mitochondrial transcriptional regulators PGC-1α, ERR-α, and nuclear respiratory factors in skeletal muscle, resulting in a decrease in mitochondrial gene expression and oxygen consumption in these cells (155). It is not precisely clear at this time why mTOR signaling induces opposing effects on mitochondrial regulation in these settings, but it may reflect tissue-specific differences or temporal events. Since both mTOR and AMPK have been shown to modulate PGC-1α activity (150, 155), the balance of how these factors regulate mitochondrial changes in T cells will need to be explored.
Concluding remarks
In recent years there has been a renewed focus on immune cell metabolism and the overall importance of particular pathways in defined immunological processes has been established. However, further investigation into when a particular metabolism or bioenergetic process is required for cellular function and survival is still needed, especially concerning how metabolic fluctuations in T cells play out during immune responses in vivo. In addition, little is known about the relative contribution of metabolic pathways in T cells to fuel energy production, provide biosynthetic precursors, or generate reducing equivalents to balance the redox status of the cell at a particular time. Defining the metabolic and energetic requirements of T cells during different phases of an immune response is critical for our understanding of T-cell development and for the future design of therapeutics.
Acknowledgments
We would like to thank Edward Pearce, Bart Everts, and David O’Sullivan for critical reading of the manuscript. Erika Pearce’s research is supported by grants from the National Institutes of Health and the Emerald Foundation. Gerritje van der Windt is supported by a fellowship from the Netherlands Organisation for Scientific Research.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 2.Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath--new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 5.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000;6:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 6.Roos D, Loos JA. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation. Exp Cell Res. 1973;77:127–135. doi: 10.1016/0014-4827(73)90561-2. [DOI] [PubMed] [Google Scholar]
- 7.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 8.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell. 2007;18:1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 10.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 11.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200:659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 14.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. Eur J Immunol. 2003;33:2223–2232. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- 17.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 18.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cham CM, Driessens G, O’Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur J Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. J Biol Chem. 1994;269:31484–31490. [PubMed] [Google Scholar]
- 24.Cham CM, Gajewski TF. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J Immunol. 2005;174:4670–4677. doi: 10.4049/jimmunol.174.8.4670. [DOI] [PubMed] [Google Scholar]
- 25.Newsholme EA, Crabtree B, Ardawi MS. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q J Exp Physiol. 1985;70:473–489. doi: 10.1113/expphysiol.1985.sp002935. [DOI] [PubMed] [Google Scholar]
- 26.Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- 27.Wellen KE, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newsholme P, Curi R, Pithon Curi TC, Murphy CJ, Garcia C, Pires de Melo M. Glutamine metabolism by lymphocytes, macrophages, and neutrophils: its importance in health and disease. J Nutr Biochem. 1999;10:316–324. doi: 10.1016/s0955-2863(99)00022-4. [DOI] [PubMed] [Google Scholar]
- 31.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eagle H, Oyama VI, Levy M, Horton CL, Fleischman R. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J Biol Chem. 1956;218:607–616. [PubMed] [Google Scholar]
- 34.Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends in immunology. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 35.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien TF, et al. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41:3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 38.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takada K, et al. Kruppel-like factor 2 is required for trafficking but not quiescence in postactivated T cells. J Immunol. 2011;186:775–783. doi: 10.4049/jimmunol.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10:618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada T, Gierach K, Lee PH, Wang X, Lacorazza HD. Cutting edge: Expression of the transcription factor E74-like factor 4 is regulated by the mammalian target of rapamycin pathway in CD8+ T cells. J Immunol. 2010;185:3824–3828. doi: 10.4049/jimmunol.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 44.Tan JT, et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc Natl Acad Sci USA. 2001;98:8732–8737. doi: 10.1073/pnas.161126098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao RR, Li Q, Bupp MR, Shrikant PA. Transcription Factor Foxo1 Represses T-bet-Mediated Effector Functions and Promotes Memory CD8(+) T Cell Differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nahle Z, et al. CD36-dependent regulation of muscle FoxO1 and PDK4 in the PPAR delta/beta-mediated adaptation to metabolic stress. J Biol Chem. 2008;283:14317–14326. doi: 10.1074/jbc.M706478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Lehtinen MK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 51.Nehme NT, et al. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T cells survival. Blood. 2012;119:3458–3468. doi: 10.1182/blood-2011-09-378364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Q, et al. The tuberous sclerosis complex-mammalian target of rapamycin pathway maintains the quiescence and survival of naive T cells. J Immunol. 2011;187:1106–1112. doi: 10.4049/jimmunol.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, et al. TSC1/2 Signaling Complex Is Essential for Peripheral Naive CD8 T Cell Survival and Homeostasis in Mice. PloS One. 2012;7:e30592. doi: 10.1371/journal.pone.0030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- 57.Hemminki A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 58.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw RJ, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woods A, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 63.Wong AK, Howie J, Petrie JR, Lang CC. AMP-activated protein kinase pathway: a potential therapeutic target in cardiometabolic disease. Clin Sci. 2009;116:607–620. doi: 10.1042/CS20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56:1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 65.Sipula IJ, Brown NF, Perdomo G. Rapamycin-mediated inhibition of mammalian target of rapamycin in skeletal muscle cells reduces glucose utilization and increases fatty acid oxidation. Metabolism. 2006;55:1637–1644. doi: 10.1016/j.metabol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Ha J, Daniel S, Broyles SS, Kim KH. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J Biol Chem. 1994;269:22162–22168. [PubMed] [Google Scholar]
