Abstract
Structural cysteine-rich Zn2+ sites that stabilize protein folds are considered to be unreactive. In this manuscript, we identified a reactive cysteine residue, Cys151, in a treble-clef zinc finger with a Cys3His coordination sphere. The protein in question is the C1B domain of Protein Kinase Cα (PKCα). Mass-tagging cysteine assays of several C1B variants were employed to ascertain the site specificity of the covalent modification. The reactivity of Cys151 in C1B also manifests itself in the structural dynamics of the Zn2+ coordination sphere where the Sγ of Cys151 alternates between the Zn2+-bound thiolate and free thiol states. We used NMR-detected pH titrations, ZZ-exchange spectroscopy, and residual dipolar coupling (RDC)-driven structure refinement to characterize the two exchanging conformations of C1B that differ in zinc coordination. Our data suggest that Cys151 serves as an entry point for the reactive oxygen species that activate PKCα in a process involving Zn2+ release.
Keywords: zinc finger, treble clef motif, C1 domain, Protein Kinase C, NMR, conformational exchange, ZZ exchange, residual dipolar coupling, cysteine reactivity, reactive oxygen species, structural zinc, zinc coordination dynamics
Zinc is an essential element required for normal growth and development in all three domains of life. A recent comparative analysis of zinc proteomes in archea, bacteria, and eukarya revealed a uniform zinc protein content of 5–7%.1 The biological roles of zinc – structural, catalytic, and regulatory – are reflected in the chemical nature of the Zn2+-binding protein motifs. These motifs have been classified based on the identity of protein ligands in the first coordination sphere2 and “minimal functional sites” that capture protein environment within 5 Å from metal-binding residues.3
One theme that emerged from the analyses of zinc protein structures is the difference between the catalytic and structural Zn2+ sites. Catalytic sites have 3 to 5 protein ligands, with a preference for His and Asp/Glu. In almost all cases, they contain an exogenous (and exchangeable) ligand, such as a water molecule.4 In contrast, structural sites – found in “zinc finger” proteins5, 6 – show little variability in their coordination number and have a tetrahedral geometry with two or more cysteine ligands. Steric repulsion between the coordinating thiolate groups and their ability to decrease the Lewis acidity of Zn2+ ensure the stability of the metal coordination sphere and hence the entire protein fold.7 The coordination environment of zinc proteins that are involved in Zn2+ storage, transport, sensing, and buffering8, 9 shows significant variability, and can include both Cys-rich and His/Asp/Glu-rich sites.3
The chemical reactivity and dynamics of catalytic and regulatory Zn2+ sites are an integral part of their function. One example is the alkyl transfer enzymes (reviewed by Penner-Hahn10) that utilize zinc for catalytic purposes. The thiol group of the substrate directly coordinates to Zn2+, thereby displacing an endogenous ligand, and then leaves the zinc site upon alkylation.
In contrast, structural Zn2+ sites have been considered unreactive until the discovery that one of the cysteines in the structural Cys4 motif of the E.coli Ada protein is methylated by methylphosphotriesters as part of the DNA repair mechanism.11 Cys38 was identified as a methyl acceptor.12, 13 The methylation serves as an electrostatic switch that enables the protein to form a tight complex with the specific DNA sequence. Another example is a structural Zn2+ site in the second zinc finger of the HIV-1 nucleocapsid p7 (NCp7) protein,14 where Cys49 is the most reactive cysteine residue.15, 16 Targeting zinc fingers of NCp7 with cysteine-modifying agents is a promising approach for the development of anti-viral therapies.17 However, there is no indication that Cys49 undergoes any biologically relevant modification in the absence of exogenous reactants. Thus, the E.coli Ada protein is currently the only example of a structural Zn2+ site, where a functionally important covalent modification resides on a specific cysteine residue.
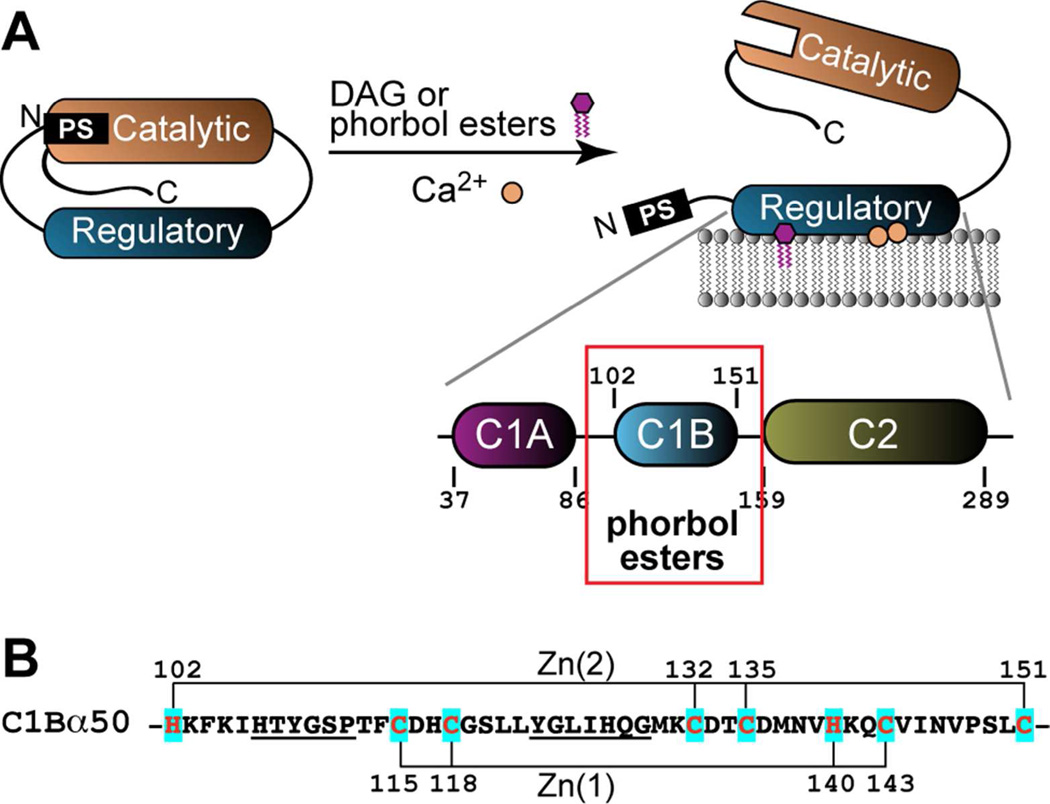
Here, we report characterization of a chemically reactive structural Zn2+ site with Cys3His coordination in a treble-clef zinc finger protein. The protein under study is the C1B domain of Protein Kinase Cα (PKCα). PKCα is a conventional isoform that belongs to the 10-member family of mammalian PKC isoenzymes.18, 19 It has a modular structure that comprises catalytic and regulatory domains. The canonical activation mechanism of the enzyme is shown in Figure 1A. The activation requires two cofactors, Ca2+ and diacylglycerol (DAG). Upon binding Ca2+ and DAG, the N-terminal regulatory domain undergoes membrane insertion, thereby releasing the pseudo-substrate region from the active site of the kinase.20 The regulatory domain comprises three independently folded modules: C1A, C1B, and C2. Both C1A and C1B can bind DAG and tumor-promoting phorbol esters (PEs). Because the affinity of C1B to PEs is higher than that of C1A,21–23 it is considered to be responsible for the PE-driven association of PKCα with membranes. This high-affinity interaction results in constitutive activation of PKCα and aberrations in the signaling response. We will refer to the C1B domain of PKCα as C1Bα in this manuscript.
Figure 1.
Parent protein context and Zn2+-coordinating residues of C1Bα. (A) Association of the regulatory domain with membranes in response to binding Ca2+ and diacylglycerol (DAG) relieves the auto-inhibition by the pseudo-substrate (PS) region. In addition to DAG, PKCα can be activated by tumor-promoting phorbol esters (PEs). C1Bα binds PEs with high affinity and is responsible for the PE-driven association of PKCα with membranes. (B) The primary structure of C1Bα with Zn2+-coordinating residues highlighted in cyan and the PE-binding loops underlined.
The core of C1Bα consists of 50 amino acids with two structural Zn2+ sites, each having a Cys3His coordination sphere (Figure 1B). The zinc ligands in C1Bα are well separated in the primary structure. Zn(1) together with the surrounding secondary structure elements forms a treble-clef motif.24 Zn(2) is coordinated by Cys132 and Cys135 that belong to a zinc “knuckle”,25 the N-terminal His102, and the C-terminal Cys151. Both zinc sites are located in the bottom half of the protein. The top half of C1Bα consists of two ligand-binding loops, β12 and β34, which are highly dynamic.26 The lipophilic ligand, such as DAG or PE, binds in between the loops thereby increasing the hydrophobicity of the complex.
In addition to the canonical activation mechanism shown in Figure 1A, PKC isoforms are activated by reactive oxygen species (ROS).27, 28 Cysteine-rich C1 domains have been suggested to be the target of ROS.29 Using NMR experiments in combination with mutagenesis and cysteine modification assays, we identified the reactive cysteine residue in the C1B domain of PKCα. One of the manifestations of this reactivity is the structural dynamics of the Zn2+ coordination sphere in the 50 amino-acid construct of C1Bα that corresponds to the minimum functional core. We report the characterization of the kinetics, thermodynamics, and structural consequences of this dynamic process.
EXPERIMENTAL PROCEDURES
Protein over-expression and purification
The E. coli codon-optimized DNA sequence of C1B domain from PKCα of Mus musculus was cloned into a pET-SUMO expression vector (Invitrogen) as a C-terminal fusion with histidine-tagged SUMO, a small ubiquitin-like protein.30 The sequence encoded a 50-residue C1B construct comprising residues His102 through Cys151. Mutagenic DNA for the C151G variant of the C1Bα50 was constructed using a Stratagene QuickChange™ site-directed mutagenesis kit and suitable PCR primers. The purification protocol for the 50-residue construct of C1Bα and C151G is given in Section S1 of the Supporting Information (SI).
Cloning, over-expression, and purification of the isolated C2 domain from PKCα have been reported previously by our laboratory.31, 32 The DNA sequence encoding a 194-residue construct of C1B-C2 domain (Ser100–Gly293) from PKCα of Mus musculus was amplified by PCR using the PKCα cDNA clone (Open Biosystems) as a template, and cloned into the pET-SUMO vector. C1B-C2 was purified using the C2 purification protocol with minor modifications.
For NMR experiments, unless otherwise stated, C1Bα and C151G were concentrated to 0.5 mM, and exchanged into the buffer containing 10 mM [2H-4]-imidazole at pH 6.5 (Cambridge Isotopes), 150 mM KCl, 8% 2H2O, 1 mM tris(2-carboxyethyl)phosphine (TCEP), and 0.02% NaN3. We will use the following abbreviations for the constructs used in this work: C1Bα50 for the 50-residue C1Bα; C151G for the Cys151-to-Gly151 variant of C1Bα50; and C1B-C2 for the construct that includes another lipid-binding domain of PKCα, the C2 domain (see Figure 1A).
Cysteine modification assays
[U-15N]-enriched C1Bα50, C151G, C1B-C2, and isolated C2 domains were treated with H2O2 solution to probe their response to ROS. The time course of the oxidation was monitored by 15N-1H hetero-nuclear single-quantum coherence (HSQC) experiments collected every 1–1.5 hours starting approximately 0.5 hours after addition of H2O2 to a final concentration of 1.25 mM. All C2 and C1B-C2 NMR experiments were carried out in the buffer containing 10 mM MES at pH 6.0, 8% 2H2O, 1 mM TCEP, 0.02% NaN3, and saturating Ca2+ at 20-fold excess with respect to protein concentration.
