Summary
Background
Fish melanocytes aggregate or disperse their melanosomes in response to the level of intracellular cAMP. The role of cAMP is to regulate both melanosome travel along microtubules and their transfer between microtubules and actin. The factors downstream of cAMP that directly modulate the motors responsible for melanosome transport are not known. To identify these factors, we are characterizing melanosome transport mutants in zebrafish.
Results
We report that a mutation in the gene encoding zebrafish melanophilin (Mlpha) interferes with melanosome dispersion downstream of cAMP. The current model of melanophilin function based on mouse genetics is that melanophilin links myosin V to melanosomes. The residues responsible for this function are conserved in the zebrafish ortholog. However, if linking myosin V to melanosomes was Mlpha’s sole function, elevated cAMP would cause mlpha mutant melanocytes to hyper-disperse their melanosomes. Yet this is not what we observe. Instead mutant melanocytes disperse their melanosomes much slower than normal and less than halfway to the cell margin. This defect is caused by a failure to suppress minus-end (dynein) motility along microtubules, as shown by tracking individual melanosomes. Disrupting the actin cytoskeleton, which causes wild-type melanocytes to hyper-disperse their melanosomes, does not affect dispersion in mutant melanocytes. Therefore, Mlpha regulates dynein independently of its putative linkage to myosin V.
Conclusions
We propose that cAMP-induced melanosome dispersion depends on the actin-independent suppression of dynein by Mlpha, and that Mlpha coordinates the early outward movement of melanosomes along microtubules and their later transfer to actin filaments.
Introduction
Fish and amphibians constantly adjust their pigmentation in order to blend with backgrounds that vary from light to dark. Specialized cells, called melanocytes, accomplish this by either aggregating or dispersing their membrane-bound pigment granules, called melanosomes, depending on the level of intracellular cAMP. Although these cells have been used for many years to investigate intracellular transport regulation [1], it is not understood how cAMP regulates the molecular motors responsible for melanosome transport. Here we have used zebrafish to identify and characterize a gene product that regulates melanosome motors downstream of cAMP.
Melanocyte microtubules are arrayed with their minus-ends at the cell center, hence the minus-end motor cytoplasmic dynein carries melanosomes inward, leading to aggregation [2, 3], while the plus-end motor kinesin-2, carries melanosomes outward, leading to dispersion [4]. Melanosomes also interact with myosin V, which switches their transport from microtubules to actin filaments [2, 5, 6]. If the transfer to actin is inhibited in fish melanocytes, e.g. by disassembling the actin cytoskeleton, melanosomes fail to disperse evenly. Instead, they “hyper-disperse”: they move to the minus-ends of microtubules without getting off and thereby accumulate at the cell periphery, leaving most of the cell pigment-free [2]. To establish an even distribution of melanosomes during dispersion, melanocytes must coordinate transport between microtubules and actin precisely.
Although melanosomes aggregate and disperse globally, the motion of individual melansomes is saltatory and bidirectional. Episodes of travel in each direction are interspersed with each other and with periods of undirected motion that reflect a switch to actin-based motility [2, 7, 8]. Biasing the frequency and/or persistence of dynein-versus kinesin-driven motility episodes is what leads to global aggregation or dispersion [7, 9]. These or similar mechanisms presumably operate in other cells to regulate the bidirectional transport of other cargoes, including chromosomes on the mitotic spindle [10] and most organelles in neurons [11]. Most other cells also coordinate microtubule- and actin-based transport: e.g. post-Golgi transport vesicles destined for the plasma membrane must leave microtubules and penetrate the cortical actin network in order to reach their final destinations [12].
A significant advantage of fish melanocytes for investigating these aspects of motor regulation is that melanosome transport is regulated by the level of intracellular cAMP. Melanin stimulating hormone (MSH) elicits a rise in cAMP that activates Protein Kinase A (PKA), leading to melanosome dispersion, whereas melanin concentrating hormone (MCH) elicits a decline in cAMP, which enhances the phosphatase activity that reverses PKA dependent phosphorylation, leading to melanosome aggregation [9, 13, 14]. The current model for how cAMP levels regulate melanosome motor activities is based on analyses of single melanosome movements in black tetra fish [9] and Xenopus [7] melanocytes. In response to MSH, cAMP levels at first rise and then decline. These kinetics correlate with those of plus-end travel, which implies that PKA phosphorylation of an as yet unknown protein leads to changes in the activity of kinesin 2. As cAMP and plus-end travel decline, the frequency of minus-end motility episodes increases. This leads to a rise in actin-based movement because melanosomes switch their transport from microtubules to actin during dynein motility events [7, 9]. The model is that myosin V and cytoplasmic dynein are in a tug-of-war, which myosin V wins [7].
The switch from aggregation to dispersion upon exposing melanocytes to MSH entails the suppression of dynein activity [7, 9]. The molecular mechanism underlying this suppression of dynein is not understood. Analyses of single melanosome velocities in latrunculin-treated melanocytes, which lack actin filaments, suggests that the rise in PKA activity at the onset of dispersion may reduce the number of active dyneins on the melanosome [15], independently of myosin V and actin. However, because the tug-of-war with myosin V is capable of shortening the persistence of dynein motility episodes [7] this might also contribute to the suppression of dynein at the onset of dispersion.
To understand how cAMP modulates melanosome transport, it would be helpful to identify the molecules that function between cAMP and the melanosome motors. Here we identify one of these molecules through the analysis of the zebrafish pigment mutant allele j120. Our studies suggest that microtubule-to-actin transfer of melanosomes may involve more than simply attaching a myosin to the organelle. Our findings support models wherein microtubule-to-actin transfer occurs through linked regulation of both microtubule- and actin-motor activities.
Results
The mutation j120 interferes with melanosome dispersion downstream of cAMP
We identified the recessive mutant allele j120 through an ENU mutagenesis screen. j120 homozygous fish are viable and appear normal apart from an inability to adapt their pigmentation to a dark background. Like other fish, wild type zebrafish aggregate and disperse their melanosomes in response to MCH and MSH, respectively (Table 1; Supplementary Figure 1). The j120 mutant fish appear normal when incubated in MCH, but upon exposure to MSH the spots of pigment in the mutant remain small whereas those in wild type fish expand dramatically (Fig. 1A). This indicates that the j120 mutation interferes with melanosome dispersion.
Table 1.
Pharmacological analysis of melanosome transport in wild type zebrafish melanocytes
| Reagent | Mechanism | Effect |
|---|---|---|
|
α-MSH (0.2-20 μM) |
Activates adenylyl cyclase via Gs-protein coupled receptor |
Dispersion |
|
MCH (0.1-1.0 μM) |
Inhibits adenylyl cyclase via Gi-protein coupled receptor |
Aggregation |
|
Caffeine (5 mM) |
A1 adenosine receptor antagonist; elevates intracellular cAMP |
Dispersion |
|
Epinephrine (0.1 mM) |
A2-adrenergic receptor agonist; inhibits adenylyl cyclase via Gi-protein coupled receptor |
Aggregation |
|
Forskolin (200 μM) |
Adenylyl cyclase activator | Dispersion |
|
sp-cAMP (30 μM) |
Membrane permeable cAMP analogue | Dispersion |
|
SQ 22,536 (100μM-1mM) |
Adenylyl cyclase inhibitor | Inhibits Dispersion |
|
Okadaic Acid (1μM) |
Serine/Threonine Phosphatase inhibitor | Inhibits Aggregation |
|
Cyclosporin A (20μM) |
Protein Phosphatase 2B inhibitor | No Effect |
|
H-89 (40μM) |
Protein Kinase A inhibitor | Aggregation |
To verify that the cAMP signaling pathways that regulate melanosome transport in other fish species [11, 12, 33] also do so in zebrafish, we exposed cultured melanocytes to reagents that activate or inhibit known players in the signaling pathways. The effects of these reagents on zebrafish melanocytes confirm the importance of cAMP modulation and the activity of PKA and a phosphatase in the regulation of zebrafish melanosome movement and suggest that the overall pathways that regulate dispersion and aggregation in zebrafish are similar to what has been established in other species of fish.
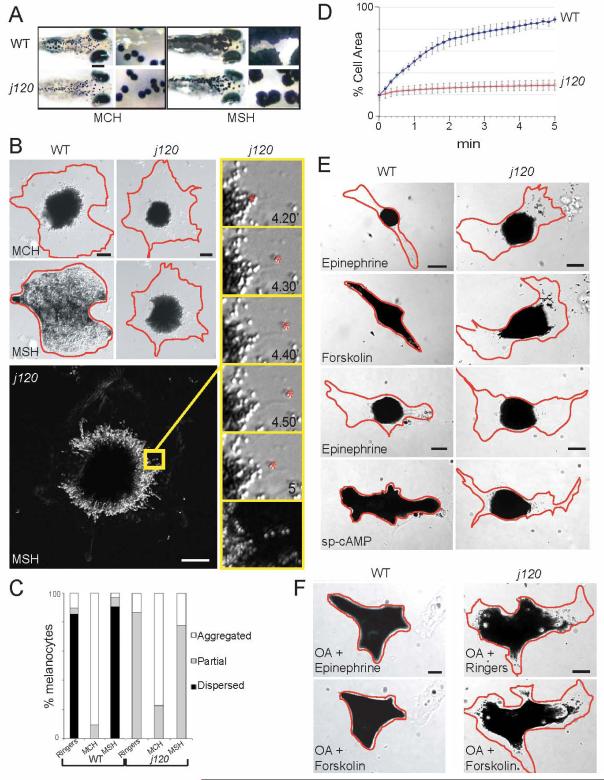
Figure 1. Characterization of melanosome transport in wild type and j120 zebrafish melanocytes.
