Abstract
The loss or injury of neurons associated with oxidative and nitrosative redox stress plays an important role in the onset of various neurodegenerative diseases. Specifically, nitric oxide (NO), can affect neuronal survival through a process called S-nitrosylation, by which the NO group undergoes a redox reaction with specific protein thiols. This in turn can lead to the accumulation of misfolded proteins, which generally form aggregates in Alzheimer’s, Parkinson’s, and other neurodegenerative diseases. Evidence suggests that S-nitrosylation can also impair mitochondrial function and lead to excessive fission of mitochondria and consequent bioenergetic compromise via effects on the activity of the fission protein dynamin-related protein 1 (Drp1). This insult leads to synaptic dysfunction and loss. Additionally, high levels of NO can S-nitrosylate a number of aberrant targets involved in neuronal survival pathways, including the antiapoptotic protein XIAP, inhibiting its ability to prevent apoptosis.
Introduction
The initial stages of many neurodegenerative diseases are characterized by injury and synaptic damage, followed by neuronal loss in specific regions of the brain. For example, the hippocampus and pars compacta of the substantia nigra are the most susceptible areas to undergo neurodegeneration in Alzheimer’s disease (AD) and Parkinson’s disease (PD), respectively. However, with disease progression, additional regions of the brain exhibit massive neuronal loss and thus undergo severe degeneration. During the injury process in a variety of neurological disorders, neurons manifest several common pathological features, including increased oxidative and nitrosative stress, mitochondrial dysfunction, activation of cell death pathways, and protein aggregation.
Although the mechanisms underlying the onset of these diseases remain largely unknown, the idea that oxidative and nitrosative stress plays a central role in the development of neurodegeneration has emerged as an attractive theory because production of toxic free radicals rises with aging, and the greatest risk factor for the onset of neurodegenerative diseases is aging. In fact, excessive generation of reactive oxygen. species (ROS), such as superoxide anion (O2·−), as well as reactive nitrogen species (RNS), including nitric oxide (NO·), can contribute to neuronal cell injury and death in experimental models of neurodegenerative diseases (Beal, 2001; Lipton, 2006; Lipton and Rosenberg, 1994). Interestingly, increasing evidence suggests that reaction of an NO group with critical cysteine thiols of target proteins results in the formation of S-nitrosoproteins (SNO-Ps) and can thus regulate protein function and hence neuronal survival (Lipton et al., 1993). Analogous to phosphorylation, this reaction was termed “S-nitrosylation,” indicating a biological effect of the chemical reaction of S-nitrosation. S-Nitrosylation can mediate either protective or neurotoxic effects depending on the action of the target protein that is affected.
In this review, we summarize recent studies on NO-mediated stress in neurodegenerative conditions and specifically attempt to delineate how activation of S-nitrosylation signaling pathways affects pathologic symptoms, particularly protein misfolding and mitochondrial function. Of note, sporadic forms of neurodegenerative diseases, rather than familial forms due to a single gene mutation, constitute the vast majority of human cases. Therefore, a secondary theme of this review is that protein misfolding, as well as other pathological changes resulting from posttranslational modifications to proteins engendered by redox stress, can mimic the rare genetic forms of disease (Zhang and Kaufman, 2006). Specifically, we have demonstrated a critical role for S-nitrosylation of a ubiquitin E3 ligase, parkin, and an endoplasmic reticulum (ER) chaperone, protein-disulfide isomerase (PDI), in accumulation of misfolded proteins in neurodegenerative diseases (Chung et al., 2004; Uehara et al., 2006; Yao et al., 2004). We also review recent findings describing S-nitrosylation of dynamin-related protein 1 (Drp1), which can contribute to the pathological fragmentation of mitochondria, and S-nitrosylation of XIAP, which influences caspase-dependent neuronal cell death. These nitrosylation reactions of specific proteins involved in neurodegenerative conditions serve to illustrate a rapidly developing field that has discovered redox-mediated modifications on many such target proteins.
Generation of ROS/RNS in the nervous system
Using flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), NADPH, and tetrahydrobiopterin as cofactors, NO synthase (NOS) produces NO from L-arginine and oxygen. Three subtypes of NOS have been identified. The two constitutive forms of NOS, neuronal NOS (nNOS or NOS1) and endothelial NOS (eNOS or NOS3), take their names from the cell type in which they were first found. The name of the third subtype, inducible NOS (iNOS or NOS2), indicates that expression of the enzyme is induced by acute inflammatory stimuli. Each NOS isoform contains an oxidase domain at its amino terminal and a reductase domain at its carboxy terminal, separated by a Ca2+/CaM binding site (Abu-Soud and Stuehr, 1993; Boucher et al., 1999; Bredt et al., 1991; Forstermann et al., 1998; Groves and Wang, 2000). Constitutive and inducible NOS are also further distinguished by CaM binding; nNOS and eNOS bind CaM in a reversible Ca2+-dependent manner and are thus activated by Ca2+. In contrast, iNOS binds CaM so tightly at resting intracellular Ca2+ concentrations that its activity does not appear to be affected by transient variations in Ca2+ concentration.
Although all three isoforms are widely distributed in the brain, nNOS provides a predominant source of NO in neurons. nNOS has been thought to be concentrated in the postsynaptic density (PSD) via binding to PSD-95, and binds to N-methyl-D-aspartate-type glutamate receptors (NMDARs) via PDZ binding domains. However, nNOS may also be present in extrasynaptic NMDA receptor complexes (Kim and Sheng, 2004; Sattler et al., 1999; Petralia et al., 2010). NMDA receptors, unlike most other glutamate receptors, are highly permeable to Ca2+. Depolarization of the neuronal cell membrane relieves blockade of NMDA receptor-coupled ion channels by Mg2+ (Mayer et al., 1984). This allows extracellular Ca2+ to enter via the channel into the cytosol, where Ca2+ binds to CaM and activates nNOS to promote NO production. The NMDA receptor/Ca2+ pathway promotes many normal intracellular signaling cascades. For instance, normal excitatory neurotransmission via NMDA receptors is essential for synaptic development and plasticity as well as learning and memory. In contrast, excessive glutamate excitation, particularly of NMDA-type glutamate receptors, plays a role in a variety of neurological disorders ranging from acute hypoxic-ischemic brain injury to chronic neurodegenerative diseases. Excessive Ca2+ influx through NMDA receptors promotes pathological signaling via production of free radicals and related molecules such as NO from nNOS activity as well as ROS from mitochondrial respiratory pathways and other enzymatic processes (see also below for further discussion of neuronal ROS generation) (Abu-Soud and Stuehr, 1993; Bonfoco et al., 1995; Bredt et al., 1991; Budd et al., 2000; Dawson et al., 1991; Lipton et al., 1993; Lipton and Rosenberg, 1994; Sattler et al., 1999). Neuronal damage caused by excessive activation of glutamate receptors is known as “Excitotoxicity” (Olney, 1969), and excessive Ca2+ influx through NMDA receptor-associated ion channels is at least partially responsible for this form of neurotoxicity (Chen and Lipton, 2006; Lipton, 2006; Lipton and Rosenberg, 1994).
NO derived from iNOS also contributes to the pathogenesis of a wide range of neurodegenerative diseases such as AD, PD, Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), and HIV-associated neurocognitive disorder. In these patients, neuroinflamatory and neurodegenerative stimuli, including cytokines, chemokines, viral infection, a secreted derivative of amyloid precursor protein, Parkinsonian toxins (e.g., MPTP), mutant huntingtin proteins, ischemia, and ROS/RNS, can all activate glial cells such as astrocytes and microglia. Such activation results in expression of iNOS and production of high levels of NO (Adamson et al., 1996; Barger and Harmon, 1997; Chen et al., 2000; Fukui and Moraes, 2008; Galea et al., 1992; Liberatore et al., 1999; Saha and Pahan, 2006). Unlike nNOS and eNOS, activation of iNOS is tightly associated with its expression levels. NF-κB and/or AP1-mediated transcription plays an important role in iNOS expression, but iNOS expression can also be controlled at translational and post-translational levels (Fukui and Moraes, 2008). NO from iNOS/glial cells is thought to contribute to neuronal damage by triggering cell death pathways. Alternatively, NO from iNOS can inhibit glutamate reuptake, potentiating excitotoxicity and thus contributing to neuronal death and injury (Li et al., 2009).