To ensure comparable signal-to-noise ratios in the NMR spectra, the protein concentration varied with the length of the protein construct: 0.1 mM C1Bα50 or C151G, 0.15 mM C2, and 0.25 mM C1B-C2. To quantify the rate of signal decay, the cross-peak intensities were plotted against the time elapsed from addition of H2O2 to the mid-point of each HSQC collected until ~20% of cross-peak intensity remained. The data were fit to an exponential decay using the Curvefit software available from Dr. Arthur G. Palmer’s laboratory at Columbia University (http://www.palmer.hs.columbia.edu/software/curvefit.html). After the cross-peak intensities decayed to less than 10% of their original cross-peak intensity, the amount of Zn2+ released into solution was quantified using the PAR assay, as described in Section S2 of the SI.
Mass-tagging33 of C1Bα50 (50 µM), C151G (35 µM), and C1B-C2 (10 µM) was accomplished by incubation with 0.25 mM polyethylene glycol maleimide 5,000 (PEG-mal, NANOCS) at room temperature for 5 hours in a buffer containing 10 mM imidazole at pH 6.5, 150 mM KCl, and 1 mM TCEP. In addition, either 0.1 mM Ca2+ or 0.1 mM ethylenediaminetetraacetic (EDTA) was added to two of the C1B-C2 samples. After 5 hours, the reactions were quenched by the addition of 50 mM dithiothreitol (DTT) and incubation at room temperature for 15 minutes. The samples were subsequently boiled for 5 minutes in an equivalent volume of 2X SDS-PAGE loading buffer (BioRad) and loaded onto a 15% SDS-PAGE. All chemicals were obtained from Fisher Scientific unless otherwise stated.
NMR experiments and data analysis
All NMR experiments were carried out on Varian Inova spectrometers operating at 1H Larmor frequencies of 500 MHz (11.7 T) and 600 MHz (14.1 T). The temperature was calibrated using methanol below 30°C and ethylene glycol above 30°C. The NMR data were processed with NMRPipe34 and assigned with Sparky.35
1. Sequence-specific resonance assignments
Assignment experiments were carried out on a [U-15N,13C]-enriched 0.5 mM sample of C1Bα50 at pH 6.5 and pH 5.7. 1HN13Cα13Cβ, and 15N resonances of C1Bα50 were assigned from gradient-enhanced three-dimensional CBCA(CO)NH, HNCACB,36 and C(CO)NH37 spectra collected at 14.1 T. The C151G assignments were transferred from those of C1Bα50. The residue-specific differences in chemical shifts between the C1Bα50 conformers were calculated according to the following equation:38
| Eq. (1) |
where ΔδH, ΔδN, ΔδCα, and ΔδCβ are the chemical shift differences between the 1HN, 15N, 13Cα, and 13Cβ nuclei, respectively. The Hδ2 protons of histidine side-chains were assigned using the 2D (HB)CB(CGCD)HD experiment.39
2. NMR-detected pH titrations of C1Bα50 and C151G
The pH of the [U-15N]-enriched C1Bα50 in the NMR buffer was adjusted using HCl or NaOH solutions to obtain a range of pH values from 5.0 to 8.5. The pH (uncorrected for the presence of 8% D2O) was measured using an Accumet AB15 pH meter (Fisher Scientific) equipped with a Hamilton Biotrode electrode. The measurement was done before and after each NMR experiment. The difference in pH values typically did not exceed 0.1. [U-15N]-enriched C151G in deionized H2O was titrated in the pH range from 5.0 to 8.3. 15N-1H HSQC experiment was used to monitor the progress of titrations.
C1Bα50 exists in two slowly exchanging conformational states that we designated a and b. Their fractional populations, fa and fb, were calculated as va/(va+vb) and vb/(va+vb), where va(b) is the residue-specific cross-peak volume of conformation a(b) at a given pH value. The cross-peak volumes were determined from the 15N-1H HSQC spectra using the integration methods implemented in Sparky.35 To obtain the apparent pKa (pKaapp) for the pH-dependent process that alters populations of a and b, the experimental data were fitted to the following equation:
| Eq. (2) |
| Eq. (3) |
The fit was done globally for all protein residues that have distinct cross-peaks in conformations a and b, and are not subject to extensive line broadening at high and low pH values.
The dependence on pH of residue-specific 15N or 1HN chemical shifts, whichever was larger, was fit to a modified Henderson-Hasselbach equation:40
| Eq. (4) |
where δ is the observed chemical shift at a given pH, offset is the chemical shift offset, and delta is the chemical shift change in the titration process with the acid dissociation constant Ka1. The chemical shifts of residues that responded to two titration events were fit to the following equation:
| Eq. (5) |
where 1 and 2 designate the parameters associated with the first and second ionization events.
3. Measurements of transverse relaxation-rate constants (R2), longitudinal relaxation-rate constants (R1), and {1H}-15N nuclear Overhauser enhancement (NOE)
The R2 and R1 values of uniformly 15N-enriched C1Bα constructs were measured on a 600 MHz Varian INOVA spectrometer at 25°C. The experiments were carried out in a pseudo-3D fashion as described previously.41 The data were collected with 9 relaxation time delays (3 of which were duplicates) ranging from 12 to 180 ms (R2) and from 30 to 600 ms (R1). R2 and R1 values were determined for all spectrally resolved cross-peaks by fitting the time dependence of peak intensities to an exponential function using Curvefit.
The NOE data were acquired in an interleaved manner, with a 3 s saturation period and a 5 s recycle delay. The uncertainties in cross-peak intensities were estimated from the root-mean-square noise level of the base planes. Spectral density J(0.87ωH), where ωH is the Larmor frequency of 1H nucleus, reports on the high-frequency motions of the N-H bond vectors. The reduced42, 43 spectral density mapping (SDM) approach44, 45 was used to calculate J(0.87ωH) from the R1 and NOE values:
| Eq. (6) |
where d = (µ0hγHγN/8π2)(1/<rXH>3); µ0 is the permeability of free space; h is Planck’s constant; γH (γN) is the gyromagnetic ratio of 1H (15N), and rXH is the length of the N-H bond.
4. NMR ZZ-exchange experiments
The kinetics of the slow conformational exchange reaction between the a and b conformers of C1Bα50 at pH = 5.75 was quantified using the ZZ-exchange experiments.46–48 In these experiments, the exchange of the 15N longitudinal magnetization during a variable mixing time generates cross-peaks between the residues subject to the slow exchange. The volumes of auto- and cross-peaks in the spectra were determined using the nlinLS routine of nmrPipe.34 The data were analyzed according to the procedure described by Miloushev et al.49 In brief, for the six well-resolved residues (103, 104 133, 147, 149, and 151), we calculated the parameter Ξ according to the following equation:
| Eq. (7) |
where Vaa and Vbb are the auto-peak volumes, Vab and Vba are the cross-peak volumes, and t is the mixing time. The experiments were carried out at 5 temperatures: 9.4, 15.0, 19.8, 25.0 and 30.2 °C. The cross-peaks of residue 149 at 30.2 °C and residue 103 at 9.4 and 19.8 °C were not used due to excessive line broadening. Residue-specific Ξ values were plotted as a function of mixing time and fitted to the following equation:
| Eq. (8) |
where k1 and k−1′ are the forward and pseudo-first order reverse rate constants for the exchange reaction:
k−1′ = k−1[H+]; and a and b are the “acidic” and “basic” conformers of C1Bα50. The data for all residues were fitted globally to obtain the product of two rate constants, k1k−1′.
The apparent equilibrium constant for the exchange reaction was defined as. Kapp was determined using an average of the ratio of the peak volumes from the two populations for 9 well-resolved residues (103, 131, 133, 134, 140, 145, 147, 149, and 151) in fully relaxed 15N-1H HSQC spectra collected with the 5 s relaxation delay. The kinetic rate constants were then calculated using the k1k−1′ and k1/k−1′ values obtained from the ZZ-exchange data and Kapp, respectively. The activation enthalpies and entropies for the forward and reverse reactions were determined from fitting the data with the Eyring equation:
| Eq. (9) |
where k is the rate constant for the reaction; kB is the Boltzmann constant; κ is the transmission coefficient assumed to be 1 in most types of analyses; h is the Planck constant; and ΔS≠ and ΔH≠ are the entropy and enthalpy of activation, respectively. Activation energies Ea were obtained from fitting the data with the Arrhenius equation:
| Eq. (10) |
where A is the pre-exponential factor.
5. Measurements of residual dipolar couplings (RDCs) for structure refinement
Partially aligned samples of wild-type C1Bα50 were made in 7% polyacrylamide stretched gels50 as previously described.26 The compressed gels51, 52 were prepared with the 20:1 acrylamide to bis-acrylamide ratio and cast at an internal diameter of 3.2 mm. The 25 mm-long gel was washed with water, dried, and then placed into a 5 mm Shighemi tube, followed by the addition of the 0.5 mM solution of [U-13C,15N]-enriched C1Bα50 at pH 5.75. The plunger of the Shighemi tube was placed such that the gel expanded to a final length of 17.5 mm to give a compression ratio of 0.7.
All NMR experiments for RDC measurements were carried out at 25°C for isotropic and partially aligned samples of C1Bα50. One-bond RDCs, 1DHN, were determined for the C1Bα50 in stretched gels using a 2D in-phase anti-phase (IPAP) experiment.53 For the compressed gels, in addition to 1DHN, we measured 1DCαHα1DNCO, and 2DHNCO using the appropriate 3D HNCO-based experiments.54, 55 The RDCs were calculated as the differences in multiplet splittings between the aligned and isotropic peak positions.
These RDC values were used to refine the structures of C1Bα50a and C1Bα50b. The refinement was carried out using the simulated annealing protocol implemented in XPLOR-NIH.56, 57 The starting structure for refinement was the recently deposited NMR structure of C1Bα (PDB ID 2ELI; RIKEN Structural Genomics/Proteomics Initiative). This structure was determined at pH 7.0, where greater than 90% of the protein has the thiolate of Cys151 coordinated to Zn2+. The refinement protocol was a modification of the procedure reported by Chou and Bax.58 It consisted of two distinct refinement steps carried out at high and low temperatures. The details of the refinement protocol are summarized in Section S9 of the SI. All images of structures were created using either Chimera59 or Visual Molecular Dynamics.60
RESULTS
Cys151 is reactive irrespective of the protein context
We used hydrogen peroxide as an oxidizing agent and PEG-mal and iodoacetamide (IAC) as alkylating agents to test the reactivity of cysteine residue(s) in three protein constructs: C1Bα50, C151G, and C1B-C2.
C1Bα50 corresponds to the minimum folded core of the C1B domain, in which the side-chains of the first (His102) and last (Cys151) residues are involved in Zn(2) coordination. C151G variant has Cys151 mutated to the Gly residue. The rationale behind constructing this variant was two-fold: (i) to gain insight into the relative reactivity of cysteine residues in C1B, and (ii) to eliminate the coordination bond between Zn(2) and Cys151. According to our NMR data (vide infra), this mutation preserves the overall fold of C1B. The C1B-C2 construct (see Figure 1) is 194 amino acids long and comprises another regulatory domain of PKCα, the C2 domain. The C2 domain is a Ca2+-responsive peripheral membrane domain lacking native cysteines. Having the C2 domain in this construct mimics the protein context of C1B in the full-length PKCα.
To enable a direct correlation of cysteine reactivity with structural and dynamical information, we used identical buffer conditions for all experiments.