A. Aggregation and dispersion in wild type and mutant five-day-old larvae. In response to MCH, pigment appears similarly aggregated in mutant and wild type fish. In response to MSH, melanocytes within wild type fish disperse their pigment fully, while melanocytes in j120 mutant fish do not. (Scale Bars = 200 μm; 50 μm in inset).
B. Aggregation and dispersion in cultured wild type and j120 mutant melanocytes visualized by DIC microscopy. Top row: wild type and mutant melanocytes fully aggregate their melanosomes after 5 min in MCH. Aggregation appears normal in the mutant melanocyte. Second row: Exposure to MSH for 5 min elicits complete dispersion in wild type, but only partial dispersion in mutant melanocytes. Bottom and right-hand column: A j120 mutant melanocyte showing that during dispersion its melanosomes load on microtubules and move bidirectionally. Digital images of this melanocyte were collected over a 5 min period following the initiation of dispersion by MSH. Each preceding frame was subtracted from each new frame to create a difference image, and the stack of difference images were summed through the recording period. Thus, this figure reveals the sum of what has moved during the recording period. Linear trajectories of many individual melanosomes are evident as white “beads-on-a-string”. One of these is within the area outlined in yellow. This area is shown at higher magnification at the bottom of the column of frames to the right. The upper five frames in the column show the direct images of the moving melanosome that produced the linear trajectory in the summed difference image. This melanosome moves bidirectionally (Scale bars = 10 μm).
C. Fraction of cultured wild type and j120 mutant melanocytes that respond to hormones fully or partially. Melanocyte cultures were treated for 10 min. with 0.5 μM MSH or 1 μM MCH. Partial dispersion (defined as melanosomes that spread from the aggregated state but did not completely fill the melanocyte within 10 minutes) or no dispersion (defined as melanosomes that remained aggregated after the dispersion stimulus) was evident in a minor fraction of wild type melanocytes. j120 homozygous mutant melanocytes were never observed to fully disperse their melanosomes (n = 96 and 75 for wild type and mutant, respectively).
D. Kinetics of global melanosome dispersion in cultured wild type and mutant melanocytes. Wild type and mutant melanocytes with melanosomes aggregated were stimulated to disperse with MSH or caffeine, and the percent of the projected cell area filled with melanosomes was measured at 10 second intervals. Data points are averages from five mutant and five wild type melanocytes (error bars = SEM).
E. Images of cultured wild type and mutant melanocytes five minutes after applying compounds that lower or elevate intracellular cAMP levels. First and third rows: melanocytes exposed to 0.1 mM epinephrine, which induces aggregation. Second and fourth rows: melanocytes exposed to forskolin (200 μM) or sp-cAMP (30 μM). Both elicit full melanosome dispersion in wild type but only slow partial dispersion in the mutant.
F. Response of cultured wild type and j120 mutant melanocytes to melanosome aggregation- and dispersion-inducing drugs applied after overnight pre-treatment with the phosphatase inhibitor okadaic acid (OA). Top left: positive control showing OA inhibits the phosphatase responsible for aggregation. Pre-treatment of wild type cultures with 1 μM OA inhibits aggregation induced by epinephrine. Top and bottom right: overnight pre-treatment of j120 mutant melanocytes with 1μM OA did not rescue partial dispersion, either in Ringers (upper) or following exposure to forskolin (lower). The small expansion of melanosomes in the j120 mutant melanocyte in F compared to those in E is a side effect of the overnight exposure to okadaic acid. The main point is that forskolin does not induce further expansion to the cell periphery, as it does in wild type melanocytes. Bottom left: Forskolin induces full dispersion in wild type melanocytes pre-treated with 1μM OA. (Scale Bar = 10 μm)
To evaluate in more detail this melanosome transport defect we cultured melanocytes from wild type or j120 mutant fish and used differential interference contrast microscopy to image and make movies of their melanosome movements (Fig. 1B). Following perfusion into the microscopy flow cell of a hormone or agonist that elicits melanosome aggregation (MCH or epinephrine) or dispersion (MSH or caffeine), we measured the time course of pigment contraction or expansion within individual cells (Fig. 1B-D). Melanocytes from j120 mutant fish aggregate their melanosomes with the same kinetics as wild type fish (Fig. 1B, C), but disperse their melanosomes much more slowly than wild type melanocytes (Fig. 1D). In addition, whereas melanosomes in wild type melanocytes spread to the cell margin within 5 min of applying dispersion-inducing drugs (Fig. 1B-D), the spreading of melanosomes in j120 mutant melanocytes plateaus at 5-8 minutes with less than half of the projected cell area filled with melanosomes (Fig 1B-D), a condition we refer to as partial dispersion.
We performed pharmacological experiments (Fig. 1E, F; Table 1) to address whether the abnormal melanosome dispersion evident in j120 melanocytes is a consequence of defective signaling through the MSH and G-protein dependent pathway that normally elicits dispersion by elevating intracellular cAMP [14]. Drugs that elevate intracellular cAMP artificially, including the activator of adenylyl cyclase, forskolin, and a membrane permeable analogue of cAMP, sp-cAMP, elicit full dispersion in wild type melanocytes (Fig. 1E), yet have no effect on the slow and partial melanosome dispersion that marks j120 melanocytes (Table 1, Fig 1E). Therefore, the defect in j120 melanocytes does not lie between MSH binding to its receptor and cAMP elevation.
To address whether elevated activity of the opposing phosphatase is responsible for the slow and partial melanosome dispersion in the mutant, we exposed wild type and j120 mutant melanocytes to 0.1 μM of the phosphatase inhibitor okadaic acid (Fig. 1F). This treatment blocks melanosome aggregation in wild type and j120 mutant melanocytes (Fig. 1F), consistent with inhibition of the phosphatase opposing PKA phosphorylation. However, this treatment does not rescue the partial dispersion characteristic of j120 mutant melanocytes: Whereas wild type melanocytes pre-treated overnight with okadaic acid and then exposed to forskolin spread their melanosomes to the cell margin, similarly treated j120 melanocytes do not (Fig. 1F).
Collectively, these pharmacological experiments indicate that the abnormal melanosome dispersion in j120 mutant melanocytes is not due to abnormal signaling through cAMP and PKA dependent phosphorylation.
Partial dispersion in j120 mutant melanocytes is independent of actin
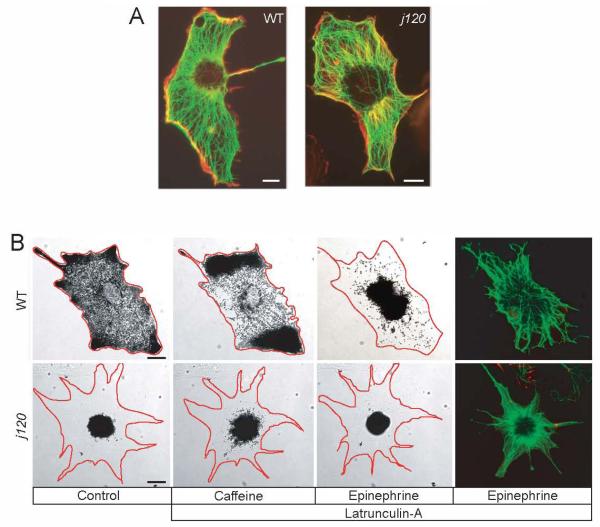
To address the possibility that partial melanosome dispersion in j120 mutant melanocytes is caused by an abnormal microtubule or actin cytoskeleton, we visualized actin filaments and microtubules within melanocytes by anti-tubulin immunofluorescence and rhodamine-phalloidin labeling (Fig. 2A). The microtubule and actin cytoskeletons of j120 melanocytes appear indistinguishable from those of wild type melanocytes. Thus, the partial dispersion of melanosomes in j120 melanocytes cannot be attributed to a failure of microtubules to reach the cell periphery, or to restriction of the melanocyte’s actin filaments to the cell center.
Figure 2. Partial dispersion in j120 melanocytes does not involve an abnormal microtubule or actin cytoskeleton and is independent of actin.
A. Fluorescent images of rhodamine-phalloidin labeled actin (red) and immuno-labeled α-tubulin (green) in cultured wild type and j120 mutant melanocytes. Microtubule and actin cytoskeletons appear normal in the mutant.