In addition to RNS, ROS levels are also elevated in degenerating brains. For instance, superoxide anion (O2·−), primarily produced from mitochondria as a byproduct of cellular respiration, can react with NO to form very toxic peroxynitrite (ONOO−). In contrast, superoxide dismutase (SOD) can convert superoxide radicals into another ROS, hydrogen peroxide, and hydrogen peroxide can react with iron (II) to produce toxic hydroxyl radicals (termed the Fenton reaction) (Fukui and Moraes, 2008). Under physiological conditions, up to 2% of the electrons in the electron transport chain can yield superoxide anion, which can be detoxified by antioxidant systems resident in cells. However, when cells such as neurons are stressed with neurodegeneration promoting insults, mitochondria may produce higher levels of ROS, contributing to pathogenesis. For example, in animal models of PD, administration of complex I inhibitors, such as MPTP (converted to MPP+), rotenone, and paraquat, recapitulates many features of sporadic PD, including degeneration of dopaminergic neurons, overproduction and aggregation of α-synuclein, accumulation of Lewy body-like intraneuronal inclusions, and impairment of behavioral function (Beal, 2001; Betarbet et al., 2000). These pesticides and other environmental toxins specifically inhibit mitochondrial complex I, generating excessive ROS as well as RNS, thus contributing to aberrant protein accumulation, as described below, and neuronal injury (Beal, 2001; Betarbet et al., 2000; Chung et al., 2004; Uehara et al., 2006; Yao et al., 2004). Furthermore, overactivation of NMDA receptors contributes to mitochondrial dysfunction, for example, by Ca2+ overload, producing ROS via the mitochondrial electron transport chain (Lafon-Cazal et al., 1993; Adam-Vizi and Starkov, 2010; Duan et al., 2007; Dugan et al., 1995; Reynolds and Hastings, 1995). Although studies such as these strongly suggest a relationship among ROS/RNS, protein misfolding, and neuronal dysfunction, the mechanism of how ROS/RNS contribute to these pathological features is only now emerging.
Additionally, via an extramitochondrial pathway, NADPH oxidase can produce free radicals, principally superoxide anion, after NMDA receptor activation (Brennan et al., 2009). NADPH oxidase consists of catalytic and regulatory subunits, and is a cytosolic protein expressed in neurons as well as astrocytes and microglia. In neurons, Ca2+ influx via NMDA receptor channels activates several isoforms of protein kinase C (PKC), which triggers assembly of an NADPH oxidase complex, thus producing high levels of superoxide anion from hyperactivated NADPH oxidase. In addition, Aβ or cytokine exposure, via PKC-, MAP kinase- or PI3 kinase-mediated pathways, increases ROS generation via glial NADPH oxidase (Bianca et al., 1999; Simonyi et al., 2010), thus contributing to neuronal damage.
NO/S-nitrosylation signaling pathways in neuronal cells
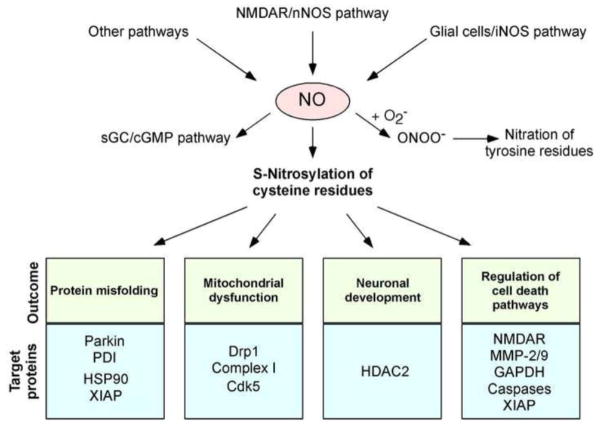
NO participates in cellular signaling pathways that regulate broad aspects of brain function, including synaptic plasticity, normal development, and neuronal cell death (Dawson et al., 1991) (Fig. 1). These effects were thought to be largely achieved by activation of guanylate cyclase to form cyclic guanosine-3′,5′-monophosphate (cGMP), but emerging evidence suggests that a more prominent reaction of NO is S-nitrosylation of regulatory protein thiol groups discussed above (Garthwaite et al., 1988; Isaacs et al., 2006; Lipton et al., 1993). S-Nitrosylation is the covalent addition of an NO group (often in the form of a nitrosonium cation, NO+), to a cysteine thiol/sulfhydryl (or more properly a thiolate anion, RS−) to form an S-nitrosothiol derivative (R-SNO). Such regulatory modifications are broadly found in mammalian, plant, and microbial proteins. As examples, we list a number of proteins found in the brain whose function is affected by S-nitrosylation (Table 1). We and our colleagues have found a consensus motif of nucleophilic residues (generally an acid and a base) surrounding a critical cysteine that facilitates the formation of a thiolate anion and thus increases the susceptibility of the sulfhydryl to S-nitrosylation (Stamler et al., 1997; Hess et al., 2005). These findings suggest that only specific subsets of cysteine residues are susceptible to this type of post-translational modification. Additionally, the recent discovery of protein-protein transnitrosylation reactions (transfer of an NO group from one protein to another) has revealed a mechanism whereby NO on one protein can S-nitrosylate a particular thiol on another specific protein [reviewed in (Anand and Stamler, 2012; Nakamura and Lipton, 2012)]. These S-nitrosylation processes are counterbalanced by denitrosylation via a number of enzymes such as thioredoxin/thioredoxin reductase, class III alcohol dehydrogenase, protein-disulfide isomerase (PDI), intracellular GSH, and other mechanisms.
Fig. 1.
Possible mechanisms whereby NO signaling regulates neuronal function. Hyperactivation of neuronal NMDARs can induce activation of neuronal NO synthase (nNOS) and thus production of NO. Glial cells (astrocytes and microglia) can also generate NO via iNOS expression or ROS-dependent inhibition of astrocytic glutamate uptake, which then activates neuronal NMDARs (Akama and Van Eldik, 2000; Lauderback et al., 1999; Li et al., 2009; Weldon et al., 1998). Additional mechanisms, such as mitochondrial dysfunction, may exist to promote NO production in the nervous system. Endothelial cells in the CNS vascular niche may also produce NO. NO thus generated can trigger formation of S-nitrosylated proteins. NO also activates soluble guanylate cyclase (sGC) to produce cGMP, which can activate cGMP-dependent protein kinase. Peroxynitrite (ONOO−), derived from reaction of NO and superoxide anion (O2−·), can nitrate tyrosine residues to form 3-nitrotyrosine. Physiological levels of NO mediate neuroprotective effects, at least in part, by S-nitrosylating the NMDAR and caspases, thus inhibiting their activity. NO can also promote neuronal development via S-nitrosylation of HDAC2. In contrast, we and others have mounted evidence that overproduction of NO can be neurotoxic via S-nitrosylation of parkin, PDI, GAPDH, MMP-2/9, Cdk5, and Drp1. For instance, S-nitrosylated parkin and PDI contribute to neuronal cell injury by triggering accumulation of misfolded proteins, and S-nitrosylation of Drp1 causes excessive mitochondrial fragmentation and thus synaptic damage in neurodegenerative conditions. Additionally, since HSP-90 is a molecular chaperone and XIAP is a ubiquitin E3 ligase, S-nitrosylation of these proteins may contribute to protein misfolding and accumulation in degenerating neurons.
Table 1.