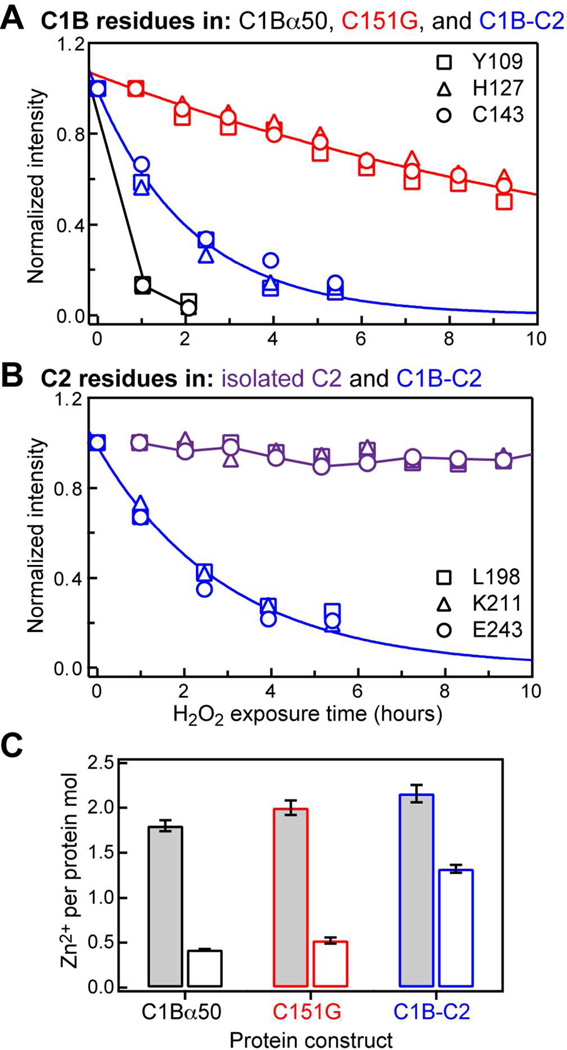
To oxidize the reactive cysteine(s), all three constructs were treated with 1.25 mM hydrogen peroxide. As a negative control, we used the isolated Ca2+-complexed C2 domain that lacks native cysteine residues. The progress of the protein reactions with H2O2 was monitored by NMR. For all C1B-containing constructs, the H2O2 treatment resulted in the gradual disappearance of NMR cross-peaks from the 15N-1H HSQC spectra. Because more than half of the protein remains in solution after the NMR signal has decayed to less than 10% of its original intensity, we attribute this phenomenon to the formation of soluble high-molecular weight aggregates that are unobservable due to slow rotational diffusion and/or aggregation processes that are intermediate on the NMR chemical shift timescale. Figure 2 shows the data for three representative residues that belong either to the C1B (A) or the C2 (B) domains.
Figure 2.
H2O2 treatment of C1B-containing proteins results in the loss of native C1B structure. (A) Representative data for three C1B residues in C1Bα50 (black), C151G (red), and C1B-C2 (blue). (B) Representative data for three C2 residues in isolated C2 (purple) and C1B-C2 (blue). Solid lines are the exponential fits to the C151G and C1B-C2 data; C1Bα50 and C2 points are connected with lines for illustration purposes. (C) The number of Zn2+ ions released per protein molecule in solution when treated with 1.25 mM H2O2 at 25 °C until 10% NMR cross-peak intensity remained (open bars) and with 4 mM H2O2 at 42 °C for one hour (shaded bars).
It is evident from Figure 2A that the stability of C1B with respect to oxidation by H2O2 depends on whether Cys is present at position 151. The signal corresponding to the native population of C1Bα50 decays within 2 hours of initiating the H2O2 treatment. However, the C151G variant lacking one coordination bond to Zn(2) (see Figures 1 and 4C) is more stable, with a quantifiable average signal decay rate of 0.06 hr−1. The residue-specific decay rates are rather uniform throughout C151G, with a standard deviation of 0.01 hr−1 (Figure S1A of the SI). Peroxide treatment of the two-domain construct C1B-C2, where Cys151 immediately precedes the linker region connecting the two domains, results in the average signal decay rate of 0.45 hr−1, about ~8-fold higher than that of the C151G variant. The residue-specific decay rates of C1B-C2 are plotted in Figure S1B of the SI.
Figure 4.
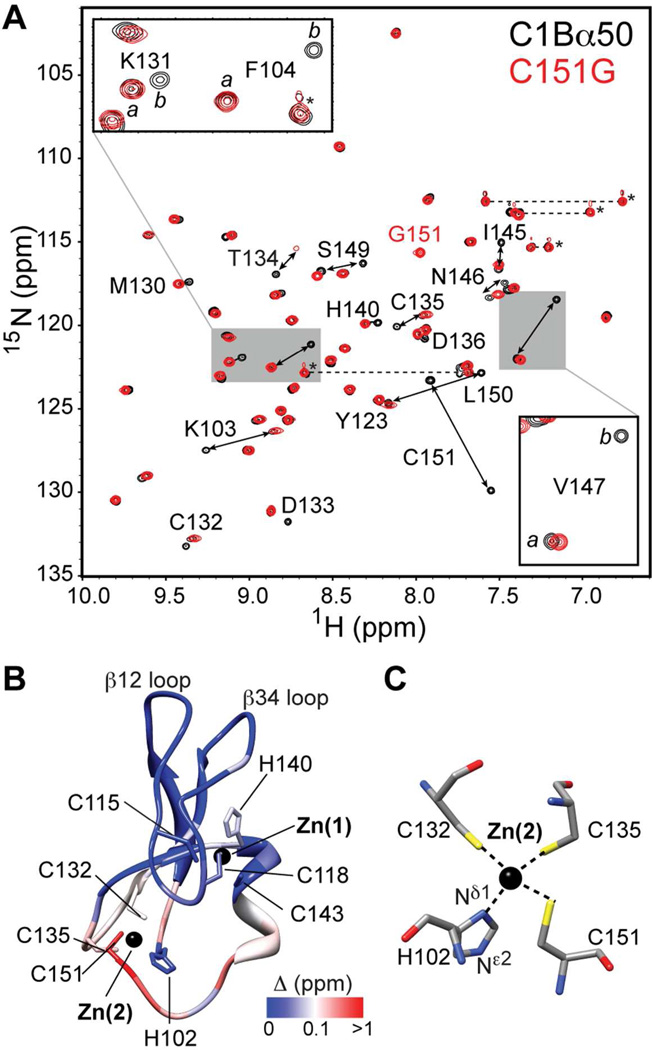
C1Bα50 exists in two slowly exchanging conformations. (A) Overlay of the 15N-1H HSQC spectra of C1Bα50 and C151G variant. The insets show the expansions for Lys131, Phe104, and Val147. (B) Chemical shift perturbation Δ mapped onto the structure of C1Bα.62 The coordinates are the courtesy of Dr. U. Hommel. (C) Coordination geometry of Zn(2) showing four protein ligands.
Figure 2B shows the results of the negative control experiment carried out on the isolated Ca2+-complexed C2 domain that lacks native cysteine residues. The signal intensity does not change significantly in the course of the NMR experiment indicating that the isolated C2 domain is not susceptible to oxidation by H2O2. In contrast, the N-HN cross-peaks corresponding to the C2 domain in the C1B-C2 construct decay with an average rate of 0.36 hr−1. These data demonstrate that H2O2 acts specifically on the cysteine-containing C1B domain.
Fully folded C1B-containing constructs, including the C151G variant, contain two Zn2+ ions per protein molecule (Figure 2C, shaded bars). These data were obtained using PAR assay under conditions favoring complete Zn2+ release – 1-hour incubation with 4 mM H2O2 at 42 °C – as described in Section S2 of the SI. PAR assay was also carried out on the supernatants of all H2O2-treated NMR samples at 25 °C, with no additional H2O2 added to the protein solutions. In all three constructs, we observed a partial release of Zn2+ ions from C1B (Figure 2C, open bars).
Given the release of structural Zn2+ ions and extensive protein precipitation observed for H2O2-treated NMR samples after ~24–48 hours, we conclude that the treatment with H2O2 results in at least partial loss of the native C1B structure.
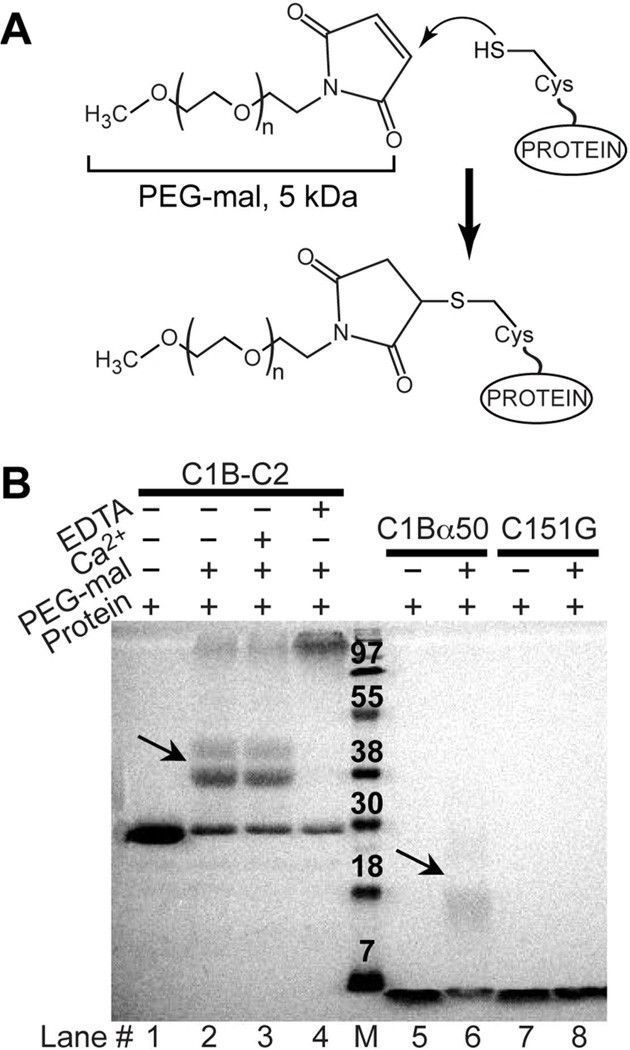
To test the protein behavior with respect to the alkylation reaction, we conducted mass-tagging assays with C1Bα50, C151G, and C1B-C2. Mass-tagging was accomplished by reacting the proteins with PEG-maleimide (PEG-mal 5,000), as shown in Figure 3A. Covalent modification of each cysteine residue adds 5 kDa of molecular weight. The mass-tagged protein species are subsequently resolved on the SDS-PAGE gel (Figure 3B), in which the 5 kDa increase in molecular weight manifests itself as an ~8–10 kDa shift on the gel.
Figure 3.
Mass-tagging assays carried out on C1Bα50, C151G, and C1B-C2. (A) Reaction between the sulfhydryl groups of exposed cysteine residues and PEG-mal. (B) SDS-PAGE results of the PEG-mal treatment of three C1Bα constructs. Arrows point to the single-Cys modified species in C1Bα50 and C1B-C2.
C1Bα50 is modified primarily at one cysteine residue (lane 6). C151G is not modified by PEG-mal (lane 8), which means that Cys151 is the most reactive residue in C1Bα50. C1B-C2 shows a “ladder” pattern of one, two, and >3 modified Cys residues, with the primary species being the single Cys-modified protein (lane 2). Addition of Ca2+ to C1B-C2 does not influence the “ladder” pattern (lane 3), indicating that Ca2+ binding by the C2 domain has little effect on the reactivity of cysteines in the C1B domain. Treating C1B-C2 with EDTA unfolds the protein by removing the structural Zn2+ ions. As a result, all cysteine residues become solvent-exposed and modified by PEG-mal with the formation of high molecular weight species (lane 4).
A similar pattern of cysteine reactivity was obtained upon treating the protein constructs with cysteine-specific alkylating agent, iodoacetamide (IAC). The reaction protocol is described in Section S4 of the SI. Modification of C1Bα50 by IAC completely unfolded the C1B domain as was evident from the 15N-1H HSQC spectra (Figure S2A). In contrast, the population of the fully folded protein was still present in the C151G variant after the completion of IAC reaction. Treatment of C1B-C2 with IAC selectively unfolded the C1B domain but left the C2 domain unaffected in the C1B-C2 construct (Figure S2B).
In summary, our oxidation and alkylation data are consistent with Cys151 being the most reactive among the six cysteines in C1Bα. Its covalent modification results in the loss of C1B native structure and subsequent covalent modification of all other cysteines accompanied by partial Zn2+ release. C1Bα50 shows unique dynamics of the Zn(2) coordination sphere that includes Cys151. We characterized this dynamics using solution NMR techniques.