B. Aggregation and dispersion of melanosomes in the absence of actin filaments. Melanocytes were treated with 5 μM latrunculin A for 15 minutes to disrupt actin filaments, and then with caffeine to induce dispersion. In latrunculin, wild type melanocytes hyper-disperse their melanosomes, whereas the j120 mutant melanocyte still only partially dispersed its melanosomes. Aggregation (induced by epinephrine) is unaffected by latrunculin. Rhodamine phalloidin staining confirms the loss of actin filaments in the latrunculin treated melanocytes. Microtubules (green) were labeled by using an antibody against α-tubulin. (Scale Bar = 10 μm).
To address whether the slow and partial melanosome transport in j120 mutants is caused by premature transfer of melanosomes from microtubules to actin early in dispersion we treated melanocytes with latrunculin A, which disassembles actin filaments (Fig. 2B). In the presence of latrunculin, wild type melanocytes respond to MSH or caffeine by hyper-dispersing their melanosomes to the cell margin (Fig. 2B), which confirms earlier work in a different species of fish [2]. By contrast, latrunculin has no effect on the slow and partial dispersion of melanosomes in j120 melanocytes (Fig. 2B). Thus, the melanosome dispersion that persists in j120 mutant melanocytes is independent of actin.
Microtubule-based transport underlies melanosome dispersion in j120 melanocytes
We addressed whether the spreading of melanosomes during MSH-induced dispersion in j120 melanocytes occurs through the transport of individual melanosomes along microtubules, as in wild type melanocytes [2, 7-9]. The alternative possibility would have been that melanosomes in j120 melanocytes fail to load onto microtubules and that they spread through diffusion or a change in cell shape. To distinguish microtubule-based transport from other modes of spreading, we ascertained whether individual melanosomes in the mutant move along linear paths. According to previous studies [2], this would imply they are transported along microtubules. One difficulty was that the partial dispersion that marks j120 melanocytes makes it difficult to identify the movements of individual melanosomes through stacks of sequential images. We therefore examined the sum of difference images, which we created by subtracting the previous frame from each new frame [16]. Summing sequences of difference images reveals only those objects that have moved, and makes linear trajectories noticeable (Fig. 1B). Our analyses reveal that many melanosomes in j120 melanocytes execute bidirectional linear movements (Fig. 1B, inset). We conclude that melanosomes in j120 mutant melanocytes do not spread by diffusion, nor are they “stuck” together or stalled on microtubules. Instead, this analysis indicates that they spread by loading onto and moving along microtubules.
The j120 mutation prevents the suppression of minus-end travel episodes during dispersion
The finding that during dispersion some melanosomes in the j120 mutant melanocytes translocate away from the main mass (Fig. 1B inset sequence) suggested we can analyze the kinetics of their movements to understand further how the mutation impacts the regulation of melanosome transport along microtubules. The motion of individual melanosomes is normally bidirectional, with a bias toward minus- or plus-end travel depending on whether the population as a whole is aggregating or dispersing [7, 9] (Figs. 1B and 3). An attenuated rate of dispersion like that seen in j120 mutant melanocytes could result from either abnormally low plus-end travel or abnormally high minus-end travel. To address which of these two possibilities underlies the j120 mutant phenotype, we undertook a kinetic analysis of melanosome dispersion based on tracking the movement of individual melanosomes (Fig. 3).
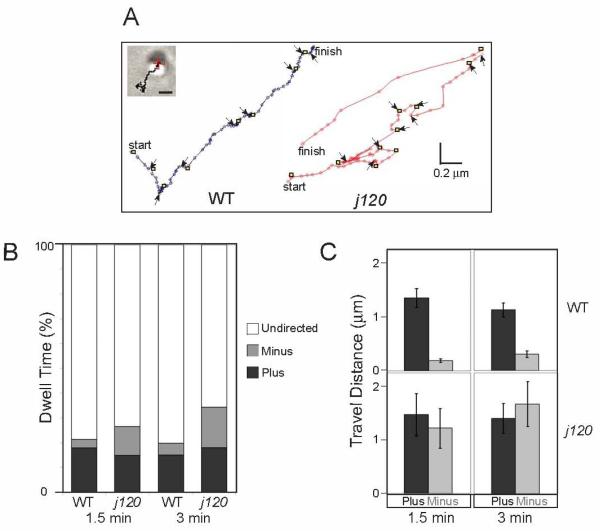
Figure 3. Analysis of individual melanosome movements during dispersion.
A. Melanosome tracks from wild type and j120 mutant melanocytes undergoing global dispersion. Track duration is 30 seconds; lines connect the successive positions. Yellow squares mark start of directed travel along microtubules; black arrows mark periods of pause or undirected movement. Inset: DIC image and track overlay for the wild type melanosome (Scale Bar = 1 μm).
B. Fraction of time melanosomes engage in the three motion states (minus-end, plus-end, or paused/undirected) during 30s observation periods made 1.5 and 3 minutes after inducing dispersion. Each bar pools the data from all tracked melanosomes (Table 2) in the indicated category (e.g. wild type, 1.5 min.). The principal difference between wild type and mutant is that the mutant spends more time engaged in travel to the minus-end.
C. Microtubule plus- and minus-end travel of wild type and j120 melanosomes during 30s observation intervals 1.5 and 3 min after initiating dispersion. Each bar is the mean ± SEM over all tracked melanosomes. Travel distances are the products of the average run lengths and episode frequencies indicated in Table 2. Whereas for wild type melanosomes plus-end exceeds minus-end travel, for mutant melanosomes travel in the two directions is balanced (error bars = SEM).
We tracked single melanosomes (Fig. 3 and Table 2) for 30s at 1.5 and 3 minutes after applying the stimulus to disperse (MSH or caffeine). Melanosome tracks (Fig. 3A) show episodes of continuous outward (plus-end) or inward (minus-end) movements whose trajectories resemble those of immunostained microtubules. These episodes are interspersed with periods of pause or random motion, which we infer reflect a stopped state on the microtubule or actin-based transport. We treated these episodes as a single motion category (“undirected motion”). Previous studies of black tetra fish and Xenopus melanocytes have documented that linear and random motion components, virtually identical to those we report here, reflect microtubule- and actin-based transport, respectively [2, 7, 9].
Table 2.
Mean Kinetic Parameters of Microtubule Based Travel
| WT | WT | j120 | j120 | WT | WT | j120 | j120 | |
|---|---|---|---|---|---|---|---|---|
| Travel direction |
Plus-
end |
Plus-
end |
Plus-
end |
Plus-
end |
Minus-
end |
Minus-
end |
Minus-
end |
Minus-
end |
| Time after dispersion stimulus (min) | 1.5 | 3 | 1.5 | 3 | 1.5 | 3 | 1.5 | 3 |
| Persistence of episodes (μm) (mean ± sem) | 0.16 (± 0.02) |
0.14 (± 0.02) |
0.22 (± 0.05) |
0.12 (± 0.03) |
0.07 (± 0.01) |
0.08 (± 0.01) |
0.20 ** (± 0.05) |
0.16 * (± 0.05) |
| Number of episodes in 30s (mean ± sem) | 4.4 (± 0.5) |
4.2 (± 0.5) |
3.5 (± 0.7) |
6.0* (± 0.9) |
1.5 (± 0.3) |
1.9 (± 0.3) |
3.1 * (± 0.6) |
5.7 ** (± 0.7) |
| # of melanocytes observed | 5 | 5 | 4 | 4 | 5 | 5 | 4 | 4 |
| # of melanosomes tracked | 86 | 76 | 36 | 39 | 86 | 76 | 36 | 39 |
The average number of plus- and minus-end motility episodes and their average persistence during 30 s observation periods 1.5 and 3 min following the dispersion stimulus. The mean frequency of minus-end episodes and their mean distance (persistence) are significantly different between WT and j120 mutants at both 1.5 and 3 minutes after the initiation of dispersion. In addition, the mean frequency of plus-end episodes are significantly different between WT and j120 mutants at 3 min. (Unpaired Student’s T-test:
, p < 0.0001;
, p < 0.05).
We first compared wild type and j120 melanosome tracks with respect to the fraction of time (dwell time) melanosomes spend in the three motion categories. To compute dwell times we pooled all tracks for wild type or j120 mutant melanosomes (Figure 3B). This analysis indicates that both wild type and j120 mutant melanosomes spend the majority of their time engaged in undirected motion, even as they disperse globally. j120 mutant melanocytes differ from wild type in that their melanosomes engage in undirected motion less often (66% versus 79% at 1.5 min; 73% versus 80% at 3 min), but engage in minus-end travel correspondingly more often (12% versus 3% at 1.5 min; 16% versus 5% at 3 min). Thus, the percentage of time melanosomes engage in plus-end travel is very similar between j120 and wild type melanocytes (15% versus 18% at 1.5 min; 18% versus 15% at 3 min). These findings suggest that excess minus-end travel may explain the slow dispersion of melanosomes in j120 mutant melanocytes.
