Summary
Sensory adaptation in bacterial chemotaxis is mediated by covalent modifications of specific glutamate and glutamine residues within the cytoplasmic domains of methyl-accepting proteins (MCPs). In Escherichia coli and Salmonella enterica, efficient methylation of MCPs depends on the localization of methyltransferase CheR to MCP clusters through an interaction between the CheR β-subdomain and a pentapeptide sequence (NWETF or NWESF) at the C terminus of the MCP. In vitro methylation analyses utilizing S. enterica and Thermotoga maritima CheR proteins and MCPs indicate that MCP methylation in T. maritima occurs independently of a pentapeptide-binding motif. Kinetic and binding measurements demonstrate that despite efficient methylation, the interaction between T. maritima CheR and T. maritima MCPs is of relatively low affinity. Comparative protein sequence analyses of CheR β-subdomains from organisms having MCPs that contain and/or lack pentapeptide-binding motifs identified key similarities and differences in residue conservation, suggesting the existence of two distinct classes of CheR proteins: pentapeptide-dependent and pentapeptide-independent methyltransferases. Analysis of MCP C-terminal ends showed that only ~10% of MCPs contain a putative C-terminal binding motif, the majority of which are restricted to the different proteobacteria classes (α, β, γ, δ). These findings suggest that tethering of CheR to MCPs is a relatively recent event in evolution and that the pentapeptide-independent methylation system is more common than the well characterized pentapeptide-dependent methylation system.
Introduction
Most proteins undergo post-translational modifications that alter their function, influencing properties such as localization, lifetime, or activity. To date more than 200 distinct post-translational modifications have been identified, most often coming in the form of proteolytic cleavage events or covalent modifications at specific amino acid residues (Blom et al., 2004; Khidekel and Hsieh-Wilson, 2004). While proteolytic cleavage is an irreversible modification, covalent modifications in many instances are reversible. In prokaryotes, two common reversible modifications are protein phosphorylation and carboxyl methylation. These modifications, performed by a protein kinase or methyltransferase, may be reversed by the action of an antagonistic enzyme, a phosphatase or methylesterase. A recent comprehensive census of signal transduction proteins from 167 completed bacterial and archaeal genomes has shown protein methylation to be a key post-translational modification in bacterial signaling, with methyl-accepting chemotaxis proteins (MCPs) being the third most abundant transmembrane signaling proteins ranking behind only histidine kinases and diguanylate cyclases (Galperin, 2005).
In the well-studied chemotaxis systems of Escherichia coli and Salmonella enterica serovar Typhimurium, several homologous transmembrane receptors (MCPs) sense extracellular stimuli, producing signals that are transmitted to their cytoplasmic domains. These domains regulate an associated two-component phosphotransfer signal transduction system that controls flagellar rotation (Bourret and Stock, 2002; Falke and Kim, 2000; Falke and Hazelbauer, 2001). Chemotaxis sensory systems are able to adapt to persistent stimuli, returning cells to their prestimulus behavior and allowing for highly sensitive detection of stimuli over a wide dynamic range. Adaptation is in part mediated by reversible covalent modifications. Specific glutamate residues within the cytoplasmic domains of receptors are methylated by AdoMet-dependent methyltransferase CheR and demethylated by methylesterase/deamidase CheB (Kehry and Dahlquist, 1982; Springer and Koshland, 1977; Stock and Koshland, 1978; Terwilliger and Koshland, 1984). The molecular mechanisms of chemotaxis signaling and adaptation have been most extensively characterized in E. coli and S. enterica (Bren and Eisenbach, 2000; Falke et al., 1997; Wadhams and Armitage, 2004). Studies of chemotaxis and MCP methylation in other organisms (Armitage and Schmitt, 1997; Astling et al., 2006; Martin et al., 2001; Perazzona and Spudich, 1999; Szurmant and Ordal, 2004; Ward and Zusman, 1997) have revealed both similarities and differences to the E. coli/S. enterica chemotaxis pathway.
One such difference is the presence of a conserved pentapeptide sequence (NWETF or NWESF) located at the extreme C-terminal end of some transmembrane receptors. While this motif is required for efficient methylation, demethylation, and deamidation in E. coli and S. enterica (Barnakov et al., 1999; Li and Hazelbauer, 2005; Wu et al., 1996), it is absent from many bacterial MCPs (Shiomi et al., 2002b). In E. coli and S. enterica, high abundance receptors Tar (aspartate), Tsr (serine), and Tcp (citrate) possess a pentapeptide sequence, while low abundance receptors Trg (ribose-galactose) and Tap (dipeptide) do not. Studies involving truncation and/or mutation of key residues within this pentapeptide motif have demonstrated lowered methylation levels and ineffective tactic responses (Feng et al., 1997; Le Moual et al., 1997; Li et al., 1997; Okumura et al., 1998). Conversely, addition of this pentapeptide sequence to low abundance receptors has been shown to greatly improve methylation (Barnakov et al., 1998; Feng et al., 1999). The crystal structure of S. enterica CheR bound to the pentapeptide NWETF identified the binding site within CheR to be the small β-subdomain that is inserted into the C-terminal methyltransferase catalytic domain (Djordjevic and Stock, 1998). This motif provides a high affinity binding interaction (~2 μM Kd) (Wu et al., 1996) that increases CheR concentration at receptor clusters, facilitating methylation of nearby high and low abundance receptors (Le Moual et al., 1997; Li et al., 1997; Li and Hazelbauer, 2005). Thus in E. coli and S. enterica, interactions between CheR and receptors involve two distinct sets of contacts: the tethering of CheR to the pentapeptide through the β-subdomain and the catalytically productive contact between the methylation regions of the receptor and the active site of CheR. The α2 helix within the N-terminal domain of CheR has been shown to be critical for the latter interaction (Perez et al., 2004; Shiomi et al., 2002b).
Thermotoga maritima is a thermophilic bacteria that thrives at high temperatures (Swanson et al., 1996), making laboratory studies of bacterial physiology difficult. Furthermore, the organism lacks a tractable genetic system. However, T. maritima has become a model system for structural characterization of chemotaxis proteins and signaling complexes. Structures of many T. maritima chemotaxis proteins have been determined, in several notable cases where structures of homologs from mesophilic organisms have been unobtainable (Bilwes et al., 1999; Bilwes et al., 2001; Chao et al., 2006; Griswold et al., 2002; Park et al., 2006; Park et al., 2004a; Park et al., 2004b; Quezada et al., 2004; Usher et al., 1998). Recently, studies addressing methylation in T. maritima showed four different receptors to be efficiently methylated by T. maritima CheR (Perez et al., 2006), despite none possessing a recognizable pentapeptide-binding sequence (Nelson et al., 1999). Receptor methylation in B. subtilis and H. salinarium, organisms that also lack the pentapeptide, has also been reported (Hanlon and Ordal, 1994; Kirby et al., 1999; Perazzona and Spudich, 1999). Thus, while CheR and CheB interactions with pentapeptide are critical for methylation, demethylation, and deamidation in E. coli and S. enterica, this is likely not the case for the majority of bacteria.
Interestingly, although MCPs of many bacteria do not contain the C-terminal binding motif, all CheR homologs possess a β-subdomain (Shiomi et al., 2002b). What is the role of the β-subdomain in organisms with receptors that all lack pentapeptide-binding motifs? Are there differences between a β-subdomain that interacts with pentapeptides and one that does not? Can CheR proteins from organisms with receptors that lack pentapeptides recognize the C-terminal binding motif in MCPs from different organisms? In this study, utilizing S. enterica and T. maritima CheR proteins and receptors, we have begun to address some of these questions. Our results demonstrate that T. maritima CheR, unlike S. enterica CheR, is not dependent upon pentapeptide binding for efficient methylation. Despite the likelihood that T. maritima CheR interacts with receptors only at the sites of methylation, a combination of binding studies and kinetic analyses revealed this to be a low affinity interaction, dissimilar to the high affinity binding exhibited between CheR and the pentapeptide. Finally, comparative protein sequence analyses of the CheR β-subdomain from organisms with receptors containing pentapeptide motifs with those from organisms with receptors lacking pentapeptide-binding motifs indicate key differences between the two groups, defining predictive features and providing further insight into CheR-pentapeptide interactions.