C1Bα50 exists in two conformations
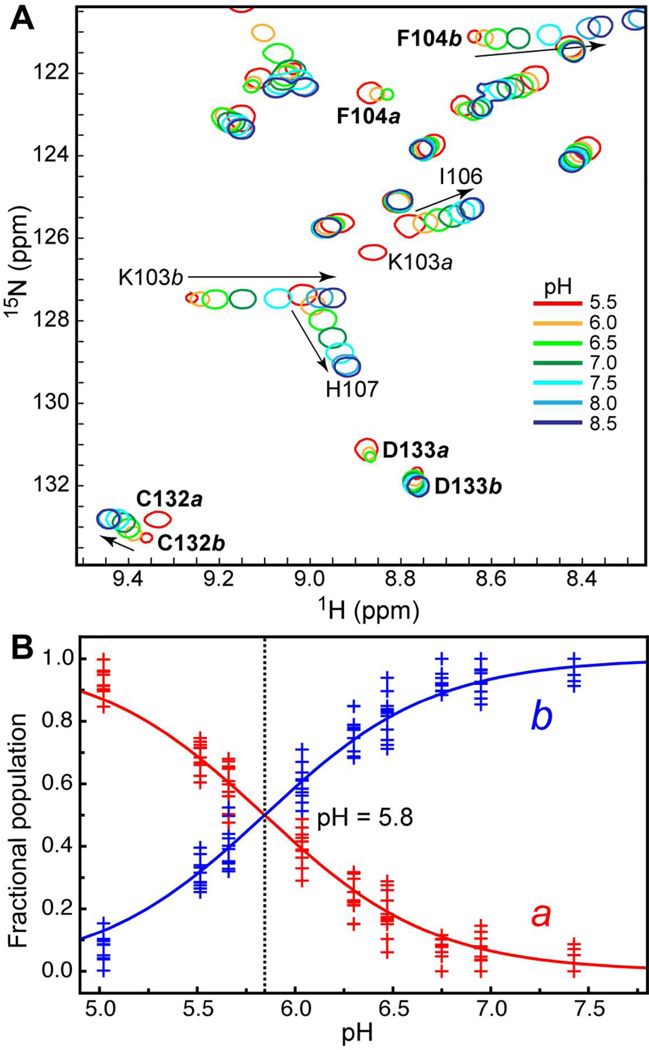
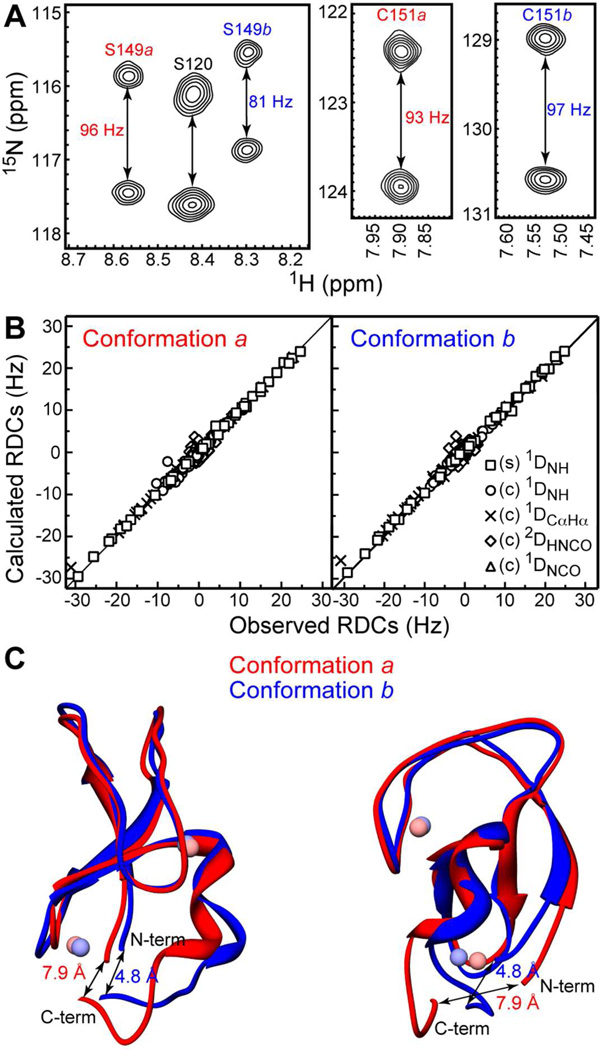
The structural and functional cores of C1 domains comprise 50 amino acid residues.61 C1Bα50 behaved as a homogeneous protein preparation in all purification steps. However, the 15N-1H HSQC spectrum of the [U-15N]-enriched C1Bα50 showed a larger number of 15N-1HN cross-peaks than was expected based on the size of the protein construct. Using triple-resonance NMR experiments, we carried out a sequence-specific assignment of [U-13C,15N]-enriched C1Bα50. Out of 48 non-proline residues, 17 gave rise to a pair of N-HN cross-peaks with distinct chemical shifts. The cross-peak pairs are labeled with the double-headed arrows in the HSQC spectrum of C1Bα50 (Figure 4A). This behavior indicates the presence of two conformations of C1Bα50 that co-exist in solution. The exchange between the conformers is slow on the NMR chemical-shift timescale.
To gain insight into the identity of the exchanging conformers, we mapped their chemical shift differences calculated according to Eq. (1) onto the ensemble-averaged NMR structure of C1Bα.62 The results are shown in Figure 4B. The residues whose electronic environment differs most between the two conformations are located in the vicinity of the Zn(2) coordination site. Cys151, the most C-terminal residue of the domain, experiences the largest combined chemical shift perturbation with the Δ value of 1.3 ppm. The Sγ atom of the Cys151 side-chain is involved in the coordination bond with Zn(2), according to the NMR structure of C1Bα and the crystal structure of a homologous C1B domain from Protein Kinase Cδ.63 The coordination geometry of Zn(2) is shown in Figure 4C.
The C151G variant of C1Bα50 presents only three protein ligands to Zn(2) instead of four. The C151G mutation was shown to preserve the functionality of a homologous C1B domain from PKCδ, as determined by the ability of the domain to bind phorbol esters.64 In Figure 4A, the HSQC spectrum of the [U-15N]-enriched C151G variant is shown in red and overlaid onto the spectrum of the wild-type protein. It is evident that C151G exists in one conformation only, and this conformation is one of the two sampled by the wild-type C1Bα50. The molar ratio of Zn2+ ions to protein is 2 in the C151G variant (see Section S2 of the SI), indicating that the protein maintains coordination to Zn(2) despite the loss of the Cys151Sγ–Zn(2) bond.
Based on this set of experiments, we concluded that the conformational exchange process in the wild-type C1Bα50 corresponds to Cys151 alternating between Zn(2)-coordinated and uncoordinated states.
The populations of C1Bα50 conformers are pH-dependent. The conformational exchange process is modulated by the pKa of the Cys151 side-chain
We observed significant changes in the fractional populations of the conformers in the C1Bα50 NMR spectra recorded at two different pH values. We then explored the pH dependence systematically in the range from 5.0 to 8.5.
At each pH value, we recorded the 15N-1H HSQC spectrum of C1Bα50. For the 17 residues that have distinct cross-peaks in the two conformers of C1Bα50, we observed a complete redistribution of conformer populations over the sampled pH range. The low-pH conformer designated a for “acidic” conditions is a dominant form at pH < 6; the high-pH conformer designated b for “basic” conditions is a dominant form at pH > 6. Conformer a corresponds to the uncoordinated Cys151, based on the near-identity between its spectrum and that of the C151G variant (Figure 4A). For several residues, including some from the aforementioned group of 17, we observed a pH-dependent shift in their N-HN cross-peak positions. The shift reflects the titration behavior of the proximal ionizable side-chain(s) and is “fast” on the NMR chemical shift timescale.
Three specific examples illustrating the pH titration behavior of C1Bα50 are shown in Figure 5A. All three residues, Phe104, Asp133, and Cys132, have two populations that give rise to distinct cross-peaks. In Asp133, the pH change results simply in the redistribution of populations a and b. In Phe104 and Cys132, this process is accompanied by the pH-dependent shift of the conformer b cross-peaks.
Figure 5.
pH dependence of the C1Bα50 HSQC spectra. (A) Expansion of the 15N-1H HSQC spectra showing examples of residues with fast and/or slow exchange behavior. (B) pH dependence of populations a and b. The fractional populations were calculated using the cross-peak volumes of all residues that have well-resolved peaks in the two conformations. The data points correspond to different exchanging residues in C1Bα50. The solid line represents a global fit of the data to Eqs. (2) and (3).
For all protein residues that are not subject to extensive line-broadening at high and low pH values, we determined the fractional populations of conformers a and b as described in the Experimental Procedures. The fractional populations are plotted against pH in Figure 5B. The solid line represents a global fit of the data to Eqs. (2) and (3). Equal populations of a and b are present at pH 5.8. Our data indicate that the loss of coordination bond between the Sγ of Cys151 and Zn(2) is a pH-dependent process with an apparent pKa of 5.8. This value either represents the side-chain pKaapp of Cys151, or the pKa of some other ionizable group in the protein. The most likely candidates would be titratable histidine residues – their average pKa value in proteins is 6.6, with a standard deviation of 1.65
To test if any of the five histidines are responsible for the pH dependence of the conformational exchange, we constructed the titration curves for all pH-responsive residues by plotting the change in 15N and 1H chemical shifts as a function of pH. The NMR-detected titration curves for His107, His117, and His127 are shown in Figure S3. The three histidines titrated with pKa values of 6.7 ± 0.1 (His107), 5.6 ± 0.1 (His117), and 6.1 ± 0.1 (His127). None of these residues are in slow exchange on the NMR chemical-shift timescale.
The two remaining histidines – His102 and His140 – coordinate Zn(2) and Zn(1), respectively. The amine group of the N-terminal His102 is not detectable in our spectra. The backbone cross-peak of His140 is in slow exchange between the populations a and b (see Figure 4A). To estimate the pKa values of His102 and His140, we carried out “fast” HSQC66 experiments tuned for the detection of side-chain resonances.67, 68 The NMR spectra at two pH values, 4.7 and 7.0, and the table of chemical shifts are given in Figure S4. The Nδ1 and Nε2 shifts at both pH values are consistent with those observed for the Zn2+-coordinated histidines in proteins.69–71 There is no indication that the protonation state of either His102 or His140 side-chain changes in the pH range from 4.7 to 7.0. This means that the pKa values of His102 and His140 are less than 4.7.
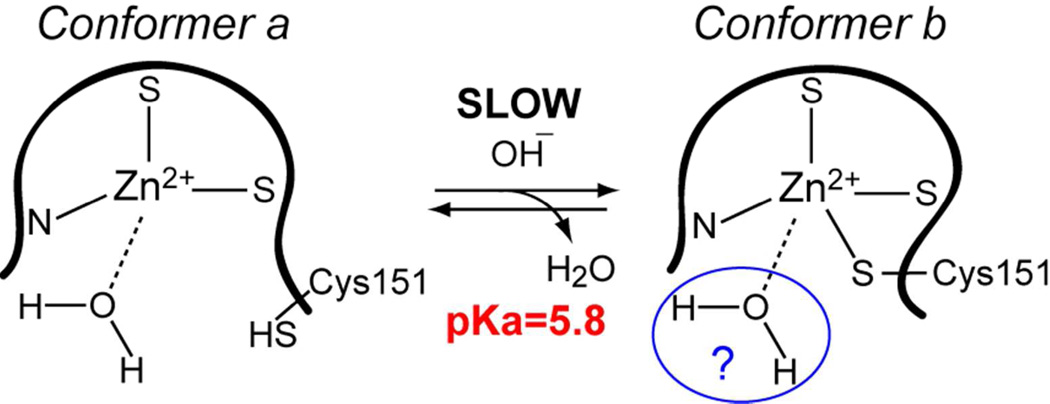
Based on these results, we ruled out histidine residue(s) as the primary ionizable group with pKaapp of 5.8. We conclude that the pH dependence of the conformational exchange process stems from the de-protonation of the Cys151 side-chain that occurs concomitantly with the loss of its coordination bond with Zn(2). Hence, the pKaapp of Cys151 in the C1Bα50 is 5.8.