To measure plus- and minus-end travel along microtubules, we analyzed the microtubule-based components of the melanosome tracks from the dispersion-stimulated cells. We computed for each direction of movement and across all movement episodes the mean frequency of occurrence and the mean distance traveled (persistence) (Table 2). The product of mean frequency and mean persistence is the mean travel distance (d) that a melanosome moves in that direction (Fig. 3C). On average, j120 mutant and wild type melanosomes travel similar distances toward microtubule plus-ends (2.9 ± 0.8 μm versus 2.7 ± 0.4 μm at 1.5 min; 2.8 ± 0.6 μm versus 2.3 ± 0.3 μm at 3 min). In contrast, j120 mutant melanosomes travel much further than wild type toward microtubule minus-ends (2.4 ± 0.7 μm versus 0.4 ± 0.1 μm at 1.5 min; 3.3 ± 0.9 μm versus 0.6 ± 0.1 μm at 3 min). The abnormally high minus-end travel of j120 melanosomes is caused by significant increases in both the frequency and persistence of minus-end motility episodes (Table 2). Persistence frequency distributions, which are described by the sum of two decaying exponentials, reflecting short and long classes of travel episodes [7, 17] (Supplementary Figure 2) indicate that an excess of long episodes is largely responsible for elevating the mean persistence of minus-end melanosome motility in j120 melanocytes.
In summary, the ratios of mean plus- to minus-end travel at the two time points are ~ 6:1 and 4:1 for wild type, consistent with rapid global dispersion, while the j120 ratios are ~ 1:1. We therefore conclude that in wild type melanocytes the function of the normal j120 gene product is to suppress minus-end travel during dispersion and that this is essential for driving the rapid outward spreading of the melanosome population as a whole. This suppression of minus-end directed travel in response to the dispersion stimulus does not occur in j120 mutant melanocytes. The observation that minus-end travel is suppressed during dispersion in wild type zebrafish melanocytes agrees with previous studies of black tetra fish and Xenopus melanocytes, which likewise show that the shift from aggregation to dispersion entails a decrease in minus-end travel [7, 9].
With respect to the kinetic parameters that underlie travel (frequency and persistence), comparisons between wild type and j120 melanocytes reveal additional facets about microtubule motor coordination. Between the 1.5 and 3 minute time points the average travel in the plus- and minus-end directions do not change dramatically. This is true for wild type and j120 melanocytes (Fig. 3C). However, as a result of the j120 mutation, both the average frequency and persistence undergo sizeable changes over time (Table 2); and these changes compensate so that for each direction, travel is constant. For plus-end travel the average frequency increases ~ two-fold (3.5 versus 6.0; Student’s T-test p < 0.020) while the average persistence decreases by ~ one-half (0.22 μm versus 0.12 μm; p < 0.0001); while for minus-end travel the frequency again increases by ~ two-fold (3.1 versus 5.7; p < 0.007) while the persistence decreases (0.20 versus 0.16 μm; p < 0.14). This is in contrast to wild type melanocytes, for which frequency and persistence do not change significantly between the 1.5 and 3 min either for plus-end (4.4 versus 4.2 events in 30s with average persistence 0.16 versus 0.14 μm) or minus-end travel (1.5 versus 1.9 events in 30s with average persistence 0.07 versus 0.08 μm). Thus, the j120 mutation unmasks a novel property of the motor coordination machinery, which is that plus- and minus-end travel distances are regulated - through changes to the frequency and/or persistence of motility episodes - so that they balance, on average (Fig. 3C). The normal function of the j120 gene product is to upset this balance between plus- and minus-end travel (by suppressing the minus-end motor) in response to the dispersion stimulus.
The j120 gene is the zebrafish ortholog of melanophilin
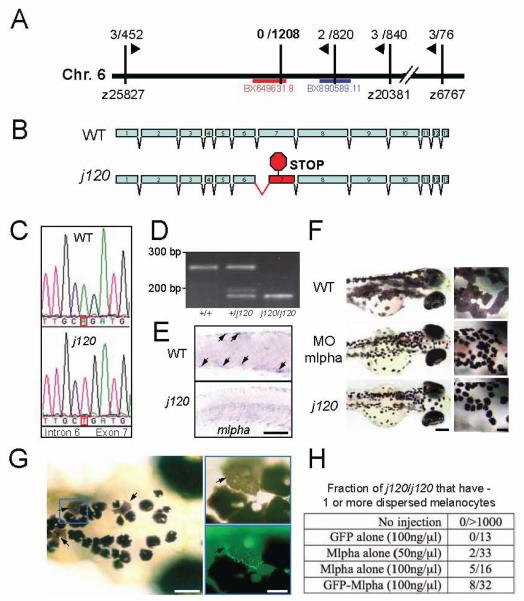
To shed light on how the j120 gene product regulates melanosome motor activities, we characterized the molecular basis of the mutation. We mapped the gene to Chromosome 6, to within 0.16 cM (0 crossovers in 1208 meioses) of an SSLP marker developed from BAC sequence BX649631 (Fig 4A). Because the ratio of physical to genetic distance in zebrafish is approximately 600 Kb/cM, this indicates that our tightly linked marker may be within 100 Kb of the j120 mutation. Accordingly, we examined a region 100 Kb on either side of our marker for candidate genes. Among 6 genes evident within this interval, the only one with a known relationship to melanosome transport is a predicted ortholog of melanophilin, which we henceforth refer to as mlpha. This acknowledges that zebrafish have a duplicate gene, which we refer to as mlphb (see below).
Figure 4. Identification of j120 as zebrafish melanophilin.
A. Linkage of j120 to SSR markers (vertical black lines) on chromosome 6. The fractions are the observed recombination rates between the mutation and that marker. Zebrafish BAC clone BX649631 contains mlpha.
B. Intron-exon organization of wild type and j120 alleles of the zebrafish mlpha gene. A mutation in the j120 allele leads to abnormal splicing of exon 7, which in turn introduces a stop codon in the coding sequence.
C. Genomic DNA sequence of wild type and j120 alleles of mlpha at the junction of intron 6 and exon 7. The mutant has an A to T mutation in the predicted splice acceptor site of intron 6.
D. RT-PCR analysis of mlpha exon 7 using RNA from wild type larvae (+/+) and larvae homozygous (j120/j120) or heterozygous (+/j120) for the mlphaj120 allele. Gel electrophoresis of the PCR products corresponding to exon 7 shows that only RNA from larvae with the mutant allele yields the shorter PCR product. RNA from homozygous mutant larvae yields only the shorter fragment and not the longer one. Normal and abnormal splicing of exon 7 predict PCR products of these sizes.
E. In situ hybridization with an anti-sense probe against mlpha mRNA performed on wild type and homozygous j120 mutant 2-day-old embryos. Arrows indicate expression in wild type melanocytes. (Scale Bar = 100μm).
F. Knockdown of Mlpha expression in 3-day-old wild type larvae. Wild type embryos were injected with a morpholino anti-sense (middle panel) or a control oligonucleotide (top panel). Larvae that received the anti-sense oligonucleotide have contracted pigment, which phenocopies the appearance of pigment in j120 mutant fish (bottom panel) (Scale Bars = 200 μm, 50 μm in inset).
G. Mosaic rescue in 5-day-old j120 mutant larvae injected at the one-cell stage with a plasmid containing Mlpha cDNA fused with GFP. Arrows indicate rescued melanocytes. Inset: A GFP-Mlpha expressing melanocyte (arrow) has even dispersion of melanosomes compared to the contracted melanosomes of adjacent melanocytes (Scale Bars = 100 μm, 25 μm in inset). The strong particulate fluorescence in the rescued cell suggests melanosomes recruit GFP-Mlpha.
H. Fraction of 5-day-old j120 mutant larvae that show mosaic rescue. Dispersed melanocytes were never observed in uninjected mutant larvae or in larvae injected with a plasmid containing GFP alone.
In mouse and human, melanophilin is genetically required for the accumulation of melanosomes at the dendritic tips of melanocytes [18-20]. Furthermore, although mammalian melanocytes differ from those of fish and frogs in that they do not engage in synchronous aggregation and dispersion of their melanosomes in response to cAMP, the mouse melanophilin mutant phenotype, called leaden [18], is nonetheless similar to that of j120 in zebrafish: in both cases melanosomes are clustered in the cell center and not dispersed throughout the cell.
To further explore whether j120 is a mutation in the mlpha gene, we compared the sequence of wild type mlpha cDNA with that of the mutant mlphaj120 cDNA. The cDNAs were obtained by performing RT-PCR on mRNA isolated from wild type or homozygous mutant larvae. This comparison, together with the genomic sequence of the wild type mlpha, reveals that the mlpha transcript from j120 mutants is mis-spliced such that it lacks the beginning of exon 7. This places the remaining 3′ portion of the transcript in the wrong reading frame, which results in a premature stop codon that would truncate the protein after residue 238 (Fig. 4B). This is consistent with the mlpha j120 genomic DNA sequence, which reveals a point mutation in the exon 7 splice acceptor site (Fig. 4C). As predicted, RT-PCR assays reveal that the mis-spliced mRNA is evident in fish homozygous and heterozygous for the mutation, but not in wild type fish (Fig. 4D). Premature stop codons often lead to mRNA degradation through nonsense mediated decay [21] which would create a null. To address this possibility, we used in situ hybridization to evaluate mlpha mRNA expression in 2-day-old embryos (Fig 4E). mlpha mRNA staining is conspicuous in the melanocytes of wild type embryos, as expected for a gene product involved in melanosome transport. Staining is weak in j120 mutant embryos, which is consistent with the possibility that the mutation leads to degradation of the mRNA.
