Abstract
Understanding the factors that drive the evolution of pathogenic fungi is central to revealing the mechanisms of virulence and host preference, as well as developing effective disease control measures. Prerequisite to these pursuits is the accurate delimitation of species boundaries. Colletotrichum gloeosporioides s.l. is a species complex of plant pathogens and endophytic fungi for which reliable species recognition has only recently become possible through a multi-locus phylogenetic approach. By adopting an intensive regional sampling strategy encompassing multiple hosts within and beyond agricultural zones associated with cranberry (Vaccinium macrocarpon Aiton), we have integrated North America strains of Colletotrichum gloeosporioides s.l. from these habitats into a broader phylogenetic framework. We delimit species on the basis of genealogical concordance phylogenetic species recognition (GCPSR) and quantitatively assess the monophyly of delimited species at each of four nuclear loci and in the combined data set with the genealogical sorting index (gsi). Our analysis resolved two principal lineages within the species complex. Strains isolated from cranberry and sympatric host plants are distributed across both of these lineages and belong to seven distinct species or terminal clades. Strains isolated from V. macrocarpon in commercial cranberry beds belong to four species, three of which are described here as new. Another species, C. rhexiae Ellis & Everh., is epitypified. Intensive regional sampling has revealed a combination of factors, including the host species from which a strain has been isolated, the host organ of origin, and the habitat of the host species, as useful indicators of species identity in the sampled regions. We have identified three broadly distributed temperate species, C. fructivorum, C. rhexiae, and C. nupharicola, that could be useful for understanding the microevolutionary forces that may lead to species divergence in this important complex of endophytes and plant pathogens.
Introduction
Delimiting species boundaries among fungi lays the groundwork for detailing the natural history and ecology of species and defines a robust framework from which further comparative studies can be designed (i.e. population genetics/genomics). This is also prerequisite to providing targeted and effective disease control measures and identifying specific pathogens against which plant breeders can focus their efforts in developing and selecting disease resistant cultivars. Strictly agro–centric studies of plant pathogens risk sampling too narrowly, overlooking important adjacent (parapatric) niches driving pathogen evolution. Extensive sampling, within, adjacent to, and beyond agricultural landscapes has the potential to provide a broader view of the natural history of pathogen species and offer insight into the evolution of differential traits among closely related lineages [1].
Colletotrichum Corda is among the most important and widespread genera of plant-associated fungi, causing disease and occurring as asymptomatic endophytes on aerial organs of a broad range of host plants [2]–[6]. Colletotrichum gloeosporioides sensu lato represents an aggregate of species frequently reported as a dominant endophyte of tropical herbaceous plants and is known as a field and post–harvest fruit pathogen of many economically important crops [7]–[11]. Morphological homoplasy and phenotypic plasticity have previously thwarted efforts to clearly define species boundaries within the species complex, necessary if we are to develop a greater understanding of the ecology and natural history of each lineage. The recent development of molecular markers suitable for resolving species limits and phylogenetic relationships within this species aggregate have been proposed and validated [12]–[16], making it possible to examine the role of geography, host preference/specificity, the nature of host–pathogen/host–endophyte associations, and other niche specialization attributes that may underlie species divergence [14]. Furthermore, the recent epitypification of C. gloeosporioides provides a much needed reference point for developing an improved taxonomic framework for the species complex [17].
The current study uses a phylogenetic approach to infer species boundaries among North American members of the C. gloeosporioides species complex, particularly those in association with the large American cranberry (Vaccinium macrocarpon Aiton) and sympatric plant species. Colletotrichum gloeosporioides s.l. has been reported as a leaf and fruit pathogen of cranberry since the late 1800s [18] and early 1900s [19] and has recently been observed to colonize stem tissue [20]. Contemporary studies of fruit pathogens have confirmed the importance of C. gloeosporioides s.l. in agricultural ecosystems throughout the cultivated range of V. macrocarpon [8], [21]–[23]. However, given the fact that C. gloeosporioides s.l. is an aggregate of species that can be difficult or impossible to distinguish morphologically, it is not clear that the strains isolated from cranberry are conspecific throughout the cultivated range of cranberry. In addition, genetic studies have focused on isolates solely from fruit and have not investigated diversity from alternate host organs or sympatric host species, despite evidence that these represent potential reservoirs of diversity for the species complex in cranberry agricultural areas [19], [24]–[27].
Vaccinium macrocarpon is one of North America’s few native, economically important crop species. Cranberry has undergone little selection from wild relatives and is cultivated in several regions alongside extant native populations [28]. It can also be found in sympatry with the closely related species V. oxycoccos L., which has been used for inter-specific hybridization in breeding programs but is not cultivated for agricultural production [29]. In addition, many of the cultivars used in past breeding regimes remain available for further experimentation. These characteristics provide cranberry breeders with excellent resources for meeting the needs of cranberry growers to improve fruit production and reduce pathogen pressure through breeding for disease resistance [30]. However, providing a refined systematic understanding of the pathogens that are the source of disease pressure will be necessary to help focus the efforts of plant breeders.
Host specificity/preference has historically been a focal criterion for species delimitation in Colletotrichum [7], [11]. Similarly, contemporary studies have indicated the presence of host specific lineages [13], [31]. However, host-fungus associations within Colletotrichum are variable. It is clear that multiple species are capable of infecting single hosts and, conversely, some Colletotrichum species are capable of infecting multiple host species [13], [14], [32]. In addition, there is indication that different plant organs may act as selective forces leading to organ specialization in