Ionizable group with pKa 7.1–7.2
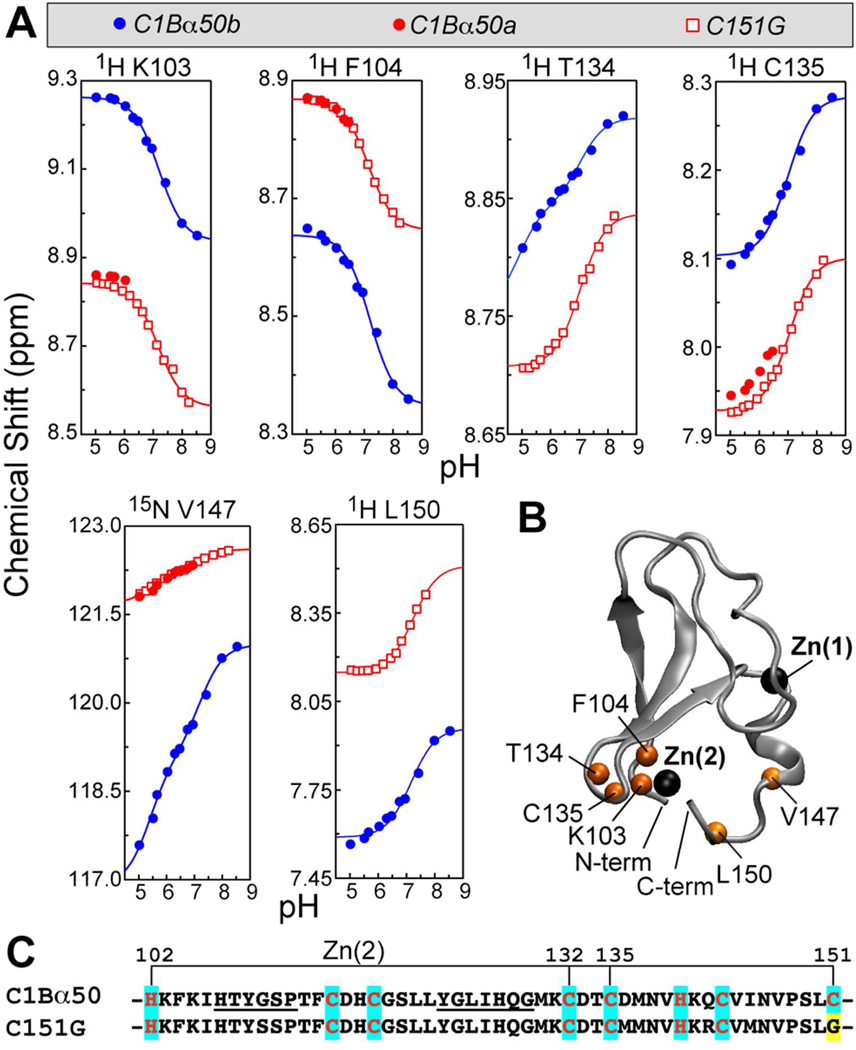
In addition to three histidine side-chains, we detected another ionizable group with an apparent pKa of 7.2 ± 0.1 in C1Bα50 and 7.1 ± 0.1 in C151G. The NMR-detected titration curves for the residues that responded to this pKa are given in Figure 6A. The parameters of the individual fit to Eqs. (4) or (5) are summarized in Table S1 of the SI. It is evident that the C151G variant mirrors the behavior of conformer a of C1Bα50 and thereby enables us to probe the “high-pH” response of conformer a, whose population in the wild-type protein becomes less than 10% at pH 7.0.
Figure 6.
(A) NMR-detected pH titration curves for the nuclei/residues that respond to the protonation event with an apparent pKa of 7.1–7.2 in C1Bα50 and C151G. Low-pH points for L150a and T134a could not be obtained because of the peak broadening/overlap. (B) N-HN groups of residues in (A) mapped onto the C1Bα structure. (C) Primary structures of the two C1Bα constructs used in NMR-monitored pH titration experiments. The mutation site is highlighted in yellow; Zn(2)-coordinating residues are highlighted in cyan; the loop residues are underlined.
The residues responding to the ionizable group with pKa 7.1–7.2 are mapped onto the ensemble-averaged NMR structure in Figure 6B. The responsive residues are located in the vicinity of the Zn(2) coordination sphere, which comprises Cys132, Cys135, His102, and Cys151 (Figure 6C).
The assignment of the functional group with pKa 7.1–7.2 is ambiguous. There are four possible candidates for this ionizable group: the C-terminal carboxyl, N-terminal amine, Cys135, and Zn2+-coordinated water molecule. The carboxyl and amine groups are the least likely candidates because they are solvent exposed and hence unlikely to have perturbed pKa values. In addition, the sidechain of the most N-terminal residue (His102) and the backbone of the most C-terminal residues, Cys151 in C1Bα50 and Gly151 in C151G, do not respond to the pKa of 7.1–7.2.
The prerequisite for Cys135 being the primary ionizable group is that its thiol – not thiolate – is involved in coordination with Zn(2). The thiol versus thiolate coordination in structural Zn2+ sites has been a subject of much discussion in the literature.72–75 According to the ab initio calculations, the structural signature of thiol coordination is the increase of the Sγ–Zn distance by 0.3–0.4 Å.75 In the same study, the Zn sites in a homologous C1B domain from PKCδ (PDB ID 1PTQ63) were identified as having a geometry compatible with thiol coordination. The longest Sγ–Zn distance, 2.51 Å, was observed in Cys264; the equivalent of this residue in C1Bα is Cys135.
Another viable candidate is a Zn2+-coordinated water molecule. A conversion of Cys151 side-chain to the thiol form in the C1Bα50a conformer or its replacement with Gly in the C151G variant creates a coordination vacancy in the Zn(2) site. Given the solvent accessibility of Zn(2), a water molecule will most likely fill this vacancy. Coordination of water by Zn2+ depresses its pKa to ~7.76, 77 The presence of water in the coordination sphere of Zn(2) in C1Bα50b would require a distortion of the tetrahedral geometry to accommodate an extra ligand. This distortion has been previously observed in our molecular dynamics simulations.26
Kinetics and thermodynamics of the Cys151Sγ–Zn(2) bond formation
The formation of the Cys151Sγ–Zn(2) bond can be represented as an inter-conversion between conformer a, in which Sγ is a thiol, and conformer b, in which Sγ is a thiolate coordinated to Zn(2). The pH dependence of the process is incorporated into the pseudo-first order rate constant to reflect the constant pH values in the sample maintained by the buffering agent.
To characterize the exchange process between a and b, we adjusted the populations of the conformers to ~0.5 by setting the pH to 5.75. At this pH, the equilibrium constant Kapp obtained by direct integration of a and b peaks in the NMR spectra, is 1.15 at 25 °C. Within the experimental error, Kapp = exp[−ΔG0/RT] does not depend on temperature in the investigated temperature range, from 10 to 30 °C. This means that the enthalpic contribution to the ΔG0 of the exchange reaction is rather small. Given a rather modest value of ΔG0 associated with Kapp ≈ 1.2, we conclude that both ΔH0 and ΔS0 make small individual contributions to ΔG0 rather than acting via an enthalpy-entropy compensatory mechanism.
We used two-dimensional ZZ-exchange NMR experiments to characterize the kinetics of C1Bα50 inter-conversion between conformations a and b. These experiments probe the slow exchange processes with kex values in the range from 0.1 to 10 s−1. kex is the sum of the forward and reverse rate constants, k1 and k−1′.
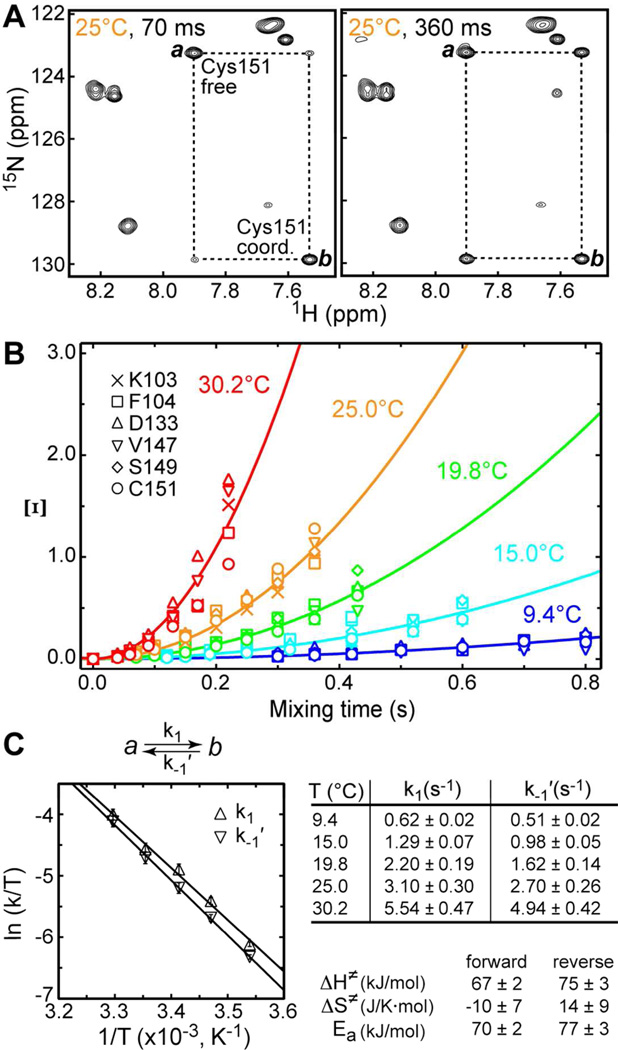
The expansions of the ZZ-exchange spectra showing the behavior of Cys151 peaks at two mixing times, 70 ms and 360 ms, are presented in Figure 7A. The gradual build-up of the cross-peaks due to exchange between populations a and b is evident. To enable the determination of activation parameters, the ZZ-exchange experiments were carried out at 5 temperature values. For each temperature and mixing time, we calculated the parameter Ξ according to Eq. (7). Ξ is plotted against the mixing time in Figure 7B, with solid lines showing the fits to Eq. (8).
Figure 7.
Kinetics of Cys151 inter-conversion between thiolate and thiol forms. (A) Expansions of the ZZ-exchange spectra showing the buildup of the Cys151 exchange cross-peaks at two mixing times, 70 and 360 ms. (B) Ξ at pH = 5.75 as a function of mixing time and temperature. Solid lines are the fits to Eq. (8). (C) Eyring plots for the forward (k1) and reverse (k−1′) kinetic rate constants that are summarized in the table. ΔH≠,ΔS≠, and Ea are the activation enthalpy, entropy, and energy, respectively.
The rate constants k1 and k−1′ calculated from Ξ and Kapp are summarized in the table of Figure 7C. We used the temperature dependence of k1 and k−1′ to construct the Eyring plots shown in the same figure. The fit of these plots generated the enthalpy and entropy of activation, ΔH≠ and ΔS≠. These values are given in Figure 7C for the forward and reverse reactions. In both cases, the ΔS≠ values are close to zero, which indicates that the total number of degrees of freedom available to the system does not change significantly between the transition state and the corresponding ground state.
The Arrhenius analysis was carried out to obtain the activation energies of Cys151Sγ–Zn(2) bond formation/breakage: 70 and 77 kJ/mol for the forward and reverse reactions, respectively. These energies are rather large, giving rise to small values of the kinetic rate constants: extrapolation of our data to 37 °C produces the rate constants of ~9.6 s-1.
