Introduction
The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) gene fusion occurs in 2–7% of NSCLC cases[1,2] and is typically identified by Vysis ALK Break-Apart fluorescent in situ hybridization (FISH) assay. Tumors expressing this fusion respond to treatment crizotinib[1].
Case Presentation
A 43 year-old never-smoker male presented with pericardial tamponade. Pericardio/thoracocentesis yielded 2.5 liters of fluid; light microscopy was consistent with lung adenocarcinoma. No epidermal growth factor receptor (EGFR) mutation or EML4-ALK rearrangement was detected.
Following talc pleurodesis, he was treated with 4 cycles of cisplatin/pemetrexed, however experienced a progression.
A specimen from the tumor was sent for comprehensive genomic profiling using a Next Generation Sequencing (NGS) assay in a CLIA laboratory (Foundation Medicine), identifying a complex ALK rearrangement. Several steps were then undertaken to better characterize the genomic abnormality. FISH testing was negative using standard criteria (Figure 1).
Figure 1.

Panel A Using a manual FISH technique, paraffin sections were hybridized with fluorescent ALK Break-Apart probe (Abbott Molecular). The specimen displayed an atypical pattern of double 3′ALK signals (red) fused with the 5′ALK signal (green), which was classified as negative for ALK gene rearrangement (< 15% of cells with split signals, 3′ ALK and 5′ ALK signals apart by >2 times signal size or single 3′ ALK signal, 50 cells scored) Panels B, C, and D. ALK IHC was performed using a primary antibody (clone D5F3, Cell Signaling Technology, Inc). All paraffin tissue sections were stained with ALK and were reviewed by two pathologists. The tumor cells have predominantly strong staining (3+) in 50% of the tumor cells (lower right). The remaining tumor cells showed (2+) in 30% (lower left) and (1+) in 20% of the tumor cells thus resulting in a histologic (H) score of 230 on a 0–300 scale. The top right is 200X and bottom two panels are 400X.
NGS analysis of genomic DNA revealed a complex rearrangement of ALK, involving breakpoints in at least 5 different genomic loci. One of the breakpoints was in ALK intron 19, the canonical breakpoint for EML4-ALK fusions. No clear fusion product was predicted; we suspected that multiple transcript products not readily identifiable existed.
We thus sequenced cDNA from the same sample to study both the expression level of ALK and the structure of any potential fusion product. The cDNA sequence demonstrated that the expressed product of the complex rearrangement was the canonical EML4-ALK fusion gene (EML4 exons 1–13, ALK exons 20–29). We hypothesized that at the genomic level the fused EML4 and ALK genes were separated by other small genomic shards, explaining why the EML4-ALK fusion was not detected by FISH. This connective region is likely to be removed during splicing and results in a functional EML4-ALK transcript. We next showed 39-fold over-expression of exons 20–29 of ALK relative to exons 1–19 (Figure 2).
Figure 2.
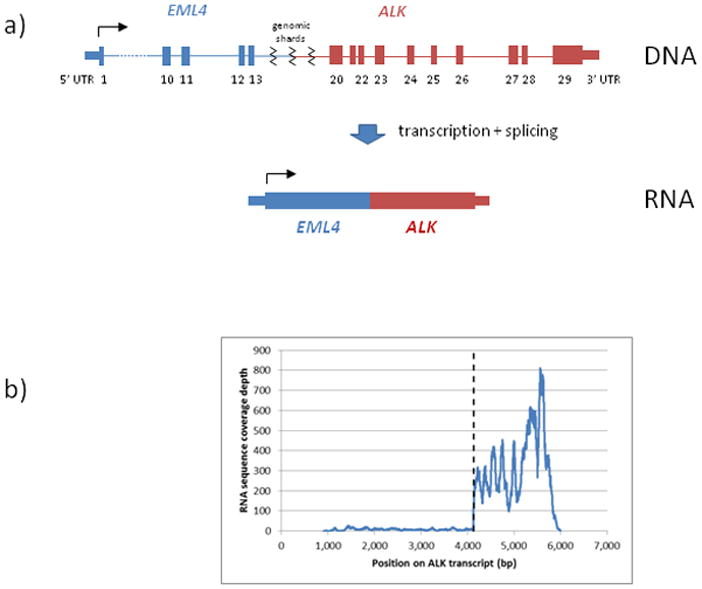
a) Hypothetical structure of rearrangement in genomic DNA (top) – complex rearrangement placing EML4 exons 1–13 upstream of ALK exons 20–29, separated by small genomic shards; RNA (bottom) – transcription and splicing remove connective region and generate a canonical EML4-ALK fusion transcript; b) Expression of ALK as measured by RNA-seq coverage, dashed line denotes boundary between exons 19 and 20.
ALK immunohistochemistry (IHC) was positive as displayed in Figure 1. Based on these results, crizotinib was begun. Within 2 weeks, the patient reported an improvement in pain in the pubic area and in exercise tolerance. Significant improvement was seen on initial CT-PET scans and after 4 months, the PET was negative and the chest CT scan showed further shrinkage of the primary lesion (RECIST 75%; Figure 3).
Figure 3.

Chest (A) and Pelvic (B) PET CT scans before, after 4 weeks and after 4 months of crizotinib.
Discussion
Recent studies demonstrate that the frequency EML4-ALK fusion transcript is detected in 2–7% of lung adenocarcinomas [1,2]; these tumors are sensitive to crizotinib.
Many ALK rearrangements have been described in NSCLC. Most of these fusion variants are comprised of various portions of the EML4 gene fused with a consistent portion of the ALK gene. In addition, non-EML4 fusion partners have also been identified, including KIF5B and TFG[3].
The novel rearrangement identified in this case is complex and was not detected by the Break-Apart FISH assay. NGS showed that the patient’s tumor harbored a complex EML4-ALK rearrangement at the genomic level. Clinical and radiographic evidence confirmed a rapid response to crizotinib. NGS should be considered in NSCLC patients with high likelihood of a driver kinase alteration when none is identified by other methods.
Footnotes
Conflict of Interest
NP, MI – nothing to declare.
GAO, GP, VM, DL, PJS, and MTC are employees and equity holders of Foundation Medicine
FRH: Consultant (advisory board) for Pfizer (compensated) and research agreement through University of Colorado with Ventana/Roche.
LSG: Employee of Oncotest-Teva Pharmaceutical Industries, Petach Tikva, Israel
References
- 1.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]