Representative examples of S-nitrosylated proteins in neurons or brains
| S-Nitrosylated proteins | Effect of S-nitrosylation | Ref. |
|---|---|---|
| Caspases | Decreased protease activity Suppression of cell death |
(Dimmeler et al., 1997; Mannick et al., 1999; Mannick et al., 2001; Tenneti et al., 1997) |
| COX-2 | Activation of activity Mediates NMDA excitotoxicity |
(Tian et al., 2008) |
| Dexras1 | Activation of GTPase Regulation of iron homeostasis |
(Cheah et al., 2006; Fang et al., 2000) |
| Drp1 | Activation of GTPase Excessive mitochondrial fragmentation Increased spine loss and neuronal injury |
(Cho et al., 2009) |
| N-Ethylmaleimide sensitive factor | Enhanced interaction with GluR2 Regulation of exocytosis |
(Huang et al., 2005; Matsushita et al., 2003) |
| GAPDH | Enhanced interaction with Siah1 Activation of p300/CBP Augmentation of cell death |
(Hara et al., 2005; Sen et al., 2008) |
| GOSPEL | Increased interaction with GAPDH Suppression of SNO-GAPDH toxicity |
(Sen et al., 2009) |
| HDAC2 | Releases HDAC2 from chromatin Enhanced dendritic growth |
(Nott et al., 2008) |
| MAP1B | Enhanced interaction with microtubles Axon retraction |
(Stroissnigg et al., 2007) |
| MMP-9 | Activation Augmentation of cell death |
(Gu et al., 2002) |
| NMDAR (NR1 and NR2A) | Inhibition of Suppression of neuronal cell death |
(Choi et al., 2000; Lipton et al., 1993) |
| Parkin | Decrease in E3 ligase activity Augmentation of cell death |
(Chung et al., 2004; Yao et al., 2004) |
| PDI | Decreased activity Accumulation of misfolded proteins Augmentation of cell death |
(Uehara et al., 2006) |
| PrxII | Decreased peroxidase activity Augmentation of cell death |
(Fang et al., 2007; Romero-Puertas et al., 2007) |
| Serine racemase | Inhibition of enzymatic activity Decrease in D-serine levels |
(Mustafa et al., 2007) |
| Stargazin | Increased binding to GluR1 Increased surface expression of AMPAR |
(Selvakumar et al., 2009) |
| TRPC5 | Activation of TRP channels Elicitation of Ca2+ entry |
(Yoshida et al., 2006) |
| XIAP | Decreased E3 ligase activity Augmented cell death |
(Nakamura et al., 2010; Tsang et al., 2009) |
Accumulating evidence suggests that S-nitrosylation is analogous to phosphorylation in regulating the biological activity of many proteins (Chung et al., 2004; Gu et al., 2002; Hara et al., 2005; Hess et al., 2005; Lipton et al., 1993; Stamler et al., 2001; Uehara et al., 2006; Yao et al., 2004). However, the chemistry of NO is much more complex. NO is often a good “leaving group,” resulting in further oxidation of the thiol to a disulfide bond between neighboring (vicinal) cysteine residues. Alternatively, as NO “leaves” because of another reaction partner, the thiol group can react with ROS to yield sulfenic (-SOH), sulfinic (-SO2H) or sulfonic (-SO3H) acid derivatives on the cysteine residue of the protein (Gu et al., 2002; Uehara et al., 2006; Yao et al., 2004). S-Nitrosylation may also produce a nitroxyl disulfide, in which the NO group is shared by proximate cysteine thiols (Houk et al., 2003).
We first identified the physiological relevance of the redox-based mechanisms by which NO and related RNS exert seemingly paradoxical effects in the CNS (Lipton et al., 1993). For example, relatively low concentrations of NO are neuroprotective through S-nitrosylation of NMDA receptors and caspases. Additionally, NO can control neuronal development via S-nitrosylation of HDAC2 (Nott et al., 2008). In contrast, high levels of NO can contribute to neurodestructive events through formation of peroxynitrite or aberrant S-nitrosylation of various proteins, including parkin, PDI, Drp1, Cdk5, XIAP, and many other targets, as discussed below (Gu et al., 2002; Hara et al., 2005; Lipton et al., 1993) (Fig. 1 and Table 1).
At low levels, NO mediates physiological signaling functions, often resulting in neuroprotection. For example, our group first identified the physiological relevance of S-nitrosylation by showing that NO reacts with the NMDA receptor to downregulate excessive activity, thus providing neuroprotective effects under excitotoxic conditions (Lipton et al., 1993). Subsequently, we found that five different cysteine residues on extracellular domains of the NMDA receptor could react with NO. One of these, located at cysteine residue #399 (Cys399) on the NR2A subunit of the NMDA receptor, mediates the predominant effect of NO under our experimental conditions (Choi et al., 2000). As deduced from recently solved crystal structures and further electrophysiological experiments, we found that NO binding to the NMDA receptor at Cys399 may induce a conformational change in the receptor protein that makes glutamate and Zn2+ bind more tightly to the receptor. The enhanced binding of glutamate and Zn2+ in turn causes the receptor to desensitize and, consequently, the ion channel to close (Lipton et al., 2002; Sobolevsky et al., 2009). As opposed to ambient air with an oxygen tension of 150 torr, a pO2 of 10–20 torr is found in normal brain, and even lower levels occur under hypoxic/ischemic conditions. We recently found that as the oxygen tension is lowered the NMDA receptor becomes more sensitive to inhibition by S-nitrosylation (Takahashi et al., 2007).
Additionally, NO can inhibit apoptotic neuronal cell death via S-nitrosylation of caspases. Caspases belong to a family of cysteine proteases, and many of them are involved in the initiation or execution of apoptosis. In fact, excitotoxic injury can be in part mediated by caspases (Budd et al., 2000; Tenneti et al., 1997). NO is known to S-nitrosylate the catalytic cysteine of most or all caspases (Dimmeler et al., 1997; Tenneti et al., 1997; Kim et al., 1997; Mannick et al., 1999; Mannick et al., 2001), inhibiting their protease activities. All caspases are initially expressed in cells as catalytically inactive zymogens (pro-forms) and undergo proteolytic activation to form active enzymes (cleaved forms) during apoptosis. Mannick et al. reported that the pro-form of caspase-3 is constitutively S-nitrosylated; however, following a variety of cell death stimuli, selective denitrosylation of the cleaved form of caspase-3 generates catalytically active enzyme (Mannick et al., 1999). Interestingly, recent findings from Stamler’s group suggest that thioredoxins can mediate denitrosylation of caspase-3, contributing to caspase-3 activation and cell death (Benhar et al., 2008). Moreover, we recently discovered that S-nitrosylated caspase-3 transnitrosylates (transfers its NO group to) XIAP, a potent antagonist of caspase activity, thus compromising XIAP’s neuroprotective function and promoting neuronal cell injury and death (Nakamura et al., 2010). These pathways are discussed in more detail in a subsequent section of this review.
In contrast, we and colleagues have also studied the consequence of excessive (pathophysiological) generation of nitrosative reaction products. For example, recent evidence suggests that the presence of excessive NO-related species may play a significant role in the process of protein misfolding. Increased nitrosative and oxidative stress are associated with chaperone and proteasomal dysfunction, resulting in accumulation of misfolded aggregates (Isaacs et al., 2006; Zhang and Kaufman, 2006). However, until recently little was known regarding the molecular and pathogenic mechanisms underlying contributions of NO to the formation of aggregates, such as amyloid plaques in AD or Lewy bodies in PD. We and others recently presented physiological and chemical evidence that S-nitrosylation modulates the ubiquitin E3 ligase activity of parkin (Chung et al., 2004; Lipton et al., 2005; Yao et al., 2004). Additionally, we found that S-nitrosylation regulates the chaperone and isomerase activities of PDI (Uehara et al., 2006), contributing to protein misfolding and neurotoxicity in models of neurodegenerative disorders. Moreover, S-nitrosylation contributes to mitochondrial dysfunction and caspase-dependent neuronal cell death via formation of SNO-Drp1 and SNO-XIAP, respectively (see below for further discussion on SNO-Parkin, SNO-PDI, SNO-Drp1, and SNO-XIAP).