Elimination of one thiolate ligand alters the C1Bα dynamics
To determine the consequences of Zn(2) loosing one of its protein ligands for the C1Bα dynamics, we measured the backbone relaxation parameters (R1, R2, and {1H}-15N NOE) for the C151G and C1Bα50 variants at pH 6.5.
Residue-specific transverse relaxation rate constants, R2, report on the sub-nanosecond (subns) dynamics of N-HN groups through their dependence on the spectral density function J sampled at three frequencies: 0, ωH, and ωN, where ωH (ωN) are the Larmor frequencies of 1H (15N). In addition, conformational exchange processes that occur on the microsecond-to-millisecond (µs-ms) timescale contribute to R2 via the Rex term. For proteins with moderate rotational diffusion anisotropy like C1Bα, elevated R2 values indicate that a particular N-HN group is subject to chemical exchange, i.e. its Rex > 0.
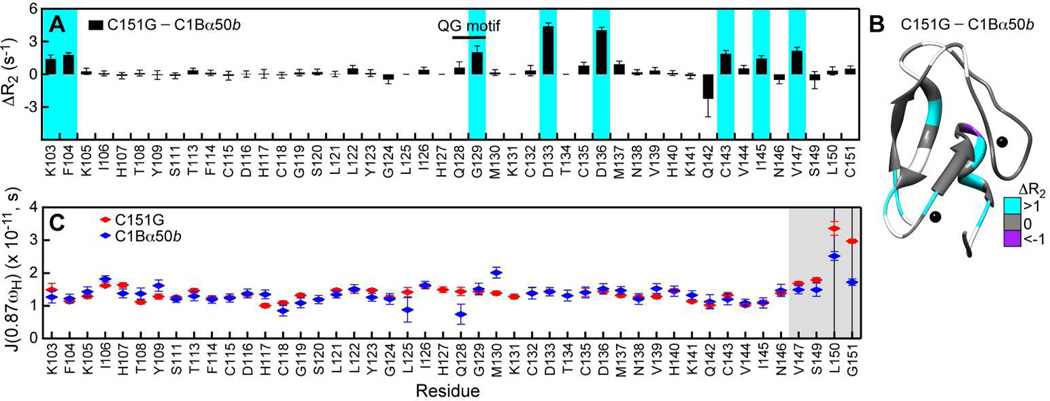
In Figure 8A, we plotted the differences in site-specific R2 values for the C151G and C1Bα50b. Many residues in C151G have elevated R2 values compared to those in C1Bα50b. This behavior indicates an increase in the µs-ms conformational dynamics of the protein backbone. The perturbed residues belong to the N-terminal region (Lys103 and Phe104), the QG motif (Gly129), Zn(2) “knuckle” and its vicinity (Asp133, Cys135, Asp136, Met137), and the C-terminal helix (Cys143, Ile145, Val147). Mapping ΔR2 values onto the C1Bα structure illustrates that most of the dynamically perturbed residues are clustered at the bottom half of C1Bα near the Zn(1) and Zn(2) structural sites (Figure 8B).
Figure 8.
Elimination of Cys151 ligand perturbs the C1Bα dynamics. (A) ΔR2, calculated as the difference between the residue-specific R2 values of C151G and those of C1Bα50b are plotted against the primary structure. Residues that experienced > 1 s−1 increase in R2 values in the C151G variant are highlighted in cyan. (B) ΔR2 for the C151G and C1Bα50b pair mapped onto the three-dimensional structure of C1Bα. Residues, for which ΔR2 values are not available, are in shown in white. (C) Spectral density values J(0.87ωH) for C151G and C1Bα50b. The C-terminal region, highlighted in grey, is flexible on the sub-ns timescale. The vertical lines mark two residues for which the differences in flexibility between the constructs are most pronounced, Leu150 and Cys151.
To compare the high-frequency motions in C1Bα constructs, we calculated the residue-specific spectral density values J(0.87ωH) using the reduced spectral density mapping (SDM) approach. ωH was 2π × 600 MHz for all relaxation experiments. J(0.87ωH) values report on the sub-nanosecond dynamics of the N-HN groups of the protein backbone. Figure 8C compares the J(0.87ωH) values for the two C1Bα constructs at pH 6.5. The spectral density profile is mostly flat throughout the primary structure and does not differ significantly between the variants. However, the C-terminal region spanning the Val147-Cys151 segment shows a high degree of flexibility. C151G, with its uncoordinated C-terminus, has the largest value of J(0.87ωH). The dynamics of Cys151 in C1Bα50b is attenuated due to its coordination to Zn(2), but Leu150 retains substantial mobility.
In summary, eliminating one protein ligand from the Zn(2) coordination sphere results in the increase of the backbone dynamics of C1Bα on at least two timescales: sub-ns and µs-ms. The C-terminal region acquires substantial flexibility on the sub-ns timescale when not constrained by the thiolate–Zn(2) bond. Residues in the vicinity of the Zn(1) and Zn(2) structural sites, including the C-terminal helix, become more dynamic on the µs-ms timescale. The effect is most evident for Asp133 and Asp136 of the Zn(2) “knuckle” motif (Figures 1 and 8A). Adding Zn2+ to the C151G NMR sample had little effect on the R2 values. This means that the conformational exchange in C151G is not due to the inter-conversion between Zn2+(2)-bound and Zn2+(2)-free forms. Determination of the identity of the conformer sampled by C151G requires a detailed NMR relaxation dispersion study and is the subject of future investigation.
Thiolate-to-thiol conversion of Cys151 “opens up” the C1Bα domain
To determine how the conversion of Cys151 into the thiol form affects the protein structure, we refined the existing NMR structure of C1Bα against the residual dipolar couplings (RDCs) measured for conformers a and b. It was evident from the 15N-1H HSQC spectra that the overall protein fold was preserved upon Cys151 protonation and that the conformational changes were not drastic. For such cases, the RDC-based approach is more efficient than the de novo structure determination.
The RDC measurements were carried out simultaneously for both conformers on the same protein sample with pH of 5.75. A total of 228 and 210 RDCs were measured for C1Bα50b and C1Bα50a, respectively, as described in the Experimental Procedures. The largest differences between the RDC values of conformers a and b were observed in the C-terminal region of the protein. An example is given in Figure 9A for the two residues, Ser149 and Cys151. The splittings between the cross-peaks are the sum of the one-bond J-coupling constant, 1JNH, and the residual dipolar coupling, 1DNH. Because the 1JNH coupling constants are fairly uniform for all amide groups, the differences in the observed splittings mostly reflect the differences in RDCs. The 1DNH values depend on the orientation of the N-HN bond vector relative to the alignment tensor, which is almost identical for both conformers (see Section S9 of the SI).
Figure 9.
RDC-based structure refinement of C1Bα50a and C1Bα50b. (A) Expansions of the 15N-1H IPAP spectra of C1Bα50 in stretched gels. The differences between the doublet splittings of Ser149 and Cys151 in conformers a and b are evident. (B) Calculated versus observed RDCs for stretched (s) and compressed (c) gels. (C) Two views of superimposed regularized average structures of C1Bα50a and C1Bα50b. The distances between the N- and C-termini are defined as the distances between the Cα atoms of His102 and Cys151.
The data in Figure 8C indicate that the C-terminal region is flexible. To properly account for this flexibility in the refinement procedure, we used half-open square well RDC potential for all dynamic residues in the protein with {1H}-15N NOE values less than 0.5. Other important details of the refinement procedure were the use of long-range NOEs to constrain the loop regions and definition of Zn2+ coordination geometry. The refinement was carried out as a two-step protocol that included high- and low-temperature simulated annealing steps. The parameters of the simulated annealing are given in Table S2 of the SI.
The agreement between calculated and observed RDCs is shown in Figure 9B. The agreement is excellent for most residues, with the RNH factor of 10% and 6% for the conformers a and b aligned in the compressed gel. The top 5% of the structures from the low-temperature simulated annealing step were used to generate the regularized average structures given in Figure 9C.
Relative to the starting model, the backbone RMSD values are 1.4 Å for both conformers. These values reduce to 1.0 Å (b) and 1.1 Å (a) if we exclude the N- and C-termini and use residues 6–44 only. Superposition of conformers a and b gives RMSD values of 1.6 Å (residues 1–50) and 1.0 Å (residues 6–44). The structures are essentially super-imposable in the top half that includes ligand-binding loops. The largest differences are observed in the N- and C-terminal regions, as well as the “knuckle” region of the Zn(2) site. The structural changes result in the bottom half of the domain opening up. The distance between the N- and C-termini, measured between the Cα atoms of His102 and Cys151 increases from 4.8 Å to 7.9 Å upon the formation of Cys151 thiol.
We speculate that the implications of this conformational change for the C1Bα structure and function are two-fold. First, any modification of the Cys151 thiolate is likely to increase the accessibility of the solvent (and other reactants) to the Zn(2) site. Second, the conformational change could be transmitted to the linker region that connects C1Bα to C2 and thereby influence the relative orientation of the two domains.
DISCUSSION
pH-dependent structural dynamics of Zn(2) coordination sphere
The chemical reactivity of Zn(2) site in C1Bα50 manifests itself in the pH-dependent structural dynamics of the coordination sphere. The dynamic process involves the Sγ atom of Cys151 alternating between the Zn2+-bound thiolate (conformer b) and free protonated thiol species (conformer a). The nature of the exchange process was established unambiguously by our finding that the NMR spectrum of conformer a that dominates at low pH is essentially super-imposable with the spectrum of the C151G C1Bα variant that lacks the fourth endogenous Zn(2) ligand.
The process of thiolate to thiol conversion, which essentially represents a competition between Zn2+ and H3O+ for the thiolate of Cys151, is slow on the NMR chemical shift timescale. ZZ exchange experiments carried out at different temperatures enabled us to characterize the kinetics and thermodynamics of the exchange process. At pH 5.75 and 25 °C, we obtained the kinetic rate constants of ~3 s−1, which are indicative of rather high activation energies in both directions.
The obtained kinetic rate constants are of the same order of magnitude as those reported for the slow exchange processes observed in two other zinc proteins: the 30-residue Cys2His2 zinc finger of a human enhancer binding protein78 and the NS3 protease.71 In both cases, the exchange processes were interpreted as one of the histidine ligands loosing its coordination bond with Zn2+; in the case of NS3 protease, the pH dependence of the process was explored to arrive at an apparent pKa of 5.9.71 A possible third example of a slow exchange process in a zinc finger protein was reported by Cornilescu et al.79 in the context of structural work, but the relevance of this process to the dynamics of metal coordination sphere was not investigated.
pKaapp of Cys151
The populations of conformers a and b are modulated by pH with an apparent pKa of 5.8. We assigned this pKa to the thiol side-chain of Cys151. How does this value compare to what has been reported for other zinc fingers? The thiol pKa values of solvent-exposed cysteine residues in proteins and non-cyclic peptides fall in the range between 7.6 and 9.4.80–82 Coordination to Zn2+ decreases the apparent pKa of the cysteine side-chain. For the coupled chemical equilibrium that involves a loss of H+ associated with Zn2+ coordination, the values of 6.3,80 5.0,81, 5.8,83, and 5.682 have been reported. Our data are in general agreement with the reported values.