To address whether a mutation in mlpha can account for the j120 mutant phenotype, we knocked-down expression of mlpha mRNA in wild type embryos with a morpholino anti-sense oligonucliotide targeting the translation initiation site of mlpha. This faithfully phenocopies the j120 mutant pigment defect (Fig 4F).
Finally, to address whether the j120 phenotype is solely due to a mutation in mlpha, we injected cDNAs encoding either Mlpha or GFP-Mlpha into one-cell mlphaj120 mutant embryos. Each plasmid contained a melanocyte specific promoter to control transcription [22]. Mosaic expression is a common feature of transient transgenics in zebrafish [23]; and accordingly, we observe a mosaic pattern of rescued melanocytes in those j120 mutant larvae that had been injected as embryos (Fig. 4G, H). The rescued melanocytes in the j120 mutants are highly conspicuous because they appear much larger than most melanocytes in the mutant and they are grey instead of black (Fig. 4G). This is because their melanosomes are distributed evenly throughout the entire cell. Furthermore, the rescued melanocytes in mutants injected with cDNA encoding GFP-Mlpha appear bright green, and when viewed at high magnification the bright green fluorescence has a granular pattern suggesting that the GFP-Mlpha is enriched on melanosomes (Fig. 4G, inset). In contrast to mutants injected with cDNAs encoding Mlpha or GFP-Mlpha, uninjected mutants or mutants injected with plasmid containing GFP alone were never observed to have large, grey melanocytes (Fig 4H). These rescue and morpholino experiments, together with the molecular analyses of j120 mutants, establish that the phenotype of partial melanosome dispersion is due to a mutation in zebrafish mlpha.
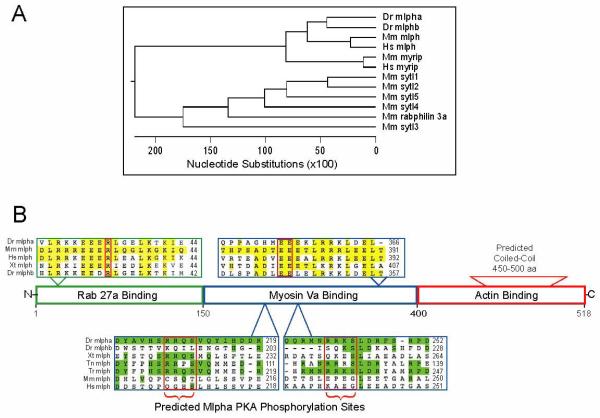
We used bioinformatics to establish that mlpha is actually the ortholog of mammalian melanophilin. Melanophilin is a member of the synaptotagmin-like protein (Slp) family, whose members are marked by a conserved N-terminal domain that interacts with GTP activated Rab27a [19]. We analyzed the phylogenetic relationships between mlpha, its predicted zebrafish paralog mlphb, and the known mammalian Slp family members (Fig. 5A). This indicates zebrafish mlpha and mlphb are both orthologs of mammalian melanophilin (Fig. 5A). Another indication that mlpha and mammalian melanophilin are orthologs is that their intron/exon structures in the region encoding the Rab27a binding domain are similar (data not shown). Synteny confirms that zebrafish mlpha is orthologous to the mouse and human melanophilin genes: all three genes are proximal to leucine rich repeat (in FLII) interacting protein 1 (LRRFIP1).
Figure 5. Relationship of mlpha and its protein product to homologs in other species.
A. Phylogenetic relationships among members of the synaptotagmin-like protein family. Among the many different members of this family known in humans (Hs) or mouse (Mm), zebrafish (Dr) Mlpha is most closely related to melanophilin (mlph). A second predicted melanophilin gene (mlphb) in zebrafish arose through gene duplication.
B. Alignments of fish, Xenopus, mouse, and human melanophilins. Between the upper and lower sequence alignments is a domain map of melanophilin, as established in studies of the mammalian ortholog. Above: alignment of sequences required for Rab27a and Myosin Va binding in mouse (Mm), zebrafish (DR), human (Hs), and Xenopus tropicalis (Xt). Yellow highlights residues that match the mouse sequence. The red boxes outline a conserved and functionally crucial Arginine within the Rab27a binding domain and a tandem pair of conserved and functionally critical glutamate residues (EE) in the Myosin Va binding domain. Below: alignment of sequences that include two regions containing predicted PKA phosphorylation sites in Mlpha (outlined in red). Green highlights residues matching zebrafish Mlpha.
The predicted PKA phosphorylation sites are conserved in Xenopus tropicalis and two other fish species (Takifugu rubripes and Tetraodon nigroviridis), but not in human, mouse, or zebrafish Mlphb, which is a paralog of Mlpha.
Zebrafish chromosomes 6 and 9 are syntenic in the regions that contain mlphb and mlpha, respectively, implying that mlpha and mlphb arose from the duplication of a chromosome segment. This is common in the zebrafish genome [31], and many genes present in single copy in mammals have two copies in zebrafish. Virtually nothing is known about the expression or function of mlphb. Because it does not compensate for the loss of mlpha, its function is not directly relevant to our conclusions about mlpha.
To shed light on whether Mlpha and its mammalian ortholog, melanophilin, are functionally similar, we compared their predicted amino acid sequences (Fig. 5B), as the current model of melanophilin function, based on studies of the mammalian protein, is reflected in the protein’s structure-function relationships. If the functionally important residues and domains were conserved in Mlpha, this would suggest that the functions of the zebrafish and mammalian proteins are related.
The current model, based on mouse genetics, is that mammalian melanophilin’s principal function is to link myosin Va to melanosomes. This is thought to facilitate the transfer of melanosomes from microtubules to cortical actin within the melanocyte dendritic tip, which is where the melanosomes accumulate prior to being taken up by the keratinocyte [18, 24, 25]. This ability of mammalian melanophilin to link myosin Va to the melanosome membrane depends on two protein-interaction domains: an N-terminal domain that binds the activated form of the membrane-anchored, GTP binding protein, Rab27a [18, 26]; and a centrally placed domain that binds myosin Va [18, 24, 27]. Within each domain are particular residues that are highly conserved and functionally crucial (Fig. 5B). One of these is R35 in the Rab 27a binding domain which, when mutated in humans, causes Griscelli Syndrome Type 3 [20]. Other crucial residues are E380 and E381 in the myosin Va binding domain, which are essential for myosin Va recruitment to melanosomes in mammals [27]. In addition, the mammalian melanophilin C-terminus contains a predicted coiled-coil region that enhances the binding of melanophilin to myosin Va in mouse melanocytes [24, 27]. Although the amino acid identity between Mlpha and the mouse or human orthologs is only 33% and 37% overall, the residues crucial for Rab27a and myosin Va binding are conserved, as is the predicted C-terminal coiled-coil (Fig. 5B). The presence in Mlpha of these conserved residues and structures suggests that Mlpha links myosin V to the melanosome membrane, as in mammals.
Unlike the mammalian orthologs, the Mlpha amino acid sequence reveals two predicted PKA phosphorylation sites within the myosin V binding domain: RRQS211 and RRKS244 [28, 29] (Fig. 5B). These are present in the melanophilin orthologs of other poikilotherms, including the frog Xenopus tropicalis, and the fish Takifugu rubripes and Tetraodon nigroviridis, but absent from mammalian melanophilins (Fig. 5B). Thus these sites are conserved in species like zebrafish, whose melanocytes engage in rapid, cAMP-dependent changes in pigmentation, but not in mammals, which lack this type of pigmentation control [30]. This correlation is consistent with the possibility that the phosphorylation state of these sites in Mlpha may be important for transducing intracellular cAMP levels into changes in the activities of the melanosome-associated motor proteins (Fig. 6). However, there is as yet no evidence that these sites are functional.
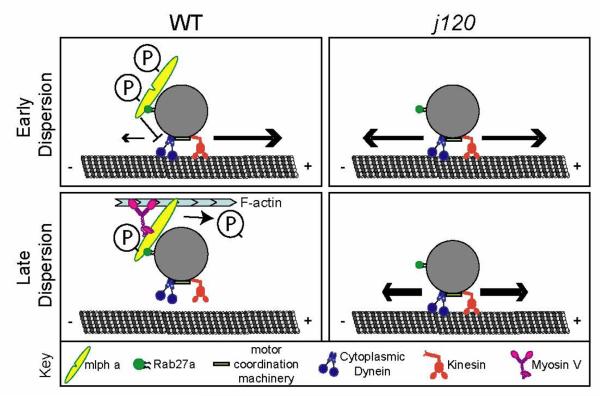
Figure 6. Model of Mlpha function during melanosome dispersion.