Protein misfolding and aggregation in neurodegenerative diseases
Misfolded proteins form aggregates in many neurodegenerative diseases, and soluble oligomers of these aberrantly folded proteins are thought to adversely affect cell function by interfering with normal cellular processes or initiating cell death signaling pathways (Muchowski and Wacker, 2005). As examples, α-synuclein and synphilin-1 often aggregate to form Lewy bodies in PD brains. Additionally, degenerating AD brains contain aberrant accumulations of misfolded, aggregated proteins – amyloid-β peptide (Aβ) and tau protein. These aggregates are recognized as either intracellular neurofibrillary tangles, which contain hyperphosphorylated tau, or extracellular plaques, which contain Aβ. β-Secretase and γ-secretase proteolytically cleave amyloid precursor protein (APP) in its transmembrane region to generate Aβ. Historically, misfolded proteins that form large aggregates were first considered to be pathogenic. However, recent evidence has suggested that macroscopic aggregates are in general an attempt by the cell to wall off these aberrant proteins, while soluble (micro-) oligomers of such proteins are the most toxic forms (although larger aggregates could potentially be toxic by location or if not contained by the proper chaperones) (Arrasate et al., 2004). Protein aggregation is also a signature of HD (a polyQ disorder), ALS, and prion disease (Ciechanover and Brundin, 2003).
Furthermore, Eissa’s group reported that microglia can redistribute iNOS to an aggresome, and that this may in part support the hypothesis that protein aggregation mediates a physiological mechanism (Kolodziejska et al., 2005; Pandit et al., 2009). This group found that sequestration of iNOS to aggresomes inactivates iNOS, thus preventing overproduction of neurotoxic NO and providing neuroprotection. CHIP and HDAC6 appeared to regulate iNOS translocation into the inclusion bodies (Sha et al., 2009). In addition, a portion of cellular nNOS may be present in aggresomes (Corso-Diaz and Krukoff). Although the molecular mechanism responsible for the inhibition of NOS activity by aggresomes remains incompletely understood, aggresome-mediated inactivation of NOS may represent a protective aspect of inclusion bodies.
In contrast to diseased neurons, healthy neurons generally show little or no accumulation of protein aggregates, indicating that the appearance of such structures is a response to pathological stress. Considerable evidence suggests that misfolded or otherwise abnormal proteins are produced even in healthy cells and yet do not form macroscopic aggregates. The difference can largely be accounted for by cellular mechanisms for quality control, such as molecular chaperones, the ubiquitin-proteasome system (UPS), and autophagy/lysosomal degradation. A reduction in molecular chaperone or proteasome activities under pathological conditions can result in deposition and accumulation of aberrant proteins either within or outside of cells in the brain. Several mutations in molecular chaperones or UPS-associated enzymes are known to contribute to neurodegeneration (Cookson, 2005; Muchowski and Wacker, 2005). For example, a reduction in proteasome activity was found in the substantia nigra of PD patients (McNaught et al., 2004), and overexpression of the molecular chaperone HSP70 prevented neurodegeneration in vivo in models of PD (Auluck et al., 2002).
S-Nitrosylation of Parkin and the UPS
Formation of polyubiquitin chains on a peptide constitutes the signal for proteasomal degradation. The cascade of activation (E1), conjugation (E2), and ubiquitin-ligase (E3)-type enzymes catalyzes conjugation of the ubiquitin chain to the proteins marked for degradation. Individual E3 ubiquitin ligases play a key role in the recognition of specific peptide substrates (Ross and Pickart, 2004).
Parkin is a member of a large family of E3 ubiquitin ligases. Parkin contains a total of 35 cysteine residues, many of which coordinate structurally important zinc atoms, which are often involved in catalysis (Marin and Ferrus, 2002). Parkin recruits for ubiquitination a substrate protein as well as one of several E2 enzymes (e.g., UbcH7, UbcH8, or UbcH13). Interestingly, mutations in the gene encoding parkin have been associated with Autosomal Recessive Juvenile PD and some even more rare forms of adult-onset PD. In general, mutations in parkin do not contribute to Lewy body formation, although there is at least one exception—familial PD patients with the R275W parkin mutation manifest Lewy bodies (Farrer et al., 2001). Mutations in both alleles of the parkin gene will cause dysfunction in its activity, although not all mutations result in loss of parkin E3 ligase activity (Cookson, 2005). Additionally, wild-type parkin can mediate the formation of non-classical and “non-degradative” lysine 63-linked polyubiquitin chains (Lim et al., 2005; Lim et al., 2006). Synphilin-1 (an α-synuclein-interacting protein) is a well-characterized substrate for parkin ubiquitination, and has been reported in Lewy body-like inclusions in cultured cells when co-expressed with α-synuclein. Accumulation of these proteins portends a poor prognosis for the survival of dopaminergic neurons in familial PD and possibly also in sporadic forms of the disease. Furthermore, recent studies have suggested that parkin can interact with PINK1 depending on mitochondrial membrane potential. The gene encoding PINK1, a mitochondrial kinase, is mutated in some forms of familial PD. Parkin functions with PINK1 to regulate mitochondrial trafficking and to remove damaged mitochondria by mitophagy (Jin et al., 2010; Wang et al., 2011).
Emerging evidence from our laboratory and others suggests that nitrosative/oxidative stress acts as a potential causal factor for protein misfolding in sporadic forms of PD. Specifically, we and others discovered that S-nitrosylation and further oxidation of parkin result in a dysfunctional E3 ligase activity and disruption of UPS function (Chung et al., 2004; Yao et al., 2004; Meng et al., 2011). We found that nitrosative stress induces S-nitrosylation of parkin (forming SNO-parkin) in rodent models of PD. Similarly, we found increased levels of SNO-parkin in brains of human patients with PD and the related α-synucleinopathy, DLBD (diffuse Lewy body disease). Initially, S-nitrosylation of parkin stimulates ubiquitin E3 ligase activity, which may contribute to Lewy body formation. Subsequently, with time we found that the E3 ligase activity of SNO-parkin decreases, resulting in UPS dysfunction (Yao et al., 2004; Lipton et al., 2005). Importantly, S-nitrosylation of parkin on critical cysteine residues can also compromise its neuroprotective activity (Chung et al., 2004).
S-Nitrosylation of PDI mediates protein misfolding and neurotoxicity
In the ER, PDI facilitates proper protein folding by introducing disulfide bonds into proteins (oxidation), breaking disulfide bonds (reduction), and catalyzing thiol/disulfide exchange (isomerization), thus facilitating disulfide bond formation, rearrangement reactions, and protein structural stability (Lyles and Gilbert, 1991). Additionally, several mammalian PDI homologues, such as ERp57 and PDIp, localize to the ER and may manifest similar functions (Conn et al., 2004). Increased expression of PDIp in neuronal cells under conditions mimicking PD suggests the possible contribution of PDIp to neuronal survival (Conn et al., 2004). In many neurodegenerative disorders and cerebral ischemia, the accumulation of immature and denatured proteins results in ER dysfunction (Conn et al., 2004), but upregulation of PDI represents an adaptive response promoting protein refolding and may offer neuroprotection (Conn et al., 2004).
Excessive NO is known to create ER stress at least in part by disruption of Ca2+ homeostasis. One possible mechanism is the increased activity of the ER Ca2+ channel-ryanodine receptor through S-nitrosylation (Xu et al., 1998). Interestingly, we have recently reported that excessive NO, as well as generation of NO after exposure to rotenone, a pesticide known to contribute to the pathogenesis of PD, can lead to S-nitrosylation of the active-site thiols of PDI (to form SNO-PDI), thus inhibiting its isomerase and chaperone activities (Uehara et al., 2006). S-Nitrosylation of PDI prevented its attenuation of neuronal cell death triggered by ER stress, misfolded proteins, or proteasome inhibition. Also, we found SNO-PDI in the brains of virtually all cases sporadic AD and PD that we examined. These results suggest that SNO-PDI may participate in protein misfolding and consequent neuronal cell injury or death.