The model of the Zn(2) site in C1Bα50 is presented in Figure 10. In conformer a, Cys151 is protonated and the vacant position is likely occupied by a water molecule. Upon de-protonation, the thiolate forms a coordination bond with Zn2+. The water molecule may or may not leave the metal coordination sphere. The presence in C1Bα50b of an ionizable group with pKa of 7.1–7.2 is consistent with the presence of water in the Zn(2) coordination sphere. This in turn would imply a deviation from the tetrahedral geometry. The existence of five-coordinate Zn2+ in peptides modeling the second zinc finger of NCp7 has been inferred from the Co2+ substitution experiments.84
Chemical reactivity of Zn(2) site
Several studies have addressed the question of why some Zn2+ sites are more chemically reactive than others. Maynard and Covell proposed that the most reactive Zn2+ cores have the least degree of steric and electrostatic shielding, with the latter provided by the S---HN hydrogen bonds.85 In their analysis of the Cys3His subclass, the C1 domain family from PKC was not identified as having labile Zn cores. Subsequently, Lee and Lim explored the indirect effects of S---HN hydrogen bonds on “unsheltered” cysteines from the same Zn2+ coordination sphere and concluded that the nucleophilicity (and hence reactivity) of those cysteines is reduced.86
We analyzed two structures for the presence of hydrogen bonds where Sγ serves as an acceptor: 1PTQ,63 which is the only crystal structure of a C1 domain from PKC isoforms (the C1B of PKCδ), and the structure of conformer b that we obtained in this work. The hydrogen bonds identified using the criteria of Zhou et al.87 are given in Table S3 of the SI. In C1Bδ, there are four S---HN hydrogen bonds per Zn2+ site. These bonds involve the Sγ atoms of the “knuckle” cysteines: Cys115 and Cys118 of Zn(1), and Cys132 and Cys135 of Zn(2), where we used the C1Bα numbering for clarity. The Sγ atoms of Cys143 and Cys151 have no hydrogen bonding partners. The same pattern is observed in the C1Bα50b structure with two exceptions. There is an additional S---HN bond that stabilizes the coordination sphere of Zn(1); it involves Cys115 and Gly119. In addition, the Sγ atoms of Cys151 and Cys132 of Zn(2) site form hydrogen bonds with the Hγ1-Oγ1 of Thr134.
For both structures, the number of S---HN bonds per structural site does not deviate significantly from 4, the average number reported for zinc fingers.85 Based on the hydrogen-bonding pattern alone, it is not immediately clear why Zn(2) is more reactive than Zn(1) or is reactive at all.
Another factor to consider in the context of chemical reactivity is the solvent exposure of the Zn(2) site. In C1Bα50b, Zn(2) joins together the N- and C-termini and is highly solvent-exposed, which enables H3O+ to successfully compete for the thiolate of Cys151. In C1B-C2, where the C2 domain is connected to the C1B by the linker region, Zn(2) site is more shielded from the solvent making Cys151 less accessible to H3O+. The higher reactivity of C1Bα50 compared to C1B-C2 was evident when both proteins were treated with H2O2 (Figure 2A).
A loss of one coordination bond by Zn(2) either due to Cys151 protonation as in C1Bα50a or its mutation as in the C151G variant opens up the Zn(2) site. This is manifested in the increased flexibility of the uncoordinated C-terminal tail (Figure 8C) and the increase in the distance between the N- and C-termini in the refined structure of C1Bα50a (Figure 9C). However, the solvent accessibility of Zn(2) in the C151G variant does not result in other cysteines of Zn(2) site being appreciably modified in the mass-tagging assays (Figure 3B). The susceptibility of C151G to alkylation by IAC (Figure S2) and oxidation by H2O2 (Figure 2) is greatly reduced compared to C1Bα50 and C1B-C2. All these observations point to Cys151 as the most chemically reactive cysteine residue in the C1B domain.
In the C1B-C2 construct, Cys151 is located at the beginning of the linker region between the two domains (Figure 1B). We expect that the solvent exposure of this residue will depend on the relative orientation of the C1B and C2 domains during the PKCα activation cycle (Figure 1). In our mass-tagging experiments with the C1B-C2 construct, we detected a substantial degree of covalent modification, which increases upon treating the protein with EDTA (Figure 3B). We subsequently carried out the same experiment at pH = 7.5, which is close to the cellular conditions, and observed the same “ladder” pattern within 1 hour of adding PEG-mal to the protein (Krystal A. Morales and Tatyana I. Igumenova, unpublished data).
Conversion of Cys151 from Zn2+-coordinated thiolate to thiol does not perturb the protein structure beyond the N- and C-terminal regions (Figure 9C). While the affinity of the protein to Zn2+ may change in the C1Bα50a conformer and the C151G variant, it is not significant enough for the domain to appreciably shed Zn2+ ions. In contrast, the treatment of C1Bα with hydrogen peroxide results in the loss of native structure with partial Zn2+ release. The reaction between hydrogen peroxide and C1Bα has specific biological implications.
Biological implications: activation of PKCα by ROS
The activation of PKC requires the rearrangement of catalytic and regulatory domains, which relieves the auto-inhibitory interaction between the pseudo-substrate region and the active site (Figure 1). Two cofactors, Ca2+ and DAG, serve as a regulatory input in this canonical activation mechanism. The first report that PKC isoforms can be activated in a cofactor-independent manner by hydrogen peroxide appeared in 1989.27 In that study, it was also shown that the presence of cofactors makes the enzyme more susceptible to oxidative modification. Subsequent work established that the treatment of PKC either by superoxide28 or hydrogen peroxide29 is accompanied by the release of Zn2+ ions. Cysteine-rich regions of PKC isoforms, the C1 domains, were suggested as the most likely targets of reactive oxygen species. These properties of PKC have made it a fully-fledged member of the “redox zinc switch” group.88
We propose that the reaction of PKCα with hydrogen peroxide (or other reactive oxygen species) is initiated at the most reactive cysteine residue, Cys151 of the C1B domain. It is currently unclear whether or not a similar pattern of cysteine reactivity exists in the C1A domain of PKCα, because so far this domain has resisted any structural characterization attempts. Previous studies showed that the cofactor-dependent activity of hippocampal PKC preparations was not affected by superoxide treatment.28 These data suggest that the DAG binding site, which resides on the C1A domains at least in PKCα and PKCβI/II isoforms, is not affected by ROS and that the C1B domain may be targeted specifically. Detailed in-vitro studies of purified PKC isoforms will be required to resolve this issue.
As a result of Cys151 being oxidized, the C1B domain looses its native structure thereby exposing other cysteines to the oxidizing agent(s). In the context of parent protein, the loss of native C1B structure is likely to disrupt the inter-molecular interactions in PKCα and relieve the auto-inhibition. This is accompanied by Zn2+ release, which can serve as a signaling species itself. Thus, the oxidation of cysteines in the C1B domain of PKCα produces a two-pronged signaling response initiated at the structural zinc site.
CONCLUSIONS
The work reported in this manuscript provides a detailed structural and dynamical view of a reactive Cys3His zinc site in one of the lipid-binding modules of PKCα, the C1B domain. Cys151 is the most reactive ligand of the four. In the 50-mer construct of C1B, this reactivity manifests itself as a pH-dependent inter-conversion of Cys151 side-chain between the Zn2+-coordinated thiolate and free thiol. The chemical reactivity pattern observed in the cysteine modification assays of the extended C1B constructs suggests that the activation of PKCα by ROS is initiated at Cys151. Our data demonstrate the remarkable potential of this Zn2+ finger to regulate the signaling response of PKCα by responding to chemical stimuli other than its membrane-embedded cofactors.
Supplementary Material
Figure 10.
Zn(2) site model in C1Bα50.
Acknowledgments
This work was supported by startup funds from Texas A&M University and Ralph E. Powe junior faculty enhancement award from Oak Ridge Associated Universities (T.I.I.); and NIH Training Grant 5-T32GM065088 (M.D.S).
ABBREVIATIONS
- DAG
diacylglycerol
- PE
phorbol ester
- ROS
reactive oxygen species
- PKCα
protein kinase Cα
- C1Bα
C1B domain from protein kinase Cα
- HSQC
heteronuclear single-quantum coherence
- NOE
nuclear Overhauser enhancement
- µs-ms
microsecond-to-millisecond
- sub-ns
sub-nanosecond
Footnotes
ASSOCIATED CONTENT
Supporting Information. Purification protocol for the C1Bα50 and its C151G variant; details of Zn2+ PAR assay; residue-specific signal decay rates for the H2O2-treated C151G and C1B-C2; NMR spectra of IAC-treated C1B-containing protein constructs; histidine pKa values; NMR spectra of histidine side-chains; fitted pKa values for residues responding to pKa 7.1–7.2; temperature coefficients of C1Bα50 amide protons; details of the RDC-driven refinement procedure; table of hydrogen bonds with Sγ of Zn2+-coordinating residues serving as an acceptor. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Decaria L, Bertini I, Williams RJ. Zinc proteomes, phylogenetics and evolution. Metallomics. 2010;2:706–709. doi: 10.1039/c0mt00024h. [DOI] [PubMed] [Google Scholar]
- 2.Karlin S, Zhu ZY. Classification of mononuclear zinc metal sites in protein structures. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14231–14236. doi: 10.1073/pnas.94.26.14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreini C, Bertini I, Cavallaro G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS One. 2011;6:e26325. doi: 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auld DS. Zinc coordination sphere in biochemical zinc sites. BioMetals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 5.Berg JM. Zinc finger domains: hypotheses and current knowledge. Annu. Rev. Biophys. Biophys. Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- 6.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 7.Lee YM, Lim C. Physical basis of structural and catalytic Zn-binding sites in proteins. J. Mol. Biol. 2008;379:545–553. doi: 10.1016/j.jmb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Maret W. Metals on the move: zinc ions in cellular regulation and in the coordination dynamics of zinc proteins. BioMetals. 2011;24:411–418. doi: 10.1007/s10534-010-9406-1. [DOI] [PubMed] [Google Scholar]
- 9.Maret W, Li Y. Coordination dynamics of zinc in proteins. Chem. Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- 10.Penner-Hahn J. Zinc-promoted alkyl transfer: a new role for zinc. Curr. Opin. Chem. Biol. 2007;11:166–171. doi: 10.1016/j.cbpa.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 11.Myers LC, Terranova MP, Nash HM, Markus MA, Verdine GL. Zinc binding by the methylation signaling domain of the Escherichia coli Ada protein. Biochemistry. 1992;31:4541–4547. doi: 10.1021/bi00134a002. [DOI] [PubMed] [Google Scholar]
- 12.He C, Hus JC, Sun LJ, Zhou P, Norman DP, Dotsch V, Wei H, Gross JD, Lane WS, Wagner G, Verdine GL. A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada. Mol. Cell. 2005;20:117–129. doi: 10.1016/j.molcel.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Takinowaki H, Matsuda Y, Yoshida T, Kobayashi Y, Ohkubo T. The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein. Protein Sci. 2006;15:487–497. doi: 10.1110/ps.051786306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summers MF, Henderson LE, Chance MR, Bess JW, Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, 3rd, Hare DR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chertova EN, Kane BP, McGrath C, Johnson DG, Sowder RC, 2nd, Arthur LO, Henderson LE. Probing the topography of HIV-1 nucleocapsid protein with the alkylating agent N-ethylmaleimide. Biochemistry. 1998;37:17890–17897. doi: 10.1021/bi980907y. [DOI] [PubMed] [Google Scholar]
- 16.Huang M, Maynard A, Turpin JA, Graham L, Janini GM, Covell DG, Rice WG. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J. Med. Chem. 1998;41:1371–1381. doi: 10.1021/jm9708543. [DOI] [PubMed] [Google Scholar]