During the early phase of dispersion, when cAMP levels are high, Mlpha suppresses cytoplasmic dynein. This leads to rapid and global outward movement of melanosomes through the action of the kinesin motor. Later in dispersion, as cAMP levels decline, Mlpha recruits Myosin V, which leads to the transfer of melanosomes from microtubules to actin filaments. We note that the Myosin V and Rab27a binding sites in Mlpha are predicted but not yet proved on the basis of experimental data. We hypothesize that the dual functions of Mlpha during dispersion may be modulated by changes in the level of PKA phosphorylation; with maximal phosphorylation early in dispersion, and declining phosphorylation later. The proposed role for PKA phosphorylation is a hypothesis: there is presently no experimental data on whether the predicted PKA sites in Mlpha are functional. We refer to our proposal that Mlpha implements the transfer of melanosomes from microtubules to actin by coordinating both microtubule- and actin-based transport as a “relay” model.
Discussion
The goal of this study was to shed light on how cytoplasmic motor proteins are regulated in vivo. We chose to investigate the problem in fish melanocytes, a well-known experimental paradigm of transport regulation, but with the new idea of using zebrafish, for its genetics. We identified the pigmentation mutant allele j120, and demonstrated that its phenotype of slow and partial melanosome dispersion is cell autonomous, downstream of cAMP signaling, and independent of actin filaments. Tracking of individual melanosomes in j120 mutant and wild type melanocytes revealed that the mutant fails to suppress minus-end travel during dispersion. This presumably explains why global melanosome dispersion in the mutant is slow and partial.
Our identification of the mutant allele j120 as mlpha, an ortholog of mammalian melanophilin, initially seemed paradoxical. The zebrafish mlpha mutant phenotype involves defective regulation of microtubule-based transport and is independent of actin (Fig. 2). Yet the current model based on studies of mammalian melanocytes is that melanophilin is an organelle associated receptor for myosin Va. It thereby regulates actin-based transport [18, 25]. The amino acid residues responsible for this function are conserved in Mlpha, hence the paradox: if Mlpha’s sole function were to attach myosin V to melanosomes, the mutant would hyper-disperse its melanosomes to the cell margin, like wild type melanocytes treated with latrunculin (Fig. 2B). Instead the mutation slows and spatially limits dispersion through a failure to suppress minus-end travel on microtubules.
To reconcile the apparent discrepancy between Mlpha’s presumed function as a myosin V receptor and our experimental finding that the mlpha mutant phenotype involves mis-regulation of dynein and is independent of actin, we invoke a previous study of black tetra fish [9]. Rodionov et al [9] quantified the microtubule- and actin-based components of melanosome transport over time during dispersion and thereby demonstrated that dispersion proceeds in two phases: an initial period of microtubule-based transport followed by a period of increasing actin-based transport. We propose that the mlphaj120 mutation interferes with the earlier microtubule-based period, and that the later actin-based phase does not occur. Therefore, treatments that would disrupt the later actin-based phase in wild type melanocytes, e.g. latrunculin, have no effect on dispersion in the mutant. The fact that wild type melanocytes disperse their melanosomes evenly implies that the initial phase of microtubule-based transport is short relative to the later phase of mixed microtubule- and actin-based transport. The previous study of black tetra melanocytes supports this prediction [9].
Our experimental findings provide evidence that Mlpha regulates microtubule-based transport independently of actin. Were it not for the observation that the j120 mutant phenotype is unaffected by latrunculin-induced actin disassembly, the failure of the mutant to suppress dynein could have been interpreted as an indirect consequence of disrupting the (putative) myosin V-melanosome linkage function of Mlpha. This alternative explanation of the mutant phenotype, now excluded, would have been based on studies of black tetra fish and frog melanocytes, which suggest that minus-end travel episodes are shortened by the tension exerted on the melanosome when myosin V walks on actin [7, 9]. Our findings do not exclude the possibility that myosin V activity also contributes to suppressing dynein motility in wild type melanocytes. However, this mode of suppression would be in addition to the more direct, actin-independent regulation of dynein that we describe in this report (Fig. 6).
Our hypothesis that during dispersion Mlpha regulates both travel on the microtubule and the initiation of actin-based transport raises the question of how these two functions of Mlpha are regulated temporally? Previous studies in black tetra melanocytes [9] suggest that to achieve an even distribution of melanosomes at the end of dispersion, the time courses of the microtubule- and actin-based components of melanosome transport must be coordinated. Rodionov et al [9] demonstrated that during dispersion plus-end travel rises and then falls and that these kinetics correlate precisely with those of the intracellular cAMP level. These and other findings have led to the current model, which is that changes in the cAMP concentration over time during dispersion leads to corresponding time-dependent changes in the activities of the melanosome associated myosin, kinesin, and dynein motors [9]. It follows that the phosphorylation state of at least one protein with one or more PKA phosphorylation sites must likewise change in accordance with the cAMP levels, and that the phosphorylation state of this protein leads to changes in the relative activities of the microtubule- and actin-based motors. In this respect it is very interesting that highly conserved, putative PKA phosphorylation motifs are present in Mlpha and its homologs in other species of fish and frog, but are not present in mouse and human melanophilin (Fig. 5B), whose melanocytes do not aggregate or disperse their melanosomes in response to cAMP. To guide future experiments, we propose the testable hypothesis shown in Fig. 6. Our working model is that the phosphorylation state of the predicted PKA sites in Mlpha change with the rise and fall of cAMP during dispersion and that this orchestrates the switch from dynein-driven transport to myosin V–driven transport (Fig. 6).
Previous studies of melanophilin or related members of the Slp family that have analogous functions in other types of cells e.g. MyRIP [32] and rabphilin 3a [33, 34] have not addressed the possibility that proteins in this family also regulate microtubule-based transport in addition to their functions of linking myosins to a organelle-bound Rab proteins. The central new idea provided by our study is that the transfer of melanosomes from microtubules to actin may involve more than simply attaching the myosin to the organelle and allowing the myosin to rip the organelle off the microtubule during dynein-driven motility episodes [7]. In contrast to this “tug- of war” model, wherein the attachment of myosin V to the organelle alone determines how far the organelle travels along the microtubule, our findings raise the possibility of a “relay model” wherein travel along the microtubule is regulated in a separate step that precedes recruitment and/or activation of myosin V. Our finding that a single protein, Mlpha, regulates melanosome travel along the microtubule and in addition may link myosin V to the melanosome supports this model.
This study raises several questions that will need to be addressed through future work. Does Mlpha actually link myosin Va to melanosomes, as suggested by Mlpha’s amino acid sequence? What is the domain within Mlpha that is responsible for suppressing dynein-driven motility? Are the predicted phosphorylation sites in Mlpha functional and does their phosphorylation state regulate microtubule- and actin-based transport, as proposed in the hypothesis of Fig. 6? Addressing these questions will help define the mechanism of dynein regulation during dispersion and shed light on whether this function is retained in the mammalian ortholog.
Experimental Procedures
Fish Stocks and mutagenesis
WIK, AB and TU lines were obtained from the Zebrafish International Resource Center (ZIRC). ENU mutagenesis was performed as described previously [35].
Larvae melanosome transport screening assay
We sedated 5-day-old larvae with a mixture of 200 ppm quinaldine and 0.1 mg ml−1 lidocaine in E3 solution (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). We avoided the sedative Tricaine because it elicits melanosome dispersion in larvae (Supplemental Fig 1). After exposing the larvae for 10 min to 5 μM MCH (Bachem, King of Prussia, PA ) diluted in sedative solution containing 0.1% DMSO, we examined them for aggregation defects. Following this we incubated the same larvae in 10-30 μM MSH (Sigma-Aldrich, St. Louis, MO) for 10 – 20 min, and then screened them for dispersion defects. We observed larvae under brightfield using a Leica MZFLII stereomicroscope equipped with a 1.0 X PlanApo lens (Leica Microsystems, Wetzlar, Germany). Digital photos were taken with a Magnifire-SP digital camera (Optronics, Goleta, California).
Primary melanocyte cultures
Prim 5 stage (24 hour) embryos were bleached and dechorionated [36], and at five to seven days of age the larvae were anesthetized and dissociated in ice-cold calcium-free Ringers solution (116 mM NaCl, 2.9 mM KCl, 5 mM HEPES; pH 7.2). Dissociated tissue was rinsed three times in 50 ml sterile calcium-free Ringers, and incubated in 0.05% Trypsin solution with EDTA (Invitrogen, Carlsbad, California) for twenty min at room temperature. The cells were centrifuged and the cell pellet rinsed with L-15 media (Cellgro, Mediatech Inc., Herndon VA) supplemented with 10% FBS, 0.2 % gentamycin, 0.5% penicillin, 0.5% streptomycin (Invitrogen, Carlsbad, California) and 0.8 mM CaCl2 1M CaCl2. The cell pellet was then resuspended in the culture media to a density of 125 −500 × 103 ml−1 and this cell suspension was then plated onto clean glass 25 mm diameter No. 1 coverslips (Fisher Scientific, Hampton, NH) that had been coated with a mixture of 20 μg ml−1 laminin and 10 μg ml−1 fibronectin (Roche, Basel, Switzerland) or 0.2 mg ml−1 E-C-L (Upstate, Charlottesville, VA). Approximately 0.25 ml of cell suspension was incubated with each coverslip for 4-8 hours. After plating, each coverslip was cultured at room temperature for up to two weeks in 2.5 ml of L15 media containing the supplements listed above.