Recently, formation of SNO-PDI and other S-nitrosothiols was also reported in the spinal cord of an ALS transgenic mouse model, (Schonhoff et al., 2006; Walker et al., 2009). This mouse model involves mutation in Cu/Zn superoxide dismutase (mtSOD1). Mutations in this enzyme are known to cause some forms of familial ALS, possibly in part by producing inclusion bodies that contain misfolded mtSOD1 as seen at postmortem examination (Arnesano et al., 2004; Doucette et al., 2004; Furukawa and O’Halloran, 2005; Rakhit et al., 2004; Tiwari and Hayward, 2003). Recent studies have shown that inhibition of PDI activity with bacitracin can increase aggregation of mtSOD1 in neuronal cells, and that regulation of endogenous PDI activity by reticulons protects against neurodegeneration (Atkin et al., 2006; Yang et al., 2009). In contrast, overexpression of PDI decreases mtSOD1 aggregation and mtSOD1-induced neuronal cell death. Taken together, these findings suggest that increased PDI activity reduces mtSOD1 aggregation and promotes neuronal survival, but SNO-PDI fails to protect motoneurons from protein misfolding toxicity (Atkin et al., 2006; Walker et al., 2009; Yang et al., 2009). Additionally, sporadic ALS patients were reported to manifest increased SNO-PDI levels, suggesting that S-nitrosylation of PDI may contribute to the pathogenesis of sporadic ALS (Walker et al., 2009). It will be important to determine whether SNO-PDI is also involved in protein aggregation and motoneuron injury in ALS in the absence of SOD1 mutations.
The activity of the UPS and molecular chaperones normally declines with age (Paz Gavilan et al., 2006). Since we and others have not found detectable levels of SNO-parkin or SNO-PDI in normal aged brain but only in disease states (Chung et al., 2004; Yao et al., 2004; Uehara et al., 2006), it is likely that S-nitrosylation of these and similar proteins is a contributing mechanism to protein misfolding in neurodegenerative conditions. Collectively, these studies raise the possibility that SNO-parkin, SNO-PDI, and S-nitrosylation of other chaperone molecules may represent potential therapeutic targets to prevent protein aggregation in a number of neurodegenerative diseases.
S-Nitrosylation of other molecular chaperones
In addition to PDI, S-nitrosylation is likely to affect critical thiol groups on other chaperones, such as HSP90 in the cytoplasm (Martinez-Ruiz et al., 2005) and possibly GRP78/GRP94 in the ER (Dall’Agnol et al., 2006). Normally, HSP90 stabilizes misfolded proteins and modulates the activity of cell signaling proteins, including NOS and calcineurin (Muchowski and Wacker, 2005). In AD brains, levels of HSP90 are increased in both the cytosolic and membranous fractions, where HSP90 is thought to maintain tau and Aβ in soluble conformations, thereby averting their aggregation (Dou et al., 2003; Kakimura et al., 2002). Martínez-Ruiz et al. (2005) recently demonstrated that S-nitrosylation of HSP90 can occur in endothelial cells, and this modification abolishes its ATPase activity, which is required for its function as a molecular chaperone. These studies raise the possibility that S-nitrosylation of neuronal HSP90 in AD brains may contribute to the accumulation of tau and Aβ aggregates.
In analogy to HSP90 in the cytoplasm, GRP78/Bip is arguably the best-characterized of the ER chaperones. Using energy from ATP hydrolysis, GRP78 facilitates translocation of nascent proteins into the ER lumen, proper folding of substrate proteins, and degradation of misfolded proteins via ERAD. Recent evidence has demonstrated that the physiological activity of GRP78 is particularly important for neuronal survival. For example, specific deletion of the grp78 gene from Purkinje cells induced apoptotic cell death (Wang et al., 2009). Using a proteomic approach, Dall’Agnol et al. (2006) found that Grp78 can be S-nitrosylated in colonic epithelial cells. It will be important to test whether or not S-nitrosylation of Grp78 regulates NO-mediated protein misfolding and neurotoxocity in neurodegenerative disorders.
Potential contribution of protein nitration to protein misfolding
NO also rapidly reacts with superoxide anion generated from both mitochondrial and non-mitochondrial sources (e.g., NADPH oxidase) to form the very toxic product peroxynitrite (ONOO−) (Beckman et al., 1990; Lipton et al., 1993). Peroxynitrite can precipitate disulfide bond formation between cysteine residues or another protein post-translational modification, i.e., nitration of tyrosine residues, which may also potentially contribute to dysfunctional protein folding and neuronal cell injury. For instance, nitration of α-synuclein and tau affects oligomer formation in vitro. Administration of nitrated α-synuclein into rat brain results in loss of dopaminergic neurons, suggesting that nitrated α-synuclein may play a role in the pathogenesis of PD (Yu et al., 2010). Moreover, it has been reported that nitrated α-synuclein and tau accumulate in inclusion bodies in PD and in neurofibrillary tangles in AD brains, respectively (Giasson et al., 2000; Reynolds et al., 2007; Reynolds et al., 2006; Uversky et al., 2005). Collectively, these findings support the proposition that S-nitrosylation of cysteine residues and possibly nitration of tyrosine residues can influence protein aggregation and neurotoxicity.
S-Nitrosylation of Drp1 mediates mitochondrial fission and synaptic loss
NO has also been reported to regulate mitochondrial function via several pathways. Under physiological conditions, the NO-cGMP pathway, through peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), induces mitochondrial biogenesis, an event comprised of fission events that produce new mitochondria (Nisoli et al., 2003). In contrast, increased nitrosative stress can result in defects in mitochondrial function. For example, NO/S-nitrosylation affects mitochondrial respiration by reversibly inhibiting complexes I and IV (Borutaite and Brown, 2006; Burwell et al., 2006; Chinta and Andersen, 2006; Chinta et al., 2007; Cleeter et al., 1994; Clementi et al., 1998; Dahm et al., 2006; Hsu et al., 2005). In turn, this effect could contribute to brain aging and pathological conditions associated with neurodegenerative diseases.
Additionally, increased nitrosative stress can elicit dysfunction of mitochondrial dynamics (comprised of normal fission and fusion events) (Barsoum et al., 2006; Bossy-Wetzel and Lipton, 2003; Yuan et al., 2007). The fission/fusion machinery proteins are known to maintain mitochondrial integrity, insuring the continued production of ATP at critical locations within neurons. Although the exact mechanism whereby NO contributes to excessive fragmentation of mitochondria remains incompletely understood, our recent findings have shed light on the molecular events underlying this relationship, particularly in AD. Specifically, we have recently discovered histological and chemical evidence that S-nitrosylation can hyperactivate the mitochondrial fission protein Drp1 (dynamin related protein 1). Overstimulation of this GTPase results in excessive mitochondrial fission/fragmentation. We also found that this degree of mitochondrial fragmentation results in bioenergetic impairment, synaptic damage, and eventually frank neuronal loss in models of AD (Cho et al., 2009).
We found that NO results in S-nitrosylation of Drp1 at Cys644 (Cho et al., 2009). Cys644 resides within the GED domain of Drp1, which influences both GTPase activity and oligomer formation of Drp1 (Low and Lowe, 2006; Pitts et al., 2004; Ramachandran et al., 2007; Zhu et al., 2004). S-Nitrosylation of Drp1 (forming SNO-Drp1) induces formation of Drp1 dimers, which contribute to tetramers and higher order structures of Drp1, and activates Drp1 GTPase activity. We also found that exposure to oligomeric Aβ peptide results in formation of SNO-Drp1 in cell culture models. Moreover, we and others have observed that Drp1 is S-nitrosylated in the brains of virtually all cases of sporadic AD (Cho et al., 2009; Wang et al., 2009). In order to determine the consequences of S-nitrosylation of Drp1 in neurons, we exposed cultured cerebrocortical neurons to the physiological NO donor, S-nitrosocysteine (SNOC), or to Aβ oligomers and found that both induced SNO-Drp1 formation and led to the accumulation of excessively fragmented mitochondria. Moreover, mutation of a specific cysteine residue in Drp1 (C644A) prevented these effects of SNOC or Aβ on mitochondrial fragmentation, consistent with the notion that SNO-Drp1 triggered excessive mitochondria fission or fragmentation. Finally, in response to Aβ, SNO-Drp1—induced mitochondrial fragmentation resulted in synaptic damage and loss, an early characteristic feature of AD; eventually this insult led to apoptotic neuronal cell death. Importantly, blockade of Drp1 nitrosylation (using the Drp1(C644A) mutant) prevented Aβ-mediated synaptic loss and neuronal cell death, implying causality of the nitrosylation event with synaptic damage. Moreover, electron-microscopic studies have described an increase in mitochondrial fragmentation in human AD brains, supporting our notion that NO-mediated excessive mitochondrial fragmentation may play a role in the pathogenesis of AD (Baloyannis, 2006; Hirai et al., 2001; Wang et al., 2009). Taken together, these studies suggest that SNO-Drp1 may represent a potential therapeutic target to protect neurons and their synapses in AD.