- 17.Adamson CS, Freed EO. Novel approaches to inhibiting HIV-1 replication. Antiviral Res. 2010;85:119–141. doi: 10.1016/j.antiviral.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton AC. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 19.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol. Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House C, Kemp BE. Protein kinase C contains a pseudosubstrate prototype in its regulatory domain. Science. 1987;238:1726–1728. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- 21.Ananthanarayanan B, Stahelin RV, Digman MA, Cho WH. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- 22.Slater SJ, Ho C, Kelly MB, Larkin JD, Taddeo FJ, Yeager MD, Stubbs CD. Protein kinase C alpha contains two activator binding sites that bind phorbol esters and diacylglycerols with opposite affinities. J. Biol. Chem. 1996;271:4627–4631. doi: 10.1074/jbc.271.9.4627. [DOI] [PubMed] [Google Scholar]
- 23.Irie K, Oie K, Nakahara A, Yanai Y, Ohigashi H, Wender PA, Fukuda H, Konishi H, Kikkawa U. Molecular basis for protein kinase C isozyme-selective binding: the synthesis, folding, and phorbol ester binding of the cysteine-rich domains of all protein kinase C isozymes. J. Am. Chem. Soc. 1998;120:9159–9167. [Google Scholar]
- 24.Grishin NV. Treble clef finger--a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Jones DN, Kaine BP, Weiss MA. High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Structure. 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 26.Stewart MD, Morgan B, Massi F, Igumenova TI. Probing the determinants of diacylglycerol binding affinity in the C1B domain of protein kinase Calpha. J. Mol. Biol. 2011;408:949–970. doi: 10.1016/j.jmb.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopalakrishna R, Anderson WB. Ca2+- and phospholipid-independent activation of protein kinase C by selective oxidative modification of the regulatory domain. Proc. Natl. Acad. Sci. U. S. A. 1989;86:6758–6762. doi: 10.1073/pnas.86.17.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knapp LT, Klann E. Superoxide-induced stimulation of protein kinase C via thiol modification and modulation of zinc content. J. Biol. Chem. 2000;275:24136–24145. doi: 10.1074/jbc.M002043200. [DOI] [PubMed] [Google Scholar]
- 29.Korichneva I, Hoyos B, Chua R, Levi E, Hammerling U. Zinc release from protein kinase C as the common event during activation by lipid second messenger or reactive oxygen. J. Biol. Chem. 2002;277:44327–44331. doi: 10.1074/jbc.M205634200. [DOI] [PubMed] [Google Scholar]
- 30.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expression Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales KA, Lasagna M, Gribenko AV, Yoon Y, Reinhart GD, Lee JC, Cho W, Li P, Igumenova TI. Pb2+ as modulator of protein-membrane interactions. J. Am. Chem. Soc. 2011;133:10599–10611. doi: 10.1021/ja2032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales KA, Igumenova TI. Synergistic effect of Pb2+ and phosphatidylinositol 4,5-bisphosphate on C2 domain-membrane interactions. Biochemistry. 2012;51:3349–3360. doi: 10.1021/bi201850h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosolapov A, Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 36.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance 3-dimensional NMR experiments with improved sensitivity. J. Magn. Reson., Ser B. 1994;103:203–216. [Google Scholar]
- 37.Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using C-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J. Am. Chem. Soc. 1992;114:10974–10975. [Google Scholar]
- 38.Schumann FH, Riepl H, Maurer T, Gronwald W, Neidig KP, Kalbitzer HR. Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR. 2007;39:275–289. doi: 10.1007/s10858-007-9197-z. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki T, Forman-Kay JD, Kay LE. 2-Dimensional NMR experiments for correlating C-13-beta and H-1-delta/epsilon chemical shifts of aromatic residues in C-13-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993;115:11054–11055. [Google Scholar]
- 40.Webb H, Tynan-Connolly BM, Lee GM, Farrell D, O'Meara F, Sondergaard CR, Teilum K, Hewage C, McIntosh LP, Nielsen JE. Remeasuring HEWL pK(a) values by NMR spectroscopy: methods, analysis, accuracy, and implications for theoretical pK(a) calculations. Proteins. 2011;79:685–702. doi: 10.1002/prot.22886. [DOI] [PubMed] [Google Scholar]
- 41.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. Characterization of the backbone dynamics of folded and denatured states of an SH3 domain. Biochemistry. 1997;36:2390–2402. doi: 10.1021/bi962548h. [DOI] [PubMed] [Google Scholar]
- 42.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 43.Farrow NA, Zhang OW, Szabo A, Torchia DA, Kay LE. Spectral density-function mapping using N-15 relaxation data exclusively. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 44.Peng JW, Wagner G. Mapping of the spectral densities of N-H bond motions in eglin-c using heteronuclear relaxation experiments. Biochemistry. 1992;31:8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- 45.Peng JW, Wagner G. Mapping of spectral density functions using heteronuclear NMR relaxation measurements. J. Magn. Reson. 1992;98:308–332. [Google Scholar]
- 46.Montelione GT, Wagner G. 2D chemical-exchange NMR spectroscopy by proton-detected heteronuclear correlation. J. Am. Chem. Soc. 1989;111:3096–3098. [Google Scholar]
- 47.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of N-15 longitudinal decay and chemical-exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 48.Palmer AG, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 49.Miloushev VZ, Bahna F, Ciatto C, Ahlsen G, Honig B, Shapiro L, Palmer AG., 3rd Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16:1195–1205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 51.Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- 52.Ishii Y, Markus MA, Tycko R. Controlling residual dipolar couplings in high-resolution NMR of proteins by strain induced alignment in a gel. J. Biomol. NMR. 2001;21:141–151. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 53.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 54.Yang DW, Venters RA, Mueller GA, Choy WY, Kay LE. TROSY-based HNCO pulse sequences for the measurement of (HN)-H-1-N-15, N-15-(CO)-C-13, (HN)-H-1-(CO)-C-13, (CO)-C-13-C-13(alpha) and (HN)-H-1-C-13(alpha) dipolar couplings in N-15, C-13, H-2-labeled proteins. J. Biomol. NMR. 1999;14:333–343. [Google Scholar]
- 55.Yang D, Tolman JR, Goto NK, Kay LE. An HNCO-based pulse scheme for the measurement of 13Calpha-1Halpha one-bond dipolar couplings in 15N, 13C labeled proteins. J. Biomol. NMR. 1998;12:325–332. doi: 10.1023/A:1008223017233. [DOI] [PubMed] [Google Scholar]
- 56.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 57.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 58.Chou JJ, Li SP, Bax A. Study of conformational rearrangement and refinement of structural homology models by the use of heteronuclear dipolar couplings. J. Biomol. NMR. 2000;18:217–227. doi: 10.1023/a:1026563923774. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hommel U, Zurini M, Luyten M. Solution structure of a cysteine-rich domain of rat protein kinase C. Nat. Struct. Biol. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- 63.Zhang GG, Kazanietz MG, Blumberg PM, Hurley JH. Crystal-structure of the Cys2 activator-binding domain of protein kinase C-delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 64.Kazanietz MG, Wang S, Milne GW, Lewin NE, Liu HL, Blumberg PM. Residues in the second cysteine-rich region of protein kinase C delta relevant to phorbol ester binding as revealed by site-directed mutagenesis. J. Biol. Chem. 1995;270:21852–21859. doi: 10.1074/jbc.270.37.21852. [DOI] [PubMed] [Google Scholar]
- 65.Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J. Biol. Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (fHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 67.Schubert M, Poon DK, Wicki J, Tarling CA, Kwan EM, Nielsen JE, Withers SG, McIntosh LP. Probing electrostatic interactions along the reaction pathway of a glycoside hydrolase: histidine characterization by NMR spectroscopy. Biochemistry. 2007;46:7383–7395. doi: 10.1021/bi700249m. [DOI] [PubMed] [Google Scholar]
- 68.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmiedeskamp M, Rajagopal P, Klevit RE. NMR chemical shift perturbation mapping of DNA binding by a zinc-finger domain from the yeast transcription factor ADR1. Protein Sci. 1997;6:1835–1848. doi: 10.1002/pro.5560060904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gooley PR, Johnson BA, Marcy AI, Cuca GC, Salowe SP, Hagmann WK, Esser CK, Springer JP. Secondary structure and zinc ligation of human recombinant short-form stromelysin by multidimensional heteronuclear NMR. Biochemistry. 1993;32:13098–13108. doi: 10.1021/bi00211a020. [DOI] [PubMed] [Google Scholar]
- 71.Urbani A, Bazzo R, Nardi MC, Cicero DO, De Francesco R, Steinkuhler C, Barbato G. The metal binding site of the hepatitis C virus NS3 protease. A spectroscopic investigation. J. Biol. Chem. 1998;273:18760–18769. doi: 10.1074/jbc.273.30.18760. [DOI] [PubMed] [Google Scholar]
- 72.Fabris D, Zaia J, Hathout Y, Fenselau C. Retention of thiol protons in two classes of protein zinc ion coordination centers. J. Am. Chem. Soc. 1996;118:12242–12243. [Google Scholar]
- 73.Larabee JL, Hocker JR, Hanas JS. Cys redox reactions and metal binding of a Cys2His2 zinc finger. Arch. Biochem. Biophys. 2005;434:139–149. doi: 10.1016/j.abb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 74.Dudev T, Lim C. Factors governing the protonation state of cysteines in proteins: an Ab initio/CDM study. J. Am. Chem. Soc. 2002;124:6759–6766. doi: 10.1021/ja012620l. [DOI] [PubMed] [Google Scholar]
- 75.Simonson T, Calimet N. Cys(x)His(y)-Zn2+ interactions: thiol vs. thiolate coordination. Proteins. 2002;49:37–48. doi: 10.1002/prot.10200. [DOI] [PubMed] [Google Scholar]
- 76.Kiefer LL, Fierke CA. Functional characterization of human carbonic anhydrase-II variants with altered zinc-binding sites. Biochemistry. 1994;33:15233–15240. doi: 10.1021/bi00255a003. [DOI] [PubMed] [Google Scholar]
- 77.Parkin G. Synthetic analogues relevant to the structure and function of zinc enzymes. Chem. Rev. 2004;104:699–767. doi: 10.1021/cr0206263. [DOI] [PubMed] [Google Scholar]
- 78.Clore GM, Omichinski JG, Gronenborn AM. Slow conformational dynamics at the metal coordination site of a zinc finger. J. Am. Chem. Soc. 1991;113:4350–4351. [Google Scholar]
- 79.Cornilescu CC, Porter FW, Zhao KQ, Palmenberg AC, Markley JL. NMR structure of the mengovirus leader protein zinc-finger domain. FEBS Lett. 2008;582:896–900. doi: 10.1016/j.febslet.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 80.Hightower KE, Huang CC, Casey PJ, Fierke CA. H-Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate. Biochemistry (Mosc) 1998;37:15555–15562. doi: 10.1021/bi981525v. [DOI] [PubMed] [Google Scholar]
- 81.Bombarda E, Morellet N, Cherradi H, Spiess B, Bouaziz S, Grell E, Roques BP, Mely Y. Determination of the pK(a) of the four Zn2+-coordinating residues of the distal finger motif of the HIV-1 nucleocapsid protein: consequences on the binding of Zn2+ J. Mol. Biol. 2001;310:659–672. doi: 10.1006/jmbi.2001.4770. [DOI] [PubMed] [Google Scholar]
- 82.Seneque O, Latour JM. Coordination properties of zinc finger peptides revisited: ligand competition studies reveal higher affinities for zinc and cobalt. J. Am. Chem. Soc. 2010;132:17760–17774. doi: 10.1021/ja104992h. [DOI] [PubMed] [Google Scholar]
- 83.Reddi AR, Gibney BR. Role of protons in the thermodynamic contribution of a Zn(II)-Cys4 site toward metalloprotein stability. Biochemistry. 2007;46:3745–3758. doi: 10.1021/bi062253w. [DOI] [PubMed] [Google Scholar]
- 84.Bombarda E, Cherradi H, Morellet N, Roques BP, Mely Y. Zn2+ binding properties of single-point mutants of the C-terminal zinc finger of the HIV-1 nucleocapsid protein: evidence of a critical role of cysteine 49 in Zn2+ dissociation. Biochemistry. 2002;41:4312–4320. doi: 10.1021/bi015956g. [DOI] [PubMed] [Google Scholar]
- 85.Maynard AT, Covell DG. Reactivity of zinc finger cores: analysis of protein packing and electrostatic screening. J. Am. Chem. Soc. 2001;123:1047–1058. doi: 10.1021/ja0011616. [DOI] [PubMed] [Google Scholar]
- 86.Lee YM, Lim C. Factors controlling the reactivity of zinc finger cores. J. Am. Chem. Soc. 2011;133:8691–8703. doi: 10.1021/ja202165x. [DOI] [PubMed] [Google Scholar]
- 87.Zhou P, Tian F, Lv F, Shang Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009;76:151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
- 88.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid. Redox. Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.