Imaging cultured melanocytes
Coverslips with attached cells were sealed into a microscope flow chamber (Model RC-21BR, Warner Instruments, Hamden, CT) and bathed with Ringers solution (116 mM NaCl, 2.9 mM KCl 5 mM, HEPES, pH 7.2, 1.8mM CaCl2). Cells were imaged by differential interference contrast or by brightfield using a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany) equipped with a custom fiber optic illuminator coupled to a Hg arc lamp. We used the following Plan-Neofuor oil immersion objectives: 40x (N.A. = 1.3); 100x (N.A. = 1.3). Digital images were acquired with a Photometrics CoolSNAP CCD camera (Roper Scientific, Trenton, NJ) and stored on a personal computer, all under the control of MetaMorph imaging software (Molecular Devices Corporation, Sunnyvale, CA).
To evaluate the organization of the melanocyte microtubule and actin cytoskeletons, cultures were fixed with 4% paraformaldehyde in 1X PBS, immunostained with a primary antibody against α-tubulin (Sigma-Aldrich), and then labeled with rhodamine phalloidin (Molecular Probes, Invitrogen, Carlsbad, California). Fluorescent cells were observed with epiillumination using the 40X oil immersion objective and either FITC or rhodamine filter sets from Chroma (Rockingham, VT). Digital images were processed using MetaMorph and Adobe Photoshop software.
Pharmacological studies of cultured melanocytes
The following reagents (from Sigma-Aldrich, St. Louis, MO unless otherwise noted) were diluted in Ringers and perfused into the microscope chamber: 0.1-10 μM MCH (Bachem, King of Prussia, PA), 0.2–20 μM MSH, 0.01 mM epinephrine, 5 mM caffeine, 200 μM forskolin, 0.1–1mM SQ 22536, 40 μM H-89, 30 μM sp-cAMP, 0.5 −10 μM okadaic acid and 5 μM latrunculin A. Melanocytes in culture media were preincubated with okadaic acid overnight prior to exposure to MCH or epinephrine.
Quantitation of global melanosome dispersion
Immediately following drug application, digital images of individual melanocytes were collected for a period of 5 – 10 min using a camera exposure time of 150 ms. To plot the time course of global melanosome dispersion, Metamorph software was used to trace the perimeter of the cell in the first frame and to compute the enclosed “projected cell area”, which did not change with time during dispersion. A threshold applied to each frame in a given sequence isolated the pixels occupied by melanosomes, and the summed projected area of the melanosomes was divided by the total projected cell area to compute the percent of the cell area filled by melanosomes. The fractional cell area occupied by melanosomes in successive frames was plotted against time.
Single melanosome tracking during dispersion
Melanocytes were first exposed to MCH or epinephrine to bring about the aggregation of their melanosomes. They were then stimulated with 5 mM caffeine or 5 μM MSH to initiate dispersion. At 1.5 and 3 minutes after initiating dispersion, images were captured at three frames per second with an exposure time of 150 or 300 ms exposure. The vast majority of image sequences lasted 30 seconds. Individual melanosomes were tracked using the centroid-tracking function provided by MetaMorph software. The method’s spatial precision was determined from the distribution of a single melanosome’s positions within a paraformaldehyde-fixed cell (mean = 2.7 nm; SD = 3.8 nm; n = 657 frames).
Melanosome tracks were manually decomposed into three motion categories: linear outward (plus-end) movement; linear inward (minus-end) movement; and episodes of non-directed movement or pause (Fig. 3). Linear melanosome displacement was defined as a displacement away from or toward the cell center and that was greater than 30 nm within an interval of 300 ms. The endpoint of a linear run was determined by a pause or reversal of movement along the microtubule axis. Pauses were defined as episodes wherein melanosome displacement was less than 30 nm over 300 ms. Periods of random motion were defined as episodes where the melanosome’s displacement was greater than 30 nm over 300 ms and did not appear to fall along a line or lines for at least 900 ms. Most random motion episodes lasted much longer than 300 ms. To measure run lengths associated with linear travel along microtubules, we used the least squares method to compute the best fit line to the trajectory and then summed the components of the motion along this line.
Positional cloning and molecular analysis of mlphaj120
j120/ j120 fish in a C32 background were out crossed with wild type WIK fish to make heterozygous j120 carriers in a C32/WIK background. These map-cross fish were mated and their progeny screened for the j120 phenotype at 4 to 5 days post fertilization. Screened larvae were fixed in methanol, the genomic DNA was isolated, and PCR reactions were performed using primers that revealed sequence polymorphisms between C32 and WIK at known loci. Linkage to chromosome 6 was established using Simple Sequence Length Polymorphisms (SSLP). Fine mapping was performed using Simple Sequence Repeat (SSR) markers documented in the following databases: The Zebrafish Information Network, http://zfin.org; Tübingen Map of Zebrafish Genome, http://wwwmap.tuebingen.mpg.de; and Zebrafish SSR Search, http://danio.mgh.harvard.edu/markers/ssr.html.
To sequence genomic DNA we PCR amplified and sequenced overlapping fragments of the mlpha gene, using information from the Zebrafish Genome-Sequencing Project (www.ensembl.org/danio_rerio/index.html) to design the PCR and sequencing primers. To make mlpha cDNA, we isolated mRNA using the (Poly(A)Purist™ Kit (Ambion Inc, Austin, TX) and then performed RT-PCR using the Advantage RT for PCR kit (BD Biosciences, San Jose, CA). The PCR primers for cloning the Mlpha protein coding sequence were TCCTGAAAGACTTGGCAACTG and CGTCTTTATGCCAGTCTGTCAAA. The primers for amplifying a 251 bp fragment that contains the premature stop codon in the mutant were TCACTCTCGCAGACAGTCTGT and TCTACCTGGACACTTCAGAGGAAGAGGATA. To identify the point mutation in j120 we amplified the genomic DNA with the primers GAAGAAAATATATTAGGAAATACGGT and CATCACTGATTAAGGCGCTTTA.
In situ Hybridization
Whole mount in situ hybridization was performed using larvae treated with 0.003% PTU [37] at 24 hours, 48 hours, 3 days and 4 days post fertilization. Digoxigenin labeled anti-sense RNA probes were synthesized using linearized plasmid DNA containing zebrafish melanophilin as the template (Open Biosystems, clone ID 8008178, Accession DT063943.1).
Morpholino Injection
Morpholino oligonucleotides (Gene Tools, Philomath, OR) were dissolved in distilled water, diluted to 1mM in 1X Danieau solution (58mM NaCl, 0.7mMKCl, 0.4mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES pH 7.6), and injected into 1-2 cell wild-type embryos with 0.05% phenol red as a marker. The sequence of the melanophilin morpholino was 5′-GACAGGTCCAACTTCTTGTCCATGT-3′. A standard control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was also injected to confirm that the phenotype observed was not due to toxicity.
Rescue
Plasmid constructs were made by inserting full length Mlpha cDNA, either with or without N-terminally fused GFP, into the BamHI/EcoRI sites of a vector containing the melanocyte specific Mitfa promoter (a gift from James Lister). The plasmids were linearized, diluted to a concentration of 50 or 100 ng μl−1, and then injected into 1-cell stage embryos. The 5-day-old larvae were sedated with 0.02% tricaine, which normally disperses their pigment, and examined through a stereo dissection microscope for rescue. For making high resolution pictures of rescued melanocytes in situ, larvae were imaged using a Zeiss Axio Imager MI microscope (Zeiss, Oberkochen, Germany) with a Plan-Neofluor 10x (N.A. = 0.3) objective and an Achroplan 63x (N.A. = 0.95) water immersion objective. Digital images were acquired with a Zeiss Axiocam.
Supplementary Material
Supplementary Figure 1: Characterization of melanosome transport in wild type zebrafish. Melanocyte pigment granules in five-day-old swimming larvae completely aggregate within five minutes of exposure to melanin concentrating hormone (MCH) and completely disperse within ten minutes of exposure to α-melanin stimulating hormone (MSH) (Supplementary Fig. 1A). The drug carrier, DMSO, does not affect melanosome distribution (data not shown). We used primary cell cultures of melanocytes from wild type larvae to confirm that the cAMP signaling pathway that regulates aggregation and dispersion in other species also directs pigment granule movements in zebrafish (Supplementary Fig. 1B; Table 1).
Supplementary Figure 2: Distribution of lengths of WT and j120 mutant melanosome travel toward the plus- or minus-ends of microtubules (0.1 μm bins). Frequency histograms shown are the run distances of melanosomes at 1.5 minutes post dispersion initiation. In wild type and j120 mutant melanocytes, both plus- and minus-end directed histograms are well fit by the sum of two decaying exponentials (solid lines), indicating that there are two states of droplet travel: short runs and long runs [38]. Top row: In wild type there are far fewer long runs (greater than 0.7 μm) toward the minus-end of microtubules (left panel) than toward the plus-end (right panel). Bottom Row: For j120 mutant melanosome travel events, the general shape of both frequency histograms is the same. Note the increase in long runs toward the minus-end of microtubules (bottom left panel) relative to WT (top left panel).