Transnitrosylation of XIAP regulates caspase-dependent cell death
The caspase family of cysteine proteases often mediates neuronal apoptotic cell death via induction of proteolysis. Caspases participate in the degradation of a variety of neuronal proteins, including APP, presenilins, and synaptic proteins, which may contribute to synaptic dysfunction and neuronal cell death (Ankarcrona et al., 1995; Bonfoco et al., 1995; Chan and Mattson, 1999; Friedlander, 2003; Lu et al., 2000; Mattson, 2000). In contrast, inhibitor of apoptosis proteins (IAPs) represent important regulators of apoptosis through their ability to repress the catalytic activity of caspases-3/7/9 (Eckelman et al., 2006; Salvesen and Duckett, 2002). XIAP can directly bind and inhibit the catalytic activity of these apoptotic caspases (Fuentes-Prior and Salvesen, 2004). Additionally, the RING domain of XIAP can act as an E3 ligase, functioning in ubiquitination and subsequent degradation of heterologous substrates that include caspases, other IAP proteins, and XIAP itself (MacFarlane et al., 2002; Schile et al., 2008; Suzuki et al., 2001; Vaux and Silke, 2005; Yang et al., 2000).
Recently, we discovered that pathophysiological levels of NO can S-nitrosylate the RING domain of XIAP (Nakamura et al., 2010), resulting in a small but definitive conformational change to the RING structure, thus decreasing E3 ubiquitin ligase activity and preventing XIAP from inhibiting apoptosis by degrading caspases. Using an advanced mass spectrometry technique, we found that Cys450 in the RING domain of XIAP is the target of NO. Critically, mutation of Cys450 in XIAP prevented accumulation of caspase-3 and enhanced neuronal survival. This finding is consistent with the notion that S-nitrosylated XIAP (or SNO-XIAP) mediates apoptotic cell death at least in part by inhibition of XIAP E3 ligase-mediated degradation of capsase-3, resulting in relatively increased levels of caspase-3. We further found that SNO-XIAP accumulates in neurons stimulated with neurotoxic levels of NMDA and in the brains of patients undergoing neurodegeneration in AD and PD. Thus, these results indicate that SNO-XIAP regulates caspase activity and contributes to neuronal injury or death in a number of neurodegenerative diseases.
Additionally, we discovered a novel mechanism whereby S-nitrosylated caspase-3 transnitrosylates XIAP to produce SNO-free/active caspase-3 and SNO-XIAP. We developed evidence that the transnitrosylation reaction from caspase-3 to XIAP would proceed under in vivo conditions by calculating the relative redox potential (ΔE°′) using a modification of the Nernst equation and calculating the associated change in Gibbs free energy (ΔG°′). XIAP is known to directly bind only to the cleaved form of caspase-3, not to the pro-form. We found that XIAP received an NO group from the cleaved form of caspase-3 but not from the proform, suggesting that the direct interaction between XIAP and active caspase-3 is required for transnitrosylation. This result is also consistent with previous findings that only cleaved caspases are selectively de-nitrosylated in apoptotic cells (Mannick et al., 1999). Thus, transnitrosylation of XIAP by caspases provides an additional mechanism for proapoptotic signaling. In general, we feel that the discovery of additional transnitrosylation reactions among various proteins will emerge as a central theme of cell regulation, and this may well represent the predominant form of nitrosylase enzyme activity in cells. For example, we recently also found that the kinase Cdk5 has dual enzyme activity, not only acting to phosphorylate substrates but also functioning as a nitrosylase to transnitrosylate specific proteins. In our case, we found that Cdk5 could be S-nitrosylated itself and then transfer its NO group to Drp1, resulting in hyperactivation of Drp1, mitochondrial fragmentation, and synaptic damage, as described above (Qu et al., 2011)
Conclusions
Excessive NMDAR activation and/or mitochondrial dysfunction, resulting in nitrosative and oxidative stress, may potentially contribute to the sporadic form of neurodegenerative diseases. These pathological processes can result in dysfunction of the UPS and molecular chaperones, thus contributing to abnormal protein accumulation and neuronal damage. In this review, we describe a mechanistic link between free radical production, protein misfolding, abnormal mitochondrial dynamics, and neuronal synaptic and cell injury in neurodegenerative disorders such as PD and AD. Although numerous studies support the importance of nitrosative stress in age-associated neurodegenerative diseases, the underlying mechanism(s) are only now emerging in in vivo models. Along these lines, our elucidation of NO-mediated S-nitrosylation of parkin, PDI, Drp1, Cdk5, and XIAP, among other proteins, in animal models of neurodegenerative diseases may be one such mechanism. This discovery may point the way to new therapeutic targets for drug development to prevent aberrant protein misfolding and excessive mitochondrial fission by targeted disruption of aberrant S-nitrosylation of specific proteins.
Acknowledgments
We thank S. McKercher and E. Holland for help with preparation of the manuscript. This work was supported in part by NIH grants P01 ES016738, P01 HD29587, R01 EY05477, R01 EY09024, and P30 NS076411, the American Parkinson’s Disease Association, San Diego Chapter, and an Ellison Senior Scholars Award in Aging (to S.A.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Soud HM, Stuehr DJ. Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc Natl Acad Sci USA. 1993;90:10769–10772. doi: 10.1073/pnas.90.22.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson DC, Wildemann B, Sasaki M, Glass JD, McArthur JC, Christov VI, Dawson TM, Dawson VL. Immunologic NO synthase: elevation in severe AIDS dementia and induction by HIV-1 gp41. Science. 1996;274:1917–1921. doi: 10.1126/science.274.5294.1917. [DOI] [PubMed] [Google Scholar]
- Adam-Vizi V, Starkov AA. Calcium and mitochondrial reactive oxygen species generation: how to read the facts. J Alzheimers Dis. 2010;20(Suppl 2):S413–426. doi: 10.3233/JAD-2010-100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama KT, Van Eldik LJ. Beta-amyloid stimulation of inducible nitric-oxide synthase in astrocytes is interleukin-1beta- and tumor necrosis factor-alpha (TNFalpha)-dependent, and involves a TNFalpha receptor-associated factor- and NFkappaB-inducing kinase-dependent signaling mechanism. J Biol Chem. 2000;275:7918–7924. doi: 10.1074/jbc.275.11.7918. [DOI] [PubMed] [Google Scholar]
- Anand P, Stamler JS. Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J Mol Med. 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankarcrona M, Dypbukt JM, Bonfoco E, Zhivotovsky B, Orrenius S, Lipton SA, Nicotera P. Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. Neuron. 1995;15:961–973. doi: 10.1016/0896-6273(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Arnesano F, Banci L, Bertini I, Martinelli M, Furukawa Y, O’Halloran TV. The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, Horne MK. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- Barger SW, Harmon AD. Microglial activation by Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature. 1997;388:878–881. doi: 10.1038/42257. [DOI] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Experimental models of Parkinson’s disease. Nat Rev Neurosci. 2001;2:325–334. doi: 10.1038/35072550. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Bianca VD, Dusi S, Bianchini E, Dal Pra I, Rossi F. beta-amyloid activates the O-2 forming NADPH oxidase in microglia, monocytes, and neutrophils. A possible inflammatory mechanism of neuronal damage in Alzheimer’s disease. J Biol Chem. 1999;274:15493–15499. doi: 10.1074/jbc.274.22.15493. [DOI] [PubMed] [Google Scholar]
- Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borutaite V, Brown GC. S-nitrosothiol inhibition of mitochondrial complex I causes a reversible increase in mitochondrial hydrogen peroxide production. Biochim Biophys Acta. 2006;1757:562–566. doi: 10.1016/j.bbabio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Lipton SA. Nitric oxide signaling regulates mitochondrial number and function. Cell Death Differ. 2003;10:757–760. doi: 10.1038/sj.cdd.4401244. [DOI] [PubMed] [Google Scholar]