Results
Characterization of cross-species methylation between S. enterica and T. maritima
Cross-species methylation and demethylation of E. coli/S. enterica receptors was previously demonstrated with CheR proteins from B. subtilis and Rhodobacter sphaeroides (Burgess-Cassler and Ordal, 1982; Martin et al., 2001) and T. maritima CheB protein (Anand and Stock, 2002). Moreover, methylation in T. maritima has recently been characterized, with determination of methylation rates for both receptor cytoplasmic domains and full-length transmembrane receptors catalyzed by T. maritima CheR (Perez et al., 2006). To further characterize T. maritima receptor methylation and to gain insight into methylation in organisms lacking the pentapeptide, we performed cross-species in vitro methylation assays utilizing S. enterica and T. maritima CheR proteins and receptors (wild-type S. enterica Tar, WT Tar; S. enterica Tar lacking the five C-terminal residues, TarΔpp; and T. maritima TM1428; TM1428). Methylation rates, determined at 30°C and 37°C, indicated that both S. enterica and T. maritima CheR efficiently methylate WT Tar, with rates for S. enterica CheR being ~3-fold higher (Table 1). Methylation rates for T. maritima CheR versus WT Tar are slightly lower than those observed with its cognate receptor, TM1428 (Perez et al., 2006). Conversely, methylation rates exhibited by S. enterica CheR versus TM1428 were significantly lower (~25-fold) compared to T. maritima CheR (Table 1). These low methylation rates for S. enterica CheR versus TM1428 appear to be due to the absence of the pentapeptide sequence in these receptors, since a similar drop in the rate of methylation was observed when the pentapeptide was eliminated from Tar (TarΔpp) (Table 1). Interestingly, methylation rates for T. maritima CheR with TarΔpp, were no different than those obtained with WT Tar (Table 1). Thus, despite possessing a β-subdomain, T. maritima CheR does not appear to interact with the NWETF motif, suggesting that methylation in T. maritima is pentapeptide independent.
TABLE 1.
Cross species methylation rates a
| Receptor | Methyltransferase | 30°C | 37°C |
|---|---|---|---|
| WT Tar | S. enterica CheR | 0.66 ± 0.07 | 0.86 ± 0.06 |
| T. maritima CheR | 0.21 ± 0.03 | 0.32 ± 0.04 | |
| TM1428fl | S. enterica CheR | 0.016 ± 0.001 | 0.018 ± 0.002 |
| T. maritima CheR | 0.35 ± 0.02b | 0.50 ± 0.04b | |
| TarΔpp | S. enterica CheR | 0.018 ± 0.002 | 0.020 ± 0.002 |
| T. maritima CheR | 0.20 ± 0.04 | 0.31 ± 0.05 |
Methyltransferase activities are expressed as mol CH3· mol CheR−1·min−1 and are averages of three independent experiments.
Values are from Perez et al. {Perez, 2006 #3187}.
Effects of C-terminal deletion of TM1428 on methylation
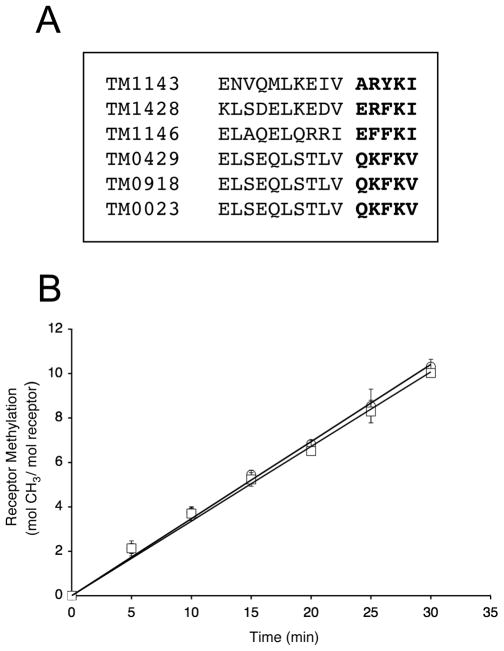
Based on primary sequence alignments, T. maritima receptors do not contain a recognizable NWETF motif at the end of a flexible linker that extends beyond the predicted coiled-coil motif. However, to investigate whether a different C-terminal motif might mediate interactions with T. maritima CheR, sequence alignments of all six T. maritima transmembrane chemoreceptors were generated, revealing strong conservation of residues located at the C termini (Fig. 1A). Previously, alanine scanning mutagenesis of the NWETF binding sequence in E. coli Tar suggested that the hydrophobicity and size of the aromatic residues Trp and Phe were critical for interaction with CheR and for efficient methylation (Shiomi et al., 2000). Close examination of the last five residues of T. maritima receptors showed the presence of two conserved hydrophobic residues (at the third and fifth positions), including one position containing a bulky aromatic residue (Trp or Tyr) (Fig. 1A). Despite some similarity between E. coli/S. enterica and T. maritima receptor C termini, the structures adopted by the residues that lead up to the C-terminal ends are different. E. coli/S. enterica high abundance receptors possess C-terminal tails (30–35 residues in length) that are predicted to be flexible and are not observed in the crystal structure (Kim et al., 1999; Le Moual and Koshland, 1996). This tail presumably acts as a flexible tether, allowing CheR to methylate neighboring receptors (Le Moual et al., 1997; Li et al., 1997; Li and Hazelbauer, 2005). Also, it has been shown that deletion of all or part of the tail reduces the activity of CheR as well as the other adaptational modifications, indicating its importance for proper chemoreceptor function (Le Moual et al., 1997; Li et al., 1997; Li and Hazelbauer, 2006). However, for T. maritima receptors, secondary structure predictions (data not shown) and the recent determination of the crystal structure of the cytoplasmic domain of TM1143 (Park et al., 2006) indicate that an alpha-helical structure is maintained throughout the cytoplasmic domain up to the C terminus, which ends prior to the 30-residue flexible extension of the major E. coli/S. enterica receptors. Although T. maritima receptors do not possess a flexible tail, the importance of the receptor C terminus for methylation by CheR was investigated. A mutant TM1428 receptor lacking five C-terminal residues, ERFKI, was generated (TM1428Δ5) and methylation rates for this truncated receptor were measured. Results showed that methylation rates for TM1428Δ5 (0.34 ± 0.04 mol CH3·min-1·mol CheR-1) were similar to those for TM1428 (0.35 ± 0.02 mol CH3·min-1·mol CheR-1) and that these residues, as expected, did not play any perceptible role in receptor methylation (Fig. 1B).
FIG. 1. Analysis of T. maritima receptor C-terminal ends and their role in methylation.
A. Sequence alignment of T. maritima transmembrane receptor C-terminal ends. A multiple sequence alignment was generated with ClustalW (Thompson et al., 1994) using the highly conserved domain (HCD) core (Zhulin, 2001) as the anchor for alignment. The last fifteen residues of each of receptor are shown, with the last five residues in bold. B. Methylation of TM1428 and TM1428Δ5. Methyltransferase activity was determined as described in Experimental Procedures using TM1428 (O) and TM1428Δ5 (□) as substrates. Assays were performed with 1.95 and 3.90 pmol of T. maritima CheR and the data shown are the average values from three independent experiments with standard errors.
Characterization of binding and kinetic parameters of methyltransferase CheR and TM1143c
A consensus methylation sequence within T. maritima MCPs has been established (Perez et al., 2006). For T. maritima and other organisms that contain receptors all lacking a C-terminal binding motif, CheR interactions with MCPs are likely limited to the conserved methylation regions, which could potentially serve dual roles, both as high affinity binding sites and as sites of modification. A precedent exists for this in the case of the T. maritima receptor-modifying deamidase/demethylase CheD. Pulldown assays utilizing His-tagged CheD and the cytoplasmic domain of TM1143 (TM1143c) demonstrated high affinity interactions, with CheD-TM1143c complex formation visualized at relatively low concentrations (1.2 μM CheD and 10 μM TM1143c) (Chao et al., 2006). CheD also forms a relatively high affinity complex (Kd = 0.9–1.4 μM) with the α2′ helix of CheC, which mimics the receptor methylation region (Park et al., 2004b, Chao, 2006 #3075). To assess binding between CheR and TM1143c, pulldown experiments were performed utilizing His-tagged TM1143c. Unlike the high affinity CheD-TM1143c interaction, complex formation of CheR with TM1143c was not observed (data not shown). Attempts to promote complex formation with higher concentrations of CheR and TM1143c (up to 60 μM of each) were unsuccessful. Based on conservative estimates of the detection limits in these assays, we conclude that CheR binding to TM1143c is a low affinity interaction with a theoretical dissociation constant (Kd) >0.6 mM.
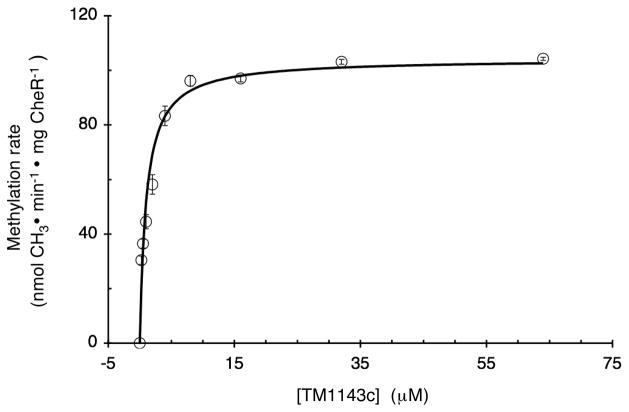
To further assess the interactions of T. maritima CheR with receptors, initial velocities of methylation were determined over a range of concentrations of TM1143c. Previous kinetic studies of S. enterica CheR methylation of Tar receptor-enriched membrane fractions exhibited typical Michaelis-Menton kinetics (Simms et al., 1987). We have shown that soluble T. maritima receptor cytoplasmic domains are methylated similarly to their full-length membrane counterparts in in vitro methylation reactions (Perez et al., 2006). The kinetic analyses performed in the present study take advantage of using a soluble substrate that can be examined at substantially higher concentrations than are achievable with receptor-enriched membrane fractions. Our results yielded Km and Vmax values of 1.14 ± 0.2 μM, and 104 nmol·min−1·mg CheR−1, which is the equivalent to a Kcat of 3.6 ± 0.4 mol CH3·min−1·mg CheR−1 for reactions at 30°C (Fig. 2). These kinetic parameters are surprisingly similar to those previously determined for S. enterica CheR and Tar, (Km = 4.2 μM, Kcat = 10 mol CH3·min−1·mg CheR−1) (Simms et al., 1987; Simms, 1991 #428). However, the Km value for CheR and TM1143c was not reflective of the Kd for CheR-receptor binding. In contrast, binding studies with S. enterica CheR and Tar receptor cytoplasmic fragments containing the pentapeptide-binding motif showed it to be a high affinity interaction with a Kd of ~2 μM, similar to the Km values for both S. enterica and T. maritima receptor methylation. Consistent with these affinity measurements, CheR was detected in Ni-NTA affinity pulldown assays utilizing a His-tagged Tar receptor cytoplasmic fragment (Wu et al., 1996), in contrast to the negative results obtained with T. maritima CheR and TM1143c. Thus, we conclude that the methylation regions of the receptors are not high affinity binding sites for T. maritima CheR. Despite weak binding, T. maritima receptor methylation proceeds with rates comparable to those observed for S. enterica.
FIG. 2. Kinetics of methylation of TM1143c catalyzed by T. maritima CheR.
Methyltransferase activity was determined as described in Experimental Procedures by incubating different concentrations of TM1143c (0 to 64 μM) with 0.20 μM methyltransferase CheR for 25 min at 30°C. Km and Kcat values of 1.14 ± 0.20 μM and 3.60 ± 0.40 mol CH3·min-1·mg CheR-1 were estimated by fitting data to the Michaelis-Menton equation using SigmaPlot 8.0. The data shown are the average values of initial velocities from three experiments with standard errors and the fitted curve used for estimation of Km and Kcat.
Screening and analysis of MCP C-terminal ends
It was previously reported that MCPs of many bacteria lack the C-terminal pentapeptide-binding motif (Shiomi et al., 2002b). To investigate which organisms possess putative pentapeptide sequences, we utilized a database of 167 completed bacterial and archaeal genomes to screen MCP C-terminal ends (Galperin, 2005). In total, 1,121 MCPs were analyzed from 78 organisms, of which only ~10% (113 MCPs total) were found to contain the putative binding motif (Table 2). Also, except for two MCPs identified in phylum Spirochetes, the overwhelming majority of the MCPs identified with a putative pentapeptide sequence (111 of 113) were restricted to the different proteobacteria classes (α, β, γ, δ) (Table 2). To examine the residues constituting the selected putative pentapeptide sequences and their conservation, a weblogo (Schneider and Stephens, 1990) was generated (inset to Table 2). Clearly, as expected based on the criteria we used to screen for putative binding motifs (see Experimental Procedures), the strongest conservation was seen at positions 2 (Trp - 83%, Phe - 17%) and 5 (Phe - 99%) of the pentapeptide. Notably, there was strict conservation of Phe at position 5 and also a paucity of Tyr at either position. Position 3 was also conserved, with 87% of residues at this position being either Glu (65%) or Gln (22%) (inset to Table 2). The crystal structure of a complex of S. enterica CheR and an NWETF pentapeptide demonstrated the structural importance of these three residues of the pentapeptide for interactions with the CheR β-subdomain (Djordjevic and Stock, 1998). The strong conservation displayed among these residues likely reflects their importance in receptors of all organisms that are reliant on CheR-pentapeptide interactions for efficient methylation.
TABLE 2.
MCPs containing putative pentapeptide sequences
| Phylum/Organisma | MCPsb | MCPs w/pp | CheR |
|---|---|---|---|
| α-proteobacteria | |||
| Agrobacterium tumefaciens | 20 | 9 | 1 |
| Caulobacter crescentus | 18 | 6 | 3 |
| Sinorhizobium meliloti | 9 | 4 | 1 |
| Zymomonas mobilis | 3 | 1 | 1 |
| β-proteobacteria | |||
| Bordetella bronchiseptica | 8 | 1 | 1 |
| Bordetella pertussis | 5 | 1 | 1 |
| Burkholderia mallei | 17 | 3 | 1 |
| Burkholderia pseudomallei | 21 | 5 | 1 |
| Chromobacterium violaceum | 42 | 3 | 3 |
| Nitrosomonas europaea | 3 | 2 | 1 |
| Ralstonia solanacearum | 22 | 4 | 1 |
| γ-proteobacteria | |||
| Erwinia carotovora | 36 | 19 | 1 |
| Escherichia coli K12 | 5 | 2 | 1 |
| Escherichia coli O157:H7 | 5 | 2 | 1 |
| Photobacterium profundu m | 39 | 1 | 2 |
| Photorhabdus luminescens | 2 | 1 | 1 |
| Pseudomonas aeruginosa | 26 | 1 | 2 |
| Pseudomonas syringae | 48 | 1 | 3 |
| Salmonella Typhimurium LT2 | 9 | 4 | 1 |
| S. enterica serovar Typhi CT18 | 6 | 3 | 1 |
| Shewanella oneidensis | 26 | 2 | 3 |
| Shigella flexneri | 4 | 1 | 1 |
| Vibrio cholerae | 45 | 2 | 3 |
| Vibrio vulnificus | 52 | 1 | 3 |
| Xanthomonas axonopodis | 21 | 12 | 3 |
| Xanthomona s campestris | 20 | 8 | 3 |
| Yersinia pestis | 6 | 2 | 1 |
| Yersinia pseudotuberculosis | 7 | 3 | 1 |
| δ-proteobacteria | |||
| Desulfovibrio vulgaris | 28 | 2 | 2 |
| Geobacter sulfurreducens | 33 | 5 | 4 |
| Spirochetes | |||
| Borrelia burgdorferi | 5 | 1 | 2 |
| Borrelia garinii | 5 | 1 | 2 |
Each specific phylum or class is underlined.
Total number of MCPs were obtain from the signal transduction
census {Galperin, 2005 #3139}.