Acknowledgements
We thank Dr. William Talbot (Department of Developmental Biology, Stanford School of Medicine) for rough mapping j120 to chromosome 6. We are grateful to Dr. Monte Westerfield (Institute of Neuroscience, University of Oregon) for helpful advice and to Dr. Gary Banker for comments on the manuscript. This work was supported by a grant from the National Institute of Health (Bruce J. Schnapp, R01 GM60045) and by a National Research Service Award predoctoral fellowship (Lavinia Sheets, F31 GM071198).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nascimento AA, Roland JT, Gelfand VI. Pigment cells: a model for the study of organelle transport. Annu Rev Cell Dev Biol. 2003;19:469–491. doi: 10.1146/annurev.cellbio.19.111401.092937. [DOI] [PubMed] [Google Scholar]
- 2.Rodionov VI, Hope AJ, Svitkina TM, Borisy GG. Functional coordination of microtubule-based and actin-based motility in melanophores. Curr Biol. 1998;8:165–168. doi: 10.1016/s0960-9822(98)70064-8. [DOI] [PubMed] [Google Scholar]
- 3.Reilein AR, Serpinskaya AS, Karcher RL, Dujardin DL, Vallee RB, Gelfand VI. Differential regulation of dynein-driven melanosome movement. Biochem Biophys Res Commun. 2003;309:652–658. doi: 10.1016/j.bbrc.2003.08.047. [DOI] [PubMed] [Google Scholar]
- 4.Tuma MC, Zill A, Le Bot N, Vernos I, Gelfand V. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J Cell Biol. 1998;143:1547–1558. doi: 10.1083/jcb.143.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers SL, Gelfand VI. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol. 1998;8:161–164. doi: 10.1016/s0960-9822(98)70063-6. [DOI] [PubMed] [Google Scholar]
- 6.Rogers SL, Karcher RL, Roland JT, Minin AA, Steffen W, Gelfand VI. Regulation of melanosome movement in the cell cycle by reversible association with myosin V. J Cell Biol. 1999;146:1265–1276. doi: 10.1083/jcb.146.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross SP, Tuma MC, Deacon SW, Serpinskaya AS, Reilein AR, Gelfand VI. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers SL, Tint IS, Fanapour PC, Gelfand VI. Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc Natl Acad Sci U S A. 1997;94:3720–3725. doi: 10.1073/pnas.94.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodionov V, Yi J, Kashina A, Oladipo A, Gross SP. Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr Biol. 2003;13:1837–1847. doi: 10.1016/j.cub.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 10.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunawardena S, Goldstein LS. Cargo-carrying motor vehicles on the neuronal highway: transport pathways and neurodegenerative disease. J Neurobiol. 2004;58:258–271. doi: 10.1002/neu.10319. [DOI] [PubMed] [Google Scholar]
- 12.Allan VJ, Schroer TA. Membrane motors. Curr Opin Cell Biol. 1999;11:476–482. doi: 10.1016/s0955-0674(99)80068-4. [DOI] [PubMed] [Google Scholar]
- 13.Rozdzial MM, Haimo LT. Bidirectional pigment granule movements of melanophores are regulated by protein phosphorylation and dephosphorylation. Cell. 1986;47:1061–1070. doi: 10.1016/0092-8674(86)90821-4. [DOI] [PubMed] [Google Scholar]
- 14.Sammak PJ, Adams SR, Harootunian AT, Schliwa M, Tsien RY. Intracellular cyclic AMP not calcium, determines the direction of vesicle movement in melanophores: direct measurement by fluorescence ratio imaging. J Cell Biol. 1992;117:57–72. doi: 10.1083/jcb.117.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi V, Serpinskaya AS, Gratton E, Gelfand V. Organelle transport along microtubules in Xenopus melanophores: evidence for cooperation between multiple motors. Biophys J. 2006;90:318–327. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnapp BJ, Crise B, Sheetz MP, Reese TS, Khan S. Delayed start-up of kinesin-driven microtubule gliding following inhibition by adenosine 5′-[beta,gamma-imido]triphosphate. Proc Natl Acad Sci U S A. 1990;87:10053–10057. doi: 10.1073/pnas.87.24.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross SP, Welte MA, Block SM, Wieschaus EF. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., 3rd Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- 20.Menasche G, Ho CH, Sanal O, Feldmann J, Tezcan I, Ersoy F, Houdusse A, Fischer A, de Saint Basile G. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1) J Clin Invest. 2003;112:450–456. doi: 10.1172/JCI18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maquat LE. Nonsense-mediated mRNA decay. Curr Biol. 2002;12:R196–197. doi: 10.1016/s0960-9822(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 22.Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 23.Meng A, Jessen JR, Lin S. Transgenesis. Methods Cell Biol. 1999;60:133–148. [PubMed] [Google Scholar]
- 24.Hume AN, Tarafder AK, Ramalho JS, Sviderskaya EV, Seabra MC. A Coiled-Coil Domain of Melanophilin Is Essential for Myosin Va Recruitment and Melanosome Transport in Melanocytes. Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XD, Ikebe R, Ikebe M. Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J Biol Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Wang F, Rao K, Sellers JR, Hammer JA., 3rd Rab27a is an essential component of melanosome receptor for myosin Va. Mol Biol Cell. 2002;13:1735–1749. doi: 10.1091/mbc.01-12-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda TS, Itoh T, Fukuda M. Functional analysis of slac2-a/melanophilin as a linker protein between Rab27A and myosin Va in melanosome transport. Methods Enzymol. 2005;403:419–431. doi: 10.1016/S0076-6879(05)03037-5. [DOI] [PubMed] [Google Scholar]
- 28.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 29.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Hammer JA., 3rd Making sense of melanosome dynamics in mouse melanocytes. Pigment Cell Res. 2000;13:241–247. doi: 10.1034/j.1600-0749.2000.130405.x. [DOI] [PubMed] [Google Scholar]
- 31.Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda TS, Fukuda M. Functional analysis of Slac2-c/MyRIP as a linker protein between melanosomes and myosin VIIa. J Biol Chem. 2005;280:28015–28022. doi: 10.1074/jbc.M501465200. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda M, Kanno E, Yamamoto A. Rabphilin and Noc2 are recruited to dense-core vesicles through specific interaction with Rab27A in PC12 cells. J Biol Chem. 2004;279:13065–13075. doi: 10.1074/jbc.M306812200. [DOI] [PubMed] [Google Scholar]
- 34.Tsuboi T, Fukuda M. The C2B domain of rabphilin directly interacts with SNAP-25 and regulates the docking step of dense core vesicle exocytosis in PC12 cells. J Biol Chem. 2005;280:39253–39259. doi: 10.1074/jbc.M507173200. [DOI] [PubMed] [Google Scholar]
- 35.Rawls JF, Frieda MR, McAdow AR, Gross JP, Clayton CM, Heyen CK, Johnson SL. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics. 2003;163:997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) 4th ed Univ. of Oregon Press; Eugene: 2000. The zebrafish book. [Google Scholar]
- 37.Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, Paffett-Lugassy N, Saganic WJ, Lim CA, Hersey C, Zhou Y, Barut BA, Lin S, Kingsley PD, Palis J, Orkin SH, Zon LI. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2:E237. doi: 10.1371/journal.pbio.0020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross SP, Guo Y, Martinez JE, Welte MA. A determinant for directionality of organelle transport in Drosophila embryos. Curr Biol. 2003;13:1660–1668. doi: 10.1016/j.cub.2003.08.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Characterization of melanosome transport in wild type zebrafish. Melanocyte pigment granules in five-day-old swimming larvae completely aggregate within five minutes of exposure to melanin concentrating hormone (MCH) and completely disperse within ten minutes of exposure to α-melanin stimulating hormone (MSH) (Supplementary Fig. 1A). The drug carrier, DMSO, does not affect melanosome distribution (data not shown). We used primary cell cultures of melanocytes from wild type larvae to confirm that the cAMP signaling pathway that regulates aggregation and dispersion in other species also directs pigment granule movements in zebrafish (Supplementary Fig. 1B; Table 1).
Supplementary Figure 2: Distribution of lengths of WT and j120 mutant melanosome travel toward the plus- or minus-ends of microtubules (0.1 μm bins). Frequency histograms shown are the run distances of melanosomes at 1.5 minutes post dispersion initiation. In wild type and j120 mutant melanocytes, both plus- and minus-end directed histograms are well fit by the sum of two decaying exponentials (solid lines), indicating that there are two states of droplet travel: short runs and long runs [38]. Top row: In wild type there are far fewer long runs (greater than 0.7 μm) toward the minus-end of microtubules (left panel) than toward the plus-end (right panel). Bottom Row: For j120 mutant melanosome travel events, the general shape of both frequency histograms is the same. Note the increase in long runs toward the minus-end of microtubules (bottom left panel) relative to WT (top left panel).