- Boucher JL, Moali C, Tenu JP. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for L-arginine utilization. Cell Mol Life Sci. 1999;55:1015–1028. doi: 10.1007/s000180050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Brennan AM, Suh SW, Won SJ, Narasimhan P, Kauppinen TM, Lee H, Edling Y, Chan PH, Swanson RA. NADPH oxidase is the primary source of superoxide induced by NMDA receptor activation. Nat Neurosci. 2009;12:857–863. doi: 10.1038/nn.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd SL, Tenneti L, Lishnak T, Lipton SA. Mitochondrial and extramitochondrial apoptotic signaling pathways in cerebrocortical neurons. Proc Natl Acad Sci USA. 2000;97:6161–6166. doi: 10.1073/pnas.100121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mattson MP. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J Neurosci Res. 1999;58:167–190. [PubMed] [Google Scholar]
- Cheah JH, Kim SF, Hester LD, Clancy KW, Patterson SE, 3rd, Papadopoulos V, Snyder SH. NMDA receptor-nitric oxide transmission mediates neuronal iron homeostasis via the GTPase Dexras1. Neuron. 2006;51:431–440. doi: 10.1016/j.neuron.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Lipton SA. The chemical biology of clinically tolerated NMDA receptor antagonists. J Neurochem. 2006;97:1611–1626. doi: 10.1111/j.1471-4159.2006.03991.x. [DOI] [PubMed] [Google Scholar]
- Chen M, Ona VO, Li M, Ferrante RJ, Fink KB, Zhu S, Bian J, Guo L, Farrell LA, Hersch SM, Hobbs W, Vonsattel JP, Cha JH, Friedlander RM. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- Chinta SJ, Andersen JK. Reversible inhibition of mitochondrial complex I activity following chronic dopaminergic glutathione depletion in vitro: implications for Parkinson’s disease. Free Radic Biol Med. 2006;41:1442–1448. doi: 10.1016/j.freeradbiomed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Chinta SJ, Kumar MJ, Hsu M, Rajagopalan S, Kaur D, Rane A, Nicholls DG, Choi J, Andersen JK. Inducible alterations of glutathione levels in adult dopaminergic midbrain neurons result in nigrostriatal degeneration. J Neurosci. 2007;27:13997–14006. doi: 10.1523/JNEUROSCI.3885-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn KJ, Gao W, McKee A, Lan MS, Ullman MD, Eisenhauer PB, Fine RE, Wells JM. Identification of the protein disulfide isomerase family member PDIp in experimental Parkinson’s disease and Lewy body pathology. Brain Res. 2004;1022:164–172. doi: 10.1016/j.brainres.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Cookson MR. The biochemistry of Parkinson’s disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- Corso-Diaz X, Krukoff TL. nNOSalpha and nNOSbeta localization to aggresome-like inclusions is dependent on HSP90 activity. J Neurochem. 114:864–872. doi: 10.1111/j.1471-4159.2010.06813.x. [DOI] [PubMed] [Google Scholar]
- Dahm CC, Moore K, Murphy MP. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J Biol Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- Dall’Agnol M, Bernstein C, Bernstein H, Garewal H, Payne CM. Identification of S-nitrosylated proteins after chronic exposure of colon epithelial cells to deoxycholate. Proteomics. 2006;6:1654–1662. doi: 10.1002/pmic.200500240. [DOI] [PubMed] [Google Scholar]
- Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc Natl Acad Sci USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmeler S, Haendeler J, Nehls M, Zeiher AM. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J Exp Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci USA. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucette PA, Whitson LJ, Cao X, Schirf V, Demeler B, Valentine JS, Hansen JC, Hart PJ. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- Duan Y, Gross RA, Sheu SS. Ca2+-dependent generation of mitochondrial reactive oxygen species serves as a signal for poly(ADP-ribose) polymerase-1 activation during glutamate excitotoxicity. J Physiol. 2007;585:741–758. doi: 10.1113/jphysiol.2007.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan LL, Sensi SL, Canzoniero LM, Handran SD, Rothman SM, Lin TS, Goldberg MP, Choi DW. Mitochondrial production of reactive oxygen species in cortical neurons following exposure to N-methyl-D-aspartate. J Neurosci. 1995;15:6377–6388. doi: 10.1523/JNEUROSCI.15-10-06377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Farrer M, Chan P, Chen R, Tan L, Lincoln S, Hernandez D, Forno L, Gwinn-Hardy K, Petrucelli L, Hussey J, Singleton A, Tanner C, Hardy J, Langston JW. Lewy bodies and parkinsonism in families with parkin mutations. Ann Neurol. 2001;50:293–300. doi: 10.1002/ana.1132. [DOI] [PubMed] [Google Scholar]
- Forstermann U, Boissel JP, Kleinert H. Expressional control of the ‘constitutive’ isoforms of nitric oxide synthase (NOS I and NOS III) FASEB J. 1998;12:773–790. [PubMed] [Google Scholar]
- Friedlander RM. Apoptosis and caspases in neurodegenerative diseases. N Engl J Med. 2003;348:1365–1375. doi: 10.1056/NEJMra022366. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci. 2008;31:251–256. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, O’Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- Galea E, Feinstein DL, Reis DJ. Induction of calcium-independent nitric oxide synthase activity in primary rat glial cultures. Proc Natl Acad Sci USA. 1992;89:10945–10949. doi: 10.1073/pnas.89.22.10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite J, Charles SL, Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- Groves JT, Wang CC. Nitric oxide synthase: models and mechanisms. Curr Opin Chem Biol. 2000;4:687–695. doi: 10.1016/s1367-5931(00)00146-0. [DOI] [PubMed] [Google Scholar]
- Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk KN, Hietbrink BN, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, Stamler JS, Toone EJ. Nitroxyl disulfides, novel intermediates in transnitrosation reactions. J Am Chem Soc. 2003;125:6972–6976. doi: 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- Hsu M, Srinivas B, Kumar J, Subramanian R, Andersen J. Glutathione depletion resulting in selective mitochondrial complex I inhibition in dopaminergic cells is via an NO-mediated pathway not involving peroxynitrite: implications for Parkinson’s disease. J Neurochem. 2005;92:1091–1103. doi: 10.1111/j.1471-4159.2004.02929.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Man HY, Sekine-Aizawa Y, Han Y, Juluri K, Luo H, Cheah J, Lowenstein C, Huganir RL, Snyder SH. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–540. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Isaacs AM, Senn DB, Yuan M, Shine JP, Yankner BA. Acceleration of amyloid beta-peptide aggregation by physiological concentrations of calcium. J Biol Chem. 2006;281:27916–27923. doi: 10.1074/jbc.M602061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimura J, Kitamura Y, Takata K, Umeki M, Suzuki S, Shibagaki K, Taniguchi T, Nomura Y, Gebicke-Haerter PJ, Smith MA, Perry G, Shimohama S. Microglial activation and amyloid-beta clearance induced by exogenous heat-shock proteins. Faseb J. 2002;16:601–603. doi: 10.1096/fj.01-0530fje. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- Kolodziejska KE, Burns AR, Moore RH, Stenoien DL, Eissa NT. Regulation of inducible nitric oxide synthase by aggresome formation. Proc Natl Acad Sci USA. 2005;102:4854–4859. doi: 10.1073/pnas.0500485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Cazal M, Pietri S, Culcasi M, Bockaert J. NMDA-dependent superoxide production and neurotoxicity. Nature. 1993;364:535–537. doi: 10.1038/364535a0. [DOI] [PubMed] [Google Scholar]