Comparative protein sequence analysis of the CheR β-subdomain
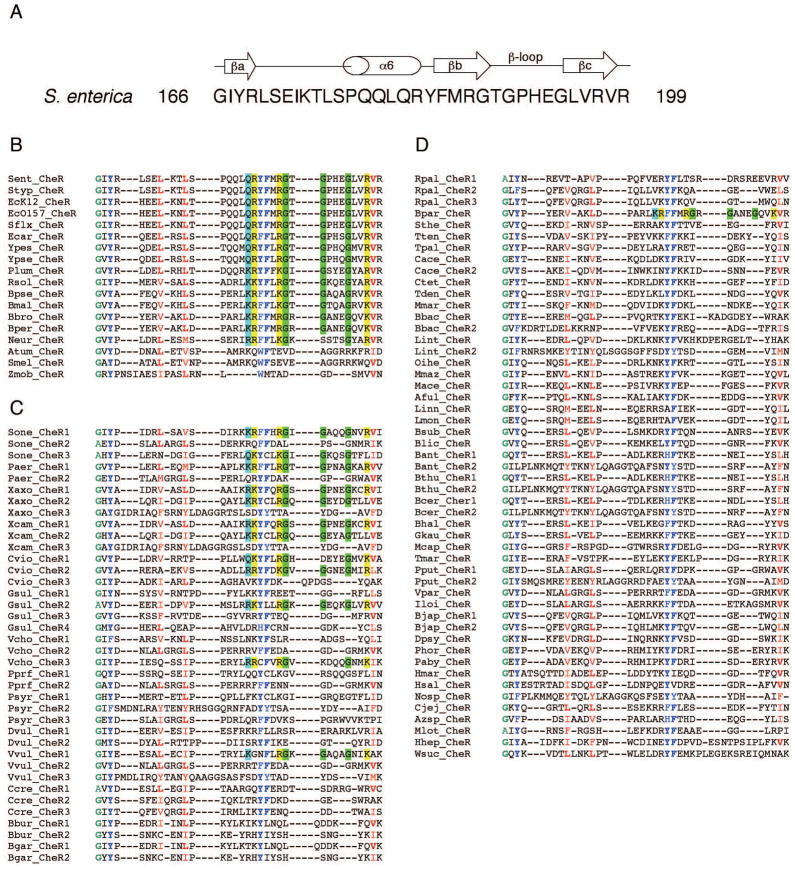
The β-subdomain is composed of an α helix and a short antiparallel three-stranded β sheet (Djordjevic and Stock, 1997) (Fig. 3A) and is present in all CheR homologs despite the absence of MCPs containing a C-terminal binding motif in the majority of organisms (Shiomi et al., 2002b). Our screen for putative pentapeptide-binding motifs not only confirmed the finding that they were absent from most MCPs, but also served as a basis for further analysis of the β-subdomain of CheR. Utilizing the TIGR database (www.tigr.org), a total of 107 homologous CheR proteins were selected from the MCP-containing bacterial genomes that were screened (MCP containing organisms from phylums Cyanobacteria and Deinoccus did not contain any CheR homologs). A multiple sequence alignment was generated from the 107 β-subdomain sequences (Fig. 3B, C, D). Each β-subdomain was then separated into one of three groups, based on the following criteria: group I consisted of CheR β-subdomains from organisms containing only 1 CheR protein and ≥1 MCP with a putative pentapeptide sequence; group II consisted of CheR β-subdomains from organisms containing ≥2 CheR proteins and ≥1 MCP with a putative pentapeptide sequence; and group III consisted of CheR β-subdomains from organisms containing ≥1 CheR and only MCPs that lacked putative pentapeptide sequences. The groups of sequences were examined for key similarities and/or differences within each group, as well as for universal conservation of residues over the entire set of β-subdomain sequences.
FIG. 3. Multiple sequence alignments of CheR β-subdomains.
A. Secondary structure diagram of the S. enterica CheR β-subdomain derived from crystal structures (Djordjevic and Stock, 1997, Djordjevic, 1998 #1644) with the amino acid sequence and residue numbers indicated. A multiple sequence alignment for all β-subdomain sequences was generated with ClustalW (Thompson et al., 1994) and the β-subdomain sequences were categorized as described in the text. B. Group I: β-subdomains from organisms containing only 1 CheR protein and ≥1 MCP with a putative pentapeptide sequence. C. Group II: β-subdomains from organisms containing ≥2 CheR proteins and ≥1 MCP with a putative pentapeptide sequence. D. Group III: β-subdomains from organisms containing ≥1 CheR and only MCPs that lacked putative pentapeptide sequences. Conserved amino acid residues in all β-subdomain sequences (≥85%) are colored: small (A, G), green; hydrophobic (L, V, I, M, F, Y, W, H), red; and aromatic (Y, F, W, H), blue; with the most common residue at each position in bold. Amino acid residues that are conserved and that are proposed to be important for CheR-pentapeptide interactions are highlighted: small (G), green; positively charged (R, K), yellow; and side chain amine/amide containing residues (Q, K, R), cyan. The organism abbreviation for each CheR homolog and the corresponding TIGR identification number are: Aful, Archaeoglobus fulgidus (AF1037); Atum, Agrobacterium tumefaciens (NT02AT0609); Azsp, Azoarcus sp. EbN1 (NT01AE1293); Bant, Bacillus anthracis Ames(BA1665, BA0995); Bbac, Bdellovibrio bacteriovorus (NT02BB3177, NT02BB2588); Bbro, Bordetella bronchiseptica (NT01BB2503); Bbur, Borrelia burgdorferi (BB0040, BB0414); Bcer, Bacillus cereus (NT01BC1535, NT01BC0909); Bgar, Borrelia garinii (NT01BG0039, NT01BG0418); Bhal, Bacillus halodurans (NT01BH1876); Bjap, Bradyrhizobium japonicum (NT01BJ0473, NT01BJ2738); Blic, Bacillus licheniformis (NT03BL2556); Bmal, Burkholderia mallei (BMA2856); Bpar, Bordetella parapertussis (NT02BP1517); Bper, Bordetella pertussis (NT03BP1055); Bpse, Burkholderia pseudomallei (ntbp3311); Bsub, Bacillus subtilis (NT01BS2884); Bthu, Bacillus thuringiensis (NT02BT1675, NT02BT1053); Cace, Clostridium acetobutylicum (NT01CA2443, NT01CA0131); Ccre, Caulobacter crescentus (CC0435, CC0598, CC3472); Cjej, Campylobacter jejuni (NT01CJ0929); Ctet, Clostridium tetani (NT02CT1848); Cvio, Chromobacterium violaceum (NT01CV2459, NT01CV3397, NT01CV3658); Dpsy, Desulfotalea psychrophila (NT01DP3085); Dvul, Desulfovibrio vulgaris (DVU1595, DVU2076); Ecar, Erwinia carotovora (NT06EC1756); EcK12, Escherichia coli K12 (NT01EC2290); Ec0157, Escherichia coli O157:H7 (NT02EC2821); Gkau, Geobacter kaustophilus (NT01GK2440); Gsul, Geobacter sulfurreducens (GSU0295, GSU1143, GSU2215, GSU3195); Hhep, Helicobacter hepaticus (NT02HP0476); Hmar, Haloarcula marismortui (NT01HMA2142); Hsal, Halobacterium salinarum (NT01HS0757); Iloi, Idiomarina loihiensis (NT01IL1191); Linn, Listeria innocua (NT01LI0715); Lint, Leptospira interrogans (NT02LI2313, NT02LI1981); Lmon, Listeria monocytogenes (NT01LM0742); Mace, Methanosarcina acetivorans (NT02MA3813); Mcap, Methylococcus capsulatus (MCA0829); Mmar, Methanococcus maripaludis (NT04MM0987); Mlot, Mesorhizobium loti (NT02MLB0021); Mmaz, Methanosarcina mazei (NT01MM1835); Neur, Nitrosomonas europaea (NT01NE2059); Nosp, Nostoc sp. PCC7120 (NT01NS2346); Oihe, Oceanobacillus iheyensis (NT01OI1946); Paby, Pyrococcus abyssi (NT01PA1746); Paer, Pseudomonas aeruginosa (NT03PA0199, NT03PA3856); Phor, Pyrococcus horikoshii (NT01PH0504); Plum, Photorhabdus luminescens (NT01PL2011); Pprf, Photobacterium profundum (NT01PP0812, NT01PP0945); Pput, Pseudomonas putida (PP4392, PP3760); Psyr, Pseudomonas syringae (PSPPH0802, PSPH2602, PSPPH3412); Rpal, Rhodopseudomonas palustris (NT02RP0142, NT02RP1682, NT02RP1731); Rsol, Ralstonia solanacearum (NT01RSA1413); Sent, Salmonella enterica serovar Typhimurium CT18 (NT03ST2156); Sflx, Shigella flexneri (NT02SF2178); Smel, Sinorhizobium meliloti (NT01SM0883); Sone, Shewanella oneidensis (SO2124, SO3251, SO2325); Sthe, Symbiobacterium thermophilum (NT07ST1815); Styp, Salmonella Typhimurium LT2 (NT05SE0993); Tden, Treponema denticola (TDE0647); Tmar, Thermotoga maritima (TM0464); Tpal, Treponema pallidum (TP0630); Tten, Thermoanaerobacter tengcongens (NT01TT1507);Vcho; Vibrio cholerae (VC1399, VC2201, VCA1091); Vpar, Vibrio parahaemolyticus (NT01VP0730); Vvul, Vibrio vulnificus (NT01VVA1160, NT01VV0231, NT01VVA0376); Wsuc, Wolinella succinogenes (NT01WS1194); Xaxo, Xanthomonas axonopodis (NT01XA2561, NT01XA3795, NT01XA1741); Xcam, Xanthomonas campestris (NT01XC2547, NT01XC3635, NT01XC1630); Ypes, Yersinia pestis (NT02YP2177); Ypse, Yersinia pseudotuberculosis (NT05YP2660); Zmob, Zymomonas mobilis (NT01ZM0078).
Group I members include CheR proteins from E. coli and S. enterica, which have been the major focus for study of β-subdomain-pentapeptide interactions. Previous analysis of the S. enterica CheR-NWETF crystal structure revealed extensive interactions between the β-subdomain and the highly conserved residues of the binding motif. Specifically, β-subdomain residue Arg187 forms a salt bridge with the Glu of the pentapeptide and His192 and Arg197 interact with the Trp residue of the NWETF binding motif (Djordjevic and Stock, 1998). In addition, the C-terminal carboxylate of the pentapeptide forms a hydrogen bond with the amide nitrogen of the side chain of Gln182 (Djordjevic and Stock, 1998). Moreover, comparison to the CheR-AdoHcy structure solved in the absence of the pentapeptide (Djordjevic and Stock, 1997) demonstrated that side chains of Gln182, Arg187, His192, and Arg197 underwent the most significant conformational changes upon pentapeptide binding (Djordjevic and Stock, 1998). Examination of β-subdomain sequences confirmed the importance of these residues for CheR binding to pentapeptide, since three of the four (excluding His192) were highly conserved in group I β-subdomain sequences (Fig. 3B) and were poorly conserved in group III β-subdomain sequences from organisms lacking receptors with pentapeptide-binding motifs (Fig. 3D). Further support for the importance of these residues in binding the pentapeptide is illustrated by a CheR His192Ala/Arg197Ala double mutant that exhibited significantly slower rates of migration in soft agar chemotaxis assays compared to wild type and that did not exhibit CheR localization to cell poles (Shiomi et al., 2002b).
In addition to Gln182, Arg187, and Arg197, our sequence analysis also identified three glycine residues (Gly188, Gly190, and Gly194) in the β-subdomain β-loop that are highly conserved in group I sequences (Fig. 3B), but are not well conserved in those of group III (Fig. 3D). The first study of sequence alignments of a small number of MCP-methytransferase pairs from different organisms, suggested that the β-subdomain possessed either a long or short β-loop, and that this difference might influence binding of the pentapeptide (Djordjevic and Stock, 1998). Subsequent studies, utilizing 8 CheR and CheR-related protein sequences, confirmed this difference in the length of the β-loop (Shiomi et al., 2002b). Furthermore, Shiomi et al. found that CheR proteins from organisms containing MCPs with pentapeptide-binding motifs possessed longer β-loops and proposed that this difference in length might reflect differences in the mode of receptor recognition by CheR (Shiomi et al., 2002b). Our analysis indicated that indeed, β-subdomain sequences from organisms containing MCPs with binding motifs had longer β-loops (Fig. 3B, 3C), while the majority of β-subdomains from organisms lacking MCPs with binding motifs had shorter β-loops (Fig. 3D). Thus the longer β-loop and the three highly conserved glycines embedded within it are likely to be important in enabling pentapeptide binding. Notably, phi/psi angles for all three glycine residues were between 110° and 115°, exhibiting backbone conformations unattainable by most other residues.
Inspection of Group II sequences and surface analysis of the β-subdomain
Structural, mutational, and sequence analyses have revealed the importance of six residues, Gln182, Arg187, Arg197, and β-loop residues Gly188, Gly190, and Gly194, for CheR-pentapeptide interactions. Utilizing these key residues we examined the group II β-subdomain sequences to investigate if they fit the profile of a CheR protein that recognized a C-terminal pentapeptide (i.e. similar to group I) or lacked these residues (i.e. similar to group III). Inspection of these β-subdomain sequences demonstrated that both types were present, with 12 of the 38 β-subdomain sequences exhibiting conservation of at least five of the six key residues (Fig. 3C). Interestingly, the eight organisms (C. violaceum, G. sulfurreducens, P. aeruginosa, S. oneidensis, V. cholerae, V. vulnificus, X. axonopodis, and X. campestris) that possessed CheR proteins with sequence conservation similar to group I, in all cases, also contained at least one other CheR protein that exhibited poor conservation of those same residues (Fig. 3C). This suggests that in these organisms, two different types of CheR proteins exist: some that utilize a pentapeptide-dependent mechanism for methylation and others that do not. For example, in S. oneidensis, based on residue conservation, CheR1 and CheR3 likely require binding to a pentapeptide for efficient methylation of MCPs, similar to the E. coli and S. enterica CheR proteins. In contrast, CheR2, similar in sequence conservation to group III CheR proteins, presumably methylates MCPs without recognizing the C-terminal binding motif, similar to the methylation system in T. maritima.
Analysis of residue conservation in the β-subdomains identified certain residues to be universally conserved and revealed differences that distinguish pentapeptide-dependent from pentapeptide-independent CheR proteins. Differences between these two groups were visualized by mapping sequence conservation onto the surface of a structural model of the β-subdomain (Djordjevic and Stock, 1998) using the Consurf server (Glaser et al., 2003). Two multiple sequence alignments were generated from sequences that were predicted to be either pentapeptide-dependent or pentapeptide-independent β-subdomains. Sequences from groups I and II that displayed high conservation of Gln182, Arg187, Arg197, Gly188, Gly190, and Gly194 were selected to represent pentapeptide-dependent β-subdomains (27 sequences). The remaining sequences from groups I and II and all sequences from group III (80 sequences) constituted the set of pentapeptide-independent β-subdomains.
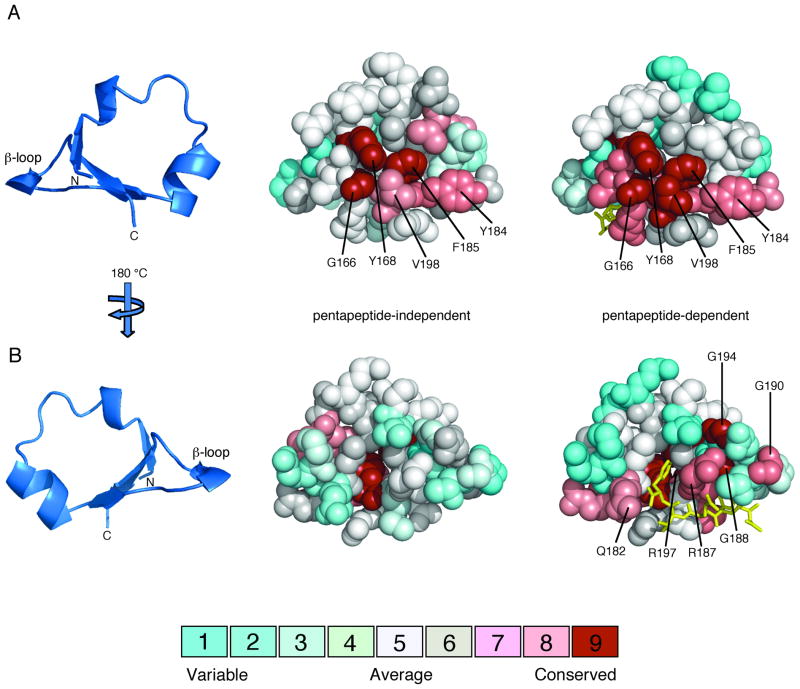
The β-subdomain can be considered to contain two distinct surfaces: an inner face that packs against the rest of the C-terminal domain of CheR and an outer face that is exposed to solvent and potentially available for interactions with other proteins. Several residues that are highly conserved in both pentapeptide-dependent and pentapeptide-independent β-subdomains (Gly166, Tyr168, Tyr184, Phe185 and Val198) all map to a subset of the inner face (Fig. 4A and 5). The outer face of pentapeptide-dependent β-subdomains contains many highly conserved residues, whereas this surface shows little conservation in pentapeptide-independent β-subdomains (Fig. 4B and 5). This patch of conserved residues (Gln182, Arg187, Arg197, Gly188, Gly190, and Gly194) in pentapeptide-dependent β-subdomains lines the pentapeptide-binding pocket. Notably, outside of the surface that interfaces with the catalytic domain of CheR and the pentapeptide-binding pocket in one class of β-subdomains, there are no other strongly conserved surfaces that might be implicated in other functions such as interactions with the conserved methylation regions of receptor substrates.
FIG. 4.
Amino acid conservation on the surfaces of pentapeptide-independent and pentapeptide-dependent β-subdomains. Multiple sequence alignments of pentapeptide-independent and pentapeptide-dependent CheR β-subdomains, classified in Figure 3, were generated using ClustalW (Thompson et al., 1994). Amino acid conservation scores for each residue were calculated using the Consurf maximum likelihood method (Glaser et al., 2003) (http://consurf.tau.ac.il) and were mapped onto space-filling models of the CheR β-subdomain (PDB accession code 1BC5) (Djordjevic and Stock, 1998) using Pymol (Delano, 2002). The coloring scheme for conservation ranges from cyan (variable) to red (conserved) and the pentapeptide (gold) is shown in stick representation. A. β-subdomain interior face. The ribbon diagram of the β-subdomain (left) indicates the orientation of the space-filling models, viewed toward the surface that packs against the catalytic domain of CheR. Residues that are highly conserved in both pentapeptide-independent (center) and pentapeptide-dependent (right) β-subdomains are labeled. B. β-subdomain exterior face. The orientation corresponds to a 180° rotation around a vertical axis relative to the view in A. Residues that are highly conserved only in pentapeptide-dependent β-subdomains (right) are labeled.
FIG. 5. Conservation of residues within the β-subdomain of CheR.
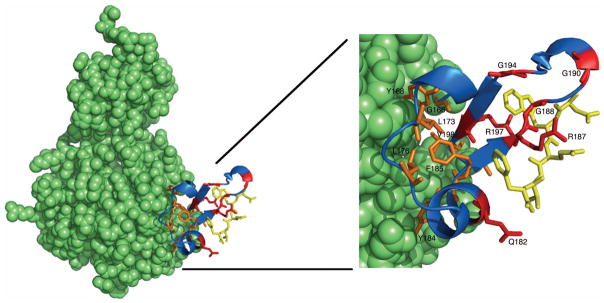
The CheR-pentapeptide structure (left) is displayed as a space-filling model (green), except for the β-subdomain (blue) and pentapeptide (gold), which are shown as a ribbon diagram and in stick representation, respectively. An enlarged view of the β-subdomain (right) shows conserved residues in stick representation. Universally conserved β-subdomain residues are shown in orange and residues that are conserved exclusively in pentapeptide-dependent β-subdomains are shown in red. Images were generated using Pymol (Delano, 2002).
Discussion
Covalent modifications of proteins have a profound impact at both the molecular and cellular level. One such example is observed in bacterial chemotaxis where methylation and demethylation of specific glutamate residues within the cytoplasmic domains of MCPs counterbalance the signaling effects of ligand binding to the sensory domain, allowing adaptive behavior in bacterial swimming. While signaling and chemotaxis have been most extensively studied in E. coli and S. enterica, studies of other organisms have identified many complex variations on the streamlined E. coli/S. enterica system (Szurmant and Ordal, 2004). Over the past decade, one organism in particular, T. maritima, has emerged as a valuable source of chemotaxis proteins and receptor signaling complexes for structural analysis (Bilwes et al., 1999; Bilwes et al., 2001; Chao et al., 2006; Griswold et al., 2002; Park et al., 2006; Park et al., 2004a; Park et al., 2004b; Quezada et al., 2004; Usher et al., 1998). As part of the characterization of in vitro methylation of T. maritima MCPs (Perez et al., 2006), we investigated a key difference between E. coli/S. enterica receptors and the majority of other bacterial and archaeal MCPs, the presence and absence of a C-terminal binding motif for methyltransferase CheR.
We have shown that T. maritima CheR efficiently methylates T. maritima MCPs and is also capable of efficiently methylating S. enterica Tar with or without the C-terminal pentapeptide. Moreover, analysis of the residues preceding the C-terminal ends of T. maritima receptors and methylation assays with TM1428 containing a truncation of the last five residues (TM1428Δ5) indicate that MCP methylation in T. maritima, unlike the E. coli/S. enterica paradigm, is performed independently of a C-terminal binding motif. Utilizing a database of transmembrane signal transduction proteins (Galperin, 2005), we screened and analyzed MCP C-terminal ends as well as CheR β-subdomains from a large number of completed bacterial and archaeal genomes. Results showed that only ~10% of MCPs possessed a putative pentapeptide-binding motif, despite all CheR proteins possessing a β-subdomain (Djordjevic and Stock, 1997; Shiomi et al., 2002b). Furthermore, key similarities and differences in residue conservation of the β-subdomain were identified when comparing pentapeptide-dependent and pentapeptide-independent CheR methyltransferases.
All CheR β-subdomains contain a set of highly conserved residues (Gly166, Tyr168, Leu173, Leu176, Tyr184, Phe185 and Val198, Fig. 3) that map to the interface formed between the β-subdomain and the rest of the CheR C-terminal catalytic domain (Fig. 4A and 5). Buried surface analysis (Brünger et al., 1998) of the CheR-NWETF pentapeptide crystal structure (Djordjevic and Stock, 1998) confirmed that all of the universally conserved residues are buried at the interface comprising 213.1 Å2 of the 596.9 Å2 total buried surface of the β-subdomain. Further examination of this interface revealed several H-bonds that bridge the interface, formed between Gly166, Tyr168, Tyr184 and residues within the catalytic domain. The β-subdomain, as its name implies, appears to be an integral part of the C-terminal domain. Attempts to delete the β-subdomain by engineering constructs that replaced the β-subdomain with a large variety of small linkers designed to join αB and αB2, helices of the catalytic domain that anchor the β-subdomain endpoints, resulted in misfolded protein (Djordjevic and Stock, unpublished data).
Thus it appears that the β-subdomain and the conserved residues at the interface with the catalytic domain play a role in folding and/or stability of the protein. However, it should be noted that the β-subdomain is unique to CheR methyltransferases and is not present within the common catalytic domain fold of any other class of methyltransferases. Thus, it is logical to speculate that the β-subdomain evolved for some function other than structural integrity. Interaction with the receptor methylation region is a plausible role, as the upper face of the β-subdomain together with a surface of the N-terminal domain form a large C-shaped cleft surrounding the active site that could potentially cradle the receptor coiled coil. However, the lack of sequence conservation in residues that comprise this surface argues against such a role.
Although bacteria lack discrete cellular compartments, many proteins and protein complexes are able to navigate the bacterial cell and ultimately localize to specific destinations (Lybarger and Maddock, 2001). For example, in E. coli/S. enterica, MCPs, CheW and CheA form a ternary complex at the cell pole (Maddock and Shapiro, 1993; Sourjik and Berg, 2000). Similar polar localization of MCPs has also been observed in C. crescentus (Alley et al., 1992), B. subtilis (Kirby et al., 2000), P. aeruginosa (Bardy and Maddock, 2005) and R. sphaeroides, that in addition, has a discrete cluster of chemotaxis proteins localizing within the cytoplasm as well (Porter et al., 2002; Wadhams et al., 2002; Wadhams et al., 2005). Proper localization of the other components of the chemotaxis pathway to receptor clusters is also necessary. Localization of response regulator CheY, CheZ phosphatase and CheB methylesterase to receptor clusters in E. coli/S. enterica, is dependent on CheA, CheAs (a short form of CheA) and the CheA P2 domain, respectively (Banno et al., 2004; Cantwell et al., 2003; Sourjik and Berg, 2000). CheR targeting to the cell pole occurs via the high affinity interaction of the CheR β-subdomain with the MCP pentapeptide. This was demonstrated with a GFP-CheR (H192/R197A) β-subdomain mutant, that unlike WT CheR, did not localize to the cell pole (Shiomi et al., 2002b). The vast majority of MCPs identified as containing a putative pentapeptide sequence (111 of 113) are almost exclusively restricted to the different proteobacteria classes (α, β, γ, δ) (Table 2). From its limited spread among bacteria, it is likely that tethering of CheR to MCPs through a β-subdomain-C-terminal pentapeptide interaction is a relatively recent event in evolution. However, the possibility that differences in CheR-MCP interactions represent a loss of function from the majority of organisms rather than a gain of function in proteobacteria cannot be excluded.
An obvious question is whether CheR localizes to receptor clusters in the absence of a C-terminal binding motif and if so, how? Does localization occur through another conserved region of the MCP cytoplasmic domain? The greatest sequence conservation among MCPs occurs at a region designated the highly conserved domain (HCD) (Zhulin, 2001), which serves as the locus of protein-protein interactions with CheA and CheW. The next most highly conserved segments correspond to regions containing the methylation sites, which are all located within a consensus sequence (Perez et al., 2006; Terwilliger and Koshland, 1984). Either of these are logical sites for possible high affinity interactions with CheR, but no high affinity binding between T. maritima CheR and TM1143c was detected in our studies. If CheR is localized to receptor clusters through high affinity interactions, other proteins must facilitate these interactions.
Two other proteins, CheC and CheD, absent from E. coli/S. enterica but present in many of the organisms that contain peptide-independent CheR (Szurmant and Ordal, 2004), have been shown to play a role in MCP methylation and adaptation (Kristich and Ordal, 2002; Rao et al., 2004; Rosario et al., 1995; Rosario and Ordal, 1996). An additional difference between methylation systems in E. coli/S. enterica and some organisms with pentapeptide-independent methylation is the specific effect of methylation on the signaling activity of the receptors. In E. coli/S. enterica receptor signaling is influenced by the total number of sites methylated (Bornhorst and Falke, 2001), while in B. subtilis and M. xanthus the receptor signaling state is dictated by the specific site methylated (Astling et al., 2006; Kirby et al., 1999; Zimmer et al., 2000). Results in B. subtilis suggest that CheC and CheD coordinate CheY-dependent selective methylation by protecting one methylation site and exposing another using phosphorylated CheY as the cue (Rao et al., 2004). This may indicate that archaea and bacteria lacking a C-terminal binding motif, instead utilize a more complex pathway combining CheC, CheD, and CheY, in conjunction with CheB and CheR to effectively methylate and demethylate specific sites in vivo to regulate CheA activity. Conversely, for the comparatively simple E. coli/S. enterica chemotaxis system, continuous localization of CheR to the receptor clusters via the pentapeptide, would seem to be more advantageous, since total receptor methylation levels and not the specific site methylated controls kinase activity.
BLAST searches using the S. enterica CheR C-terminal catalytic domain identified several CheR-related proteins FrzF (M. xanthus), SMb20515 (S. meliloti) and CheW3 (B. burgdorferi) that also contained a β-subdomain (Shiomi et al., 2002b). These β-subdomains have sequences similar to the pentapeptide-independent type. Interestingly all of these proteins contain additional domains that are capable of facilitating protein-protein interactions; FrzF contains several tetratrico peptide repeats (TPR), SMb20515 possesses several PAS domains, and CheW3 has a CheW domain in its N terminus (Blatch and Lassle, 1999; Das et al., 1998; Lamb et al., 1995; Liu and Parkinson, 1989; Shiomi et al., 2002b; Taylor and Zhulin, 1999). Furthermore, recent studies with FrzF have demonstrated that the TPR motifs mediate multiple interactions with several other chemotaxis proteins (Bustamante et al., 2004). The presence of other protein-protein interaction motifs in pentapeptide-independent CheR proteins suggests that other mechanisms of molecular recognition may have been utilized by CheR prior to the evolution of the pentapeptide-binding function of the β-subdomain. Whether the β-subdomain ever played, or currently plays, a role outside of structural integrity of the methyltransferase catalytic domain in pentapeptide-independent CheR proteins remains a mystery.
CheR localization to receptor clusters through a C-terminal binding motif has been extensively studied, yet appears to be used by only a very small percentage of bacterial organisms. Moreover, with the current array of completed genomes over-representing proteobacteria classes and under-representing organisms from other phylums, the percentage of organisms with pentapeptide-independent methylation systems is likely to be even greater. In this study, we have utilized T. maritima and S. enterica to begin to address the fundamental differences in the mechanisms utilized by CheR for MCP methylation. We anticipate that future studies in organisms more suitable for in vivo studies will provide further insight into the more prevalent pentapeptide-independent methylation systems.
Experimental Procedures
Bacterial Strains and Plasmids
Construction of plasmids pEP01 encoding T. maritima CheR, pTM1428 encoding full-length T. maritima receptor TM1428, and pTM1143c encoding the C-terminal cytoplasmic domain of T. maritima receptor TM1143 (residues 225–530) was described previously (Perez et al., 2006). S. enterica CheR was expressed in an E. coli salt-inducible strain GJ1158 (Bhandari and Gowrishankar, 1997) from pT7CheR, generated by PCR amplification of the cheR gene from pME43 (Simms et al., 1987) and insertion of an NdeI-HindIII cheR fragment into complementary sites in vector PJES307 (Tabor and Richardson, 1985). Plasmid pME98 (Simms et al., 1985), encoding the S. enterica tar gene, was used as a vector to express tar in E. coli HCB437 (Δ(tsr)7021Δ(trg)100 zbd::Tn5Δ(cheA-cheZ)2209 metF159(Am)) (Wolfe et al., 1987).
Mutagenesis of tm1428 and tar
TM1428 C-terminal truncation (TM1428Δ5) was constructed using a 3′ primer designed to anneal to GTG (V561) of pTM1428 and introduce a termination codon directly following nucleotides encoding V561, truncating the last five residues (ERFKI) of TM1428. Following amplification of the truncated gene flanked by restriction sites SalI and NotI, the SalI-NotI tm1428 fragment was inserted into complementary sites in the pET28b vector (Novagen), generating plasmid pTM1428Δ5. The TarΔpp construct (residues 1–560) was generated using the QuikChange Site-directed Mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Briefly, using tar containing pME98 plasmid DNA as template, a stop codon was introduced at the fifth to last codon by altering AAC to TAA, yielding tarΔpp, which was transformed into HCB437 for expression. Mutations for both constructs were confirmed by DNA sequencing (Genewiz, Inc., North Brunswick, NJ).
Expression and Purification of CheR Proteins and Receptors
Expression and purification of S. enterica and T. maritima CheR were performed as previously described (Perez et al., 2004; Perez et al., 2006). Salt-washed membrane fractions containing full-length chemoreceptors (WT Tar, TarΔpp, TM1428, and TM1428Δ5) were prepared as described previously (Perez et al., 2004), as was preparation and purification of TM1143c (Perez et al., 2006). TM1143c was estimated to be ~90% pure and CheR proteins ~70% pure. Concentrations of CheR and TM1143 were estimated by comparison to protein standards on Coomassie Blue-stained SDS polyacrylamide gel electrophoretograms.
Receptor Methylation Assays
Receptor methylation assays to determine initial rates of methylation were performed as described previously (Perez et al., 2006). Each 100-μl reaction mixture contained 40–50 μl of either salt-washed membranes or receptor cytoplasmic domains (each containing ~6 μM receptor), and CheR protein preparation in 100 mM potassium phosphate, 50 μM [3H]-S-adenosylmethionine at 162 Ci/mol (specific activity, 15 Ci/mmol; NEN Life Science Products, Inc.), pH 7.0. Samples were pre-equilibrated for 10 min at either 30°C or 37°C, and reactions were initiated by addition of 10 μl of CheR (ranging from 0.94 to 37.7 pmol of S. enterica CheR and 1.95 to 4.97 pmol of T. maritima CheR to achieve linear rates). For typical assays, five to six time points were taken at 3–6 min intervals, and initial rates were estimated using linear regression analysis. Each CheR-receptor pair was assayed at two different CheR concentrations to confirm linearity with respect to CheR and all methylation rates reported were derived from assays repeated three times.
T. maritima TM1143c-CheR Pulldown Assays
Pulldown assays were performed according to the manufacturer’s protocol utilizing Ni Sepharose 6 fast flow beads (Amersham Biosciences). Briefly, assays were performed by incubating 30 μM or 60 μM CheR at 30°C for 1 h with 30 μM or 60 μM His-tagged TM1143c bound to 20 μl of Ni beads previously equilibrated in binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, pH 7.4). Wash steps for the beads were bypassed to avoid dissociation of a potentially weak CheR-TM1143c complex. Beads were mixed with 4X SDS loading buffer and heated at 100°C for 10 min prior to analysis by SDS-PAGE electrophoresis. Controls without TM1143c were performed to estimate the limits of detection in these assays. Gel bands were quantified by Coomassie blue staining, followed by densitometry with ImageJ software.
Data Base Search and Protein Sequence and Structure Analysis
Utilizing the census data, available at http://www.ncbi.nlm.nih.gov/Complete_Genomes/SignalCensus.html (Galperin, 2005), C-terminal ends of MCP sequences were screened for the presence of putative pentapeptide-binding motifs. MCPs containing this motif were selected based on the following criteria: MCPs were required to possess the xZxxZ putative binding motif (Shiomi et al., 2002a) (where Z represents either F, W, or Y at positions 2 and 5 of the pentapeptide) and contain a C-terminal tail of at least ten residues in length extending beyond the alignment of C-terminal ends of receptors lacking pentapeptides and preceding the xZxxZ motif. Secondary structure predictions were performed using Jpred (Cuff et al., 1998). The TIGR database (www.tigr.org) was used to select homologous CheR proteins from all MCP-containing bacterial genomes included in the M. Y. Galperin census. The β-subdomain for each CheR homolog was identified and multiple sequence alignments (LxxxYxF sequence downstream of the β-subdomain was used as an anchor) were generated using ClustalW (Thompson et al., 1994). Buried surface areas were calculated using CNS 1.1 (Brünger et al., 1998) using a probe radius of 1.4 Å. Interface contacts were examined using iMoltalk (Diemand and Scheib, 2004) and Pymol (Delano, 2002).
Acknowledgments
This work was supported by grant R37GM047958 from the National Institutes of Health. E.P. was supported by an NIH supplement grant for underrepresented minorities (3 R37 GM047958-1051). A.M.S is an investigator of the Howard Hughes Medical Institute. We thank Bryan Beel and Brian Crane for plasmid constructs and Roger Alexander for valuable discussions and contributions to the genomic analyses of MCPs and the CheR β-subdomain.
References
- Alley MRK, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes and Development. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Anand GS, Stock AM. Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry. 2002;41:6752–6760. doi: 10.1021/bi012102n. [DOI] [PubMed] [Google Scholar]
- Armitage JP, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti-variations on a theme? Microbiology. 1997;143 (Pt 12):3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- Astling DP, Lee JY, Zusman DR. Differential effects of chemoreceptor methylation-domain mutations on swarming and development in the social bacterium Myxococcus xanthus. Mol Microbiol. 2006;59:45–55. doi: 10.1111/j.1365-2958.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- Banno S, Shiomi D, Homma M, Kawagishi I. Targeting of the chemotaxis methylesterase/deamidase CheB to the polar receptor-kinase cluster in an Escherichia coli cell. Mol Microbiol. 2004;53:1051–1063. doi: 10.1111/j.1365-2958.2004.04176.x. [DOI] [PubMed] [Google Scholar]
- Bardy SL, Maddock JR. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J Bacteriol. 2005;187:7840–7844. doi: 10.1128/JB.187.22.7840-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P, Gowrishankar J. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J Bacteriol. 1997;179:4403–4406. doi: 10.1128/jb.179.13.4403-4406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bornhorst JA, Falke JJ. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J Gen Physiol. 2001;118:693–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- Bren A, Eisenbach M. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J Bacteriol. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burgess-Cassler A, Ordal GW. Functional homology of Bacillus subtilis methyltransferase II and Escherichia coli cheR protein. J Biol Chem. 1982;257:12835–12838. [PubMed] [Google Scholar]
- Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol. 2004;53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao X, Muff T, Park S, Zhang S, Pollard AM, Ordal G, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. The Pymol Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- Diemand AV, Scheib H. iMolTalk: an interactive, internet-based protein structure analysis server. Nucl Acids Res. 2004;32:W512–W516. doi: 10.1093/nar/gkh403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Chemotaxis receptor recognition by methyltransferase CheR. Nat Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Kim SH. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr Opin Struct Biol. 2000;10:462–469. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Baumgartner JW, Hazelbauer GL. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Lilly AA, Hazelbauer GL. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J Bacteriol. 1999;181:3164–3171. doi: 10.1128/jb.181.10.3164-3171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Griswold IJ, Zhou H, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- Hanlon DW, Ordal GW. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem. 1994;269:14038–14046. [PubMed] [Google Scholar]
- Kehry MR, Dahlquist FW. The methyl-accepting chemotaxis proteins of Escherichia coli Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J Biol Chem. 1982;257:10378–10386. [PubMed] [Google Scholar]
- Khidekel N, Hsieh-Wilson LC. A ‘molecular switchboard’ - covalent modifications to proteins and their impact on transcription. Org Biomol Chem. 2004;2:1–7. doi: 10.1039/b312466e. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Kirby JR, Saulmon MM, Kristich CJ, Ordal GW. CheY-dependent methylation of the asparagine receptor, McpB, during chemotaxis in Bacillus subtilis. J Biol Chem. 1999;274:11092–12100. doi: 10.1074/jbc.274.16.11092. [DOI] [PubMed] [Google Scholar]
- Kirby JR, Niewold TB, Maloy S, Ordal GW. CheB is required for behavioural responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol Microbiol. 2000;35:44–57. doi: 10.1046/j.1365-2958.2000.01676.x. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Ordal GW. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem. 2002;277:25356–25362. doi: 10.1074/jbc.M201334200. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Le Moual H, Koshland DE., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- Le Moual H, Quang T, Koshland DE., Jr Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Weis RM. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. The carboxyl-terminal linker is important for chemoreceptor function. Mol Microbiol. 2006;60:469–479. doi: 10.1111/j.1365-2958.2006.05108.x. [DOI] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Martin AC, Wadhams GH, Shah DS, Porter SL, Mantotta JC, Craig TJ, Verdult PH, Jones H, Armitage JP. CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides. J Bacteriol. 2001;183:7135–7144. doi: 10.1128/JB.183.24.7135-7144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleishmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Okumura H, Nishiyama S, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:382–384. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci USA. 2004a;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Chao X, Gonzalez-Bonet G, Beel BD, Bilwes AM, Crane BR. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell. 2004b;16:563–574. doi: 10.1016/j.molcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Perazzona B, Spudich JL. Identification of methylation sites and effects of phototaxis stimuli on transducer methylation in Halobacterium salinarum. J Bacteriol. 1999;181:5676–5683. doi: 10.1128/jb.181.18.5676-5683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E, West AH, Stock AM, Djordjevic S. Discrimination between different methylation states of chemotaxis receptor Tar by receptor methyltransferase CheR. Biochemistry. 2004;43:9539–9561. doi: 10.1021/bi035455q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E, Zheng H, Stock AM. Identification of methylation sites in Thermotoga maritima chemotaxis receptors. J Bacteriol. 2006;188:4093–4100. doi: 10.1128/JB.00181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Warren AV, Martin AC, Armitage JP. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol Microbiol. 2002;46:1081–1094. doi: 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- Rao CV, Kirby JR, Arkin AP. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2004;2:E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario MM, Kirby JR, Bochar DA, Ordal GW. Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry. 1995;34:3823–3831. doi: 10.1021/bi00011a040. [DOI] [PubMed] [Google Scholar]
- Rosario MML, Ordal GW. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol Microbiol. 1996;21:511–518. doi: 10.1111/j.1365-2958.1996.tb02560.x. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Okumura H, Homma M, Kawagishi I. The aspartate chemorectptor Tar is effectively methylated by binding to the methyltransferase mainly through hydrophobic interaction. Mol Microbiol. 2000;36:132–140. doi: 10.1046/j.1365-2958.2000.01834.x. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Homma M, Kawagishi I. Intragenic suppressors of a mutation in the aspartate chemoreceptor gene that abolishes binding of the receptor to methyltransferase. Microbiology. 2002a;148:3265–3275. doi: 10.1099/00221287-148-10-3265. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem. 2002b;277:42325–42333. doi: 10.1074/jbc.M202001200. [DOI] [PubMed] [Google Scholar]
- Simms SA, Keane MG, Stock J. Multiple forms of the CheB methylesterase in bacterial chemosensing. J Biol Chem. 1985;260:10161–10168. [PubMed] [Google Scholar]
- Simms SA, Stock AM, Stock JB. Purification and characterization of the S-adenosylmethionine: glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987;262:8537–8543. [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Springer WR, Koshland DE., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JB, Koshland DE., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci USA. 1978;75:3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RV, Sanna MG, Simon MI. Thermostable chemotaxis proteins from the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 1996;178:484–489. doi: 10.1128/jb.178.2.484-489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;84:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Koshland DE., Jr Sites of methyl-esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984;259:7719–7725. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher KC, de la Cruz AF, Dahlquist FW, Swanson RV, Simon MI, Remington SJ. Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability. Protein Sci. 1998;7:403–412. doi: 10.1002/pro.5560070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Martin AC, Porter SL, Maddock JR, Mantotta JC, King HM, Armitage JP. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol Microbiol. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Martin AC, Warren AV, Armitage JP. Requirements for chemotaxis protein localization in Rhodobacter sphaeroides. Mol Microbiol. 2005;58:895–902. doi: 10.1111/j.1365-2958.2005.04880.x. [DOI] [PubMed] [Google Scholar]
- Ward MJ, Zusman DR. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Conley MP, Kramer TJ, Berg HC. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- Zhulin IB. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv Microb Physiol. 2001;45:157–198. doi: 10.1016/s0065-2911(01)45004-1. [DOI] [PubMed] [Google Scholar]
- Zimmer MA, Tiu J, Collins MA, Ordal GW. Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation. J Biol Chem. 2000;275:24264–24272. doi: 10.1074/jbc.M004001200. [DOI] [PubMed] [Google Scholar]