- 68.Kerner J, Hoppel C. Fatty acid import into mitochondria. Biochim Biophys Acta. 2000;1486:1–17. doi: 10.1016/s1388-1981(00)00044-5. [DOI] [PubMed] [Google Scholar]
- 69.Cao Y, Li H, Liu H, Zheng C, Ji H, Liu X. The serine/threonine kinase LKB1 controls thymocyte survival through regulation of AMPK activation and Bcl-XL expression. CellRes. 2010;20:99–108. doi: 10.1038/cr.2009.141. [DOI] [PubMed] [Google Scholar]
- 70.Tamas P, et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur J Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacIver NJ, et al. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J Immunol. 2011;187:4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hand TW, et al. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci USA. 2010;107:16601–16606. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood. 2006;108:600–608. doi: 10.1182/blood-2005-12-4827. [DOI] [PubMed] [Google Scholar]
- 74.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol Cell Biol. 2009;29:5952–5962. doi: 10.1128/MCB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Macintyre AN, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 82.Schaffer JE. Lipotoxicity: when tissues overeat. Current opinion in lipidology. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 83.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 84.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of Immune Responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laplante M, Sabatini DM. mTOR signaling at a glance. JJ Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocrine Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 92.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Michalek RD, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci USA. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D’Souza AD, Parikh N, Kaech SM, Shadel GS. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci USA. 2008;105:7821–7826. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gatza E, et al. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Science Trans Med. 2011;3:67ra68. doi: 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 102.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Banerjee A, et al. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van der Windt GJ, et al. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8(+) T Cell Memory Development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109:1179–1191. doi: 10.1111/j.1471-4159.2009.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37:1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- 107.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27:7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 110.Zaugg K, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramsay RR, Zammit VA. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol Aspects Med. 2004;25:475–493. doi: 10.1016/j.mam.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 112.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 113.Angelini G, et al. Antigen-presenting dendritic cells provide the reducing extracellular microenvironment required for T lymphocyte activation. Proc Natl Acad Sci USA. 2002;99:1491–1496. doi: 10.1073/pnas.022630299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Edinger AL, Thompson CB. Antigen-presenting cells control T cell proliferation by regulating amino acid availability. Proc Natl Acad Sci USA. 2002;99:1107–1109. doi: 10.1073/pnas.042707999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 116.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 117.McGill J, Van Rooijen N, Legge KL. IL-15 trans-presentation by pulmonary dendritic cells promotes effector CD8 T cell survival during influenza virus infection. J Exp Med. 2010;207:521–534. doi: 10.1084/jem.20091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stonier SW, Ma LJ, Castillo EF, Schluns KS. Dendritic cells drive memory CD8 T-cell homeostasis via IL-15 transpresentation. Blood. 2008;112:4546–4554. doi: 10.1182/blood-2008-05-156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Colpitts SL, Stoklasek TA, Plumlee CR, Obar JJ, Guo C, Lefrancois L. Cutting Edge: The Role of IFN-alpha Receptor and MyD88 Signaling in Induction of IL-15 Expression In Vivo. J Immunol. 2012;188:2483–2487. doi: 10.4049/jimmunol.1103609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fabre S, et al. Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J Immunol. 2005;174:4161–4171. doi: 10.4049/jimmunol.174.7.4161. [DOI] [PubMed] [Google Scholar]
- 122.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 123.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 124.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Siegel AM, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 127.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 128.Pellegrini M, et al. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell. 2011;144:601–613. doi: 10.1016/j.cell.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 129.Wegrzyn J, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Savy M, et al. Landscape analysis of interactions between nutrition and vaccine responses in children. J Nutr. 2009;139:2154S–2218S. doi: 10.3945/jn.109.105312. [DOI] [PubMed] [Google Scholar]
- 132.Scrimshaw NS, SanGiovanni JP. Synergism of nutrition, infection, and immunity: an overview. The American journal of clinical nutrition. 1997;66:464S–477S. doi: 10.1093/ajcn/66.2.464S. [DOI] [PubMed] [Google Scholar]
- 133.Iyer SS, et al. Protein energy malnutrition impairs homeostatic proliferation of memory CD8 T cells. J Immunol. 2012;188:77–84. doi: 10.4049/jimmunol.1004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26:495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- 136.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kapahi P, Zid B. TOR pathway: linking nutrient sensing to life span. Science of aging knowledge environment: SAGE KE. 2004;2004:PE34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 140.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 141.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 142.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 143.Greer EL, Banko MR, Brunet A. AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann NY Acad Sci. 2009;1170:688–692. doi: 10.1111/j.1749-6632.2009.04019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Anisimov VN, Semenchenko AV, Yashin AI. Insulin and longevity: antidiabetic biguanides as geroprotectors. Biogerontology. 2003;4:297–307. doi: 10.1023/a:1026299318315. [DOI] [PubMed] [Google Scholar]
- 145.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Anderson RM, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Finley LW, et al. Skeletal muscle transcriptional coactivator PGC-1alpha mediates mitochondrial, but not metabolic, changes during calorie restriction. Proc Natl Acad Sci USA. 2012;109:2931–2936. doi: 10.1073/pnas.1115813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Asami DK, et al. Effect of aging, caloric restriction, and uncoupling protein 3 (UCP3) on mitochondrial proton leak in mice. Exp Gerontol. 2008;43:1069–1076. doi: 10.1016/j.exger.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rera M, et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]