- Lauderback CM, Harris-White ME, Wang Y, Pedigo NW, Jr, Carney JM, Butterfield DA. Amyloid beta-peptide inhibits Na+-dependent glutamate uptake. Life Sci. 1999;65:1977–1981. doi: 10.1016/s0024-3205(99)00459-2. [DOI] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberatore GT, Jackson-Lewis V, Vukosavic S, Mandir AS, Vila M, McAuliffe WG, Dawson VL, Dawson TM, Przedborski S. Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat Med. 1999;5:1403–1409. doi: 10.1038/70978. [DOI] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A, Bankston LA. Cysteine regulation of protein function--as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Nakamura T, Yao D, Shi ZQ, Uehara T, Gu Z. Comment on “S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function”. Science. 2005;308:1870. doi: 10.1126/science.1110353. author reply 1870. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- MacFarlane M, Merrison W, Bratton SB, Cohen GM. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J Biol Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I, Ferrus A. Comparative genomics of the RBR family, including the Parkinson’s disease-related gene parkin and the genes of the ariadne subfamily. Mol Biol Evol. 2002;19:2039–2050. doi: 10.1093/oxfordjournals.molbev.a004029. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci USA. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O’Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Perl DP, Brownell AL, Olanow CW. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson’s disease. Ann Neurol. 2004;56:149–162. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- Meng F, Yao D, Shi Y, Kabakoff J, Wu W, Reicher J, Ma Y, Moosmann B, Masliah E, Lipton SA, Gu Z. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegen. 2011;6:34–49. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Kumar M, Selvakumar B, Ho GP, Ehmsen JT, Barrow RK, Amzel LM, Snyder SH. Nitric oxide S-nitrosylates serine racemase, mediating feedback inhibition of D-serine formation. Proc Natl Acad Sci USA. 2007;104:2950–2955. doi: 10.1073/pnas.0611620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S, Salvesen GS, Riek R, Yates JR, 3rd, Lipton SA. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Lipton SA. Emerging Role of Protein-Protein Transnitrosylation in Cell Signaling Pathways. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4703. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Olney JW. Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science. 1969;164:719–721. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- Pandit L, Kolodziejska KE, Zeng S, Eissa NT. The physiologic aggresome mediates cellular inactivation of iNOS. Proc Natl Acad Sci USA. 2009;106:1211–1215. doi: 10.1073/pnas.0810968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz Gavilan M, Vela J, Castano A, Ramos B, del Rio JC, Vitorica J, Ruano D. Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol Aging. 2006;27:973–982. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge1 L, Stephenson FA, Wenthold RJ. Organization of NMDA receptors at extrasynaptic locations. Neuroscience. 2010;167:68–87. doi: 10.1016/j.neuroscience.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts KR, McNiven MA, Yoon Y. Mitochondria-specific function of the dynamin family protein DLP1 is mediated by its C-terminal domains. J Biol Chem. 2004;279:50286–50294. doi: 10.1074/jbc.M405531200. [DOI] [PubMed] [Google Scholar]
- Qu J, Nakamura T, Cao G, Holland EA, McKercher SR, Lipton SA. S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc Natl Acad Sci USA. 2011;108:14330–14335. doi: 10.1073/pnas.1105172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu, Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds MR, Berry RW, Binder LI. Nitration in neurodegeneration: deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- Reynolds MR, Reyes JF, Fu Y, Bigio EH, Guillozet-Bongaarts AL, Berry RW, Binder LI. Tau nitration occurs at tyrosine 29 in the fibrillar lesions of Alzheimer’s disease and other tauopathies. J Neurosci. 2006;26:10636–10645. doi: 10.1523/JNEUROSCI.2143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds IJ, Hastings TG. Glutamate induces the production of reactive oxygen species in cultured forebrain neurons following NMDA receptor activation. J Neurosci. 1995;15:3318–3327. doi: 10.1523/JNEUROSCI.15-05-03318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Matte A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell. 2007;19:4120–4130. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Saha RN, Pahan K. Regulation of inducible nitric oxide synthase gene in glial cells. Antioxid Redox Signal. 2006;8:929–947. doi: 10.1089/ars.2006.8.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284:1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff CM, Matsuoka M, Tummala H, Johnson MA, Estevez AG, Wu R, Kamaid A, Ricart KC, Hashimoto Y, Gaston B, Macdonald TL, Xu Z, Mannick JB. S-nitrosothiol depletion in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:2404–2409. doi: 10.1073/pnas.0507243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar B, Huganir RL, Snyder SH. S-nitrosylation of stargazin regulates surface expression of AMPA-glutamate neurotransmitter receptors. Proc Natl Acad Sci USA. 2009;106:16440–16445. doi: 10.1073/pnas.0908949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, Agrawal N, Hester L, Dore S, Snyder SH, Sawa A. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63:81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y, Pandit L, Zeng S, Eissa NT. A critical role for CHIP in the aggresome pathway. Mol Cell Biol. 2009;29:116–128. doi: 10.1128/MCB.00829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyi A, He Y, Sheng W, Sun AY, Wood WG, Weisman GA, Sun GY. Targeting NADPH oxidase and phospholipases A2 in Alzheimer’s disease. Mol Neurobiol. 2010;41:73–86. doi: 10.1007/s12035-010-8107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Stroissnigg H, Trancikova A, Descovich L, Fuhrmann J, Kutschera W, Kostan J, Meixner A, Nothias F, Propst F. S-nitrosylation of microtubule-associated protein 1B mediates nitric-oxide-induced axon retraction. Nat Cell Biol. 2007;9:1035–1045. doi: 10.1038/ncb1625. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Shin Y, Cho SJ, Zago WM, Nakamura T, Gu Z, Ma Y, Furukawa H, Liddington R, Zhang D, Tong G, Chen HS, Lipton SA. Hypoxia Enhances S-Nitrosylation-Mediated NMDA Receptor Inhibition via a Thiol Oxygen Sensor Motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenneti L, D’Emilia DM, Lipton SA. Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci Lett. 1997;236:139–142. doi: 10.1016/s0304-3940(97)00780-5. [DOI] [PubMed] [Google Scholar]
- Tian J, Kim SF, Hester L, Snyder SH. S-nitrosylation/activation of COX-2 mediates NMDA neurotoxicity. Proc Natl Acad Sci USA. 2008;105:10537–10540. doi: 10.1073/pnas.0804852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Hayward LJ. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J Biol Chem. 2003;278:5984–5992. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM, Chung KK. S-nitrosylation of XIAP compromises neuronal survival in Parkinson’s disease. Proc Natl Acad Sci USA. 2009;106:4900–4905. doi: 10.1073/pnas.0810595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Yamin G, Munishkina LA, Karymov MA, Millett IS, Doniach S, Lyubchenko YL, Fink AL. Effects of nitration on the structure and aggregation of alpha-synuclein. Brain Res Mol Brain Res. 2005;134:84–102. doi: 10.1016/j.molbrainres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD. Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis. Brain. 2009 doi: 10.1093/brain/awp267. [DOI] [PubMed] [Google Scholar]
- Wang M, Ye R, Barron E, Baumeister P, Mao C, Luo S, Fu Y, Luo B, Dubeau L, Hinton DR, Lee AS. Essential role of the unfolded protein response regulator GRP78/BiP in protection from neuronal apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon DT, Rogers SD, Ghilardi JR, Finke MP, Cleary JP, O’Hare E, Esler WP, Maggio JE, Mantyh PW. Fibrillar beta-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J Neurosci. 1998;18:2161–2173. doi: 10.1523/JNEUROSCI.18-06-02161.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yang YS, Harel NY, Strittmatter SM. Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J Neurosci. 2009;29:13850–13859. doi: 10.1523/JNEUROSCI.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Inoue R, Morii T, Takahashi N, Yamamoto S, Hara Y, Tominaga M, Shimizu S, Sato Y, Mori Y. Nitric oxide activates TRP channels by cysteine S-nitrosylation. Nat Chem Biol. 2006;2:596–607. doi: 10.1038/nchembio821. [DOI] [PubMed] [Google Scholar]
- Yu Z, Xu X, Xiang Z, Zhou J, Zhang Z, Hu C, He C. Nitrated alpha-synuclein induces the loss of dopaminergic neurons in the substantia nigra of rats. PLoS One. 2010;5:e9956. doi: 10.1371/journal.pone.0009956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Gerencser AA, Liot G, Lipton SA, Ellisman M, Perkins GA, Bossy-Wetzel E. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–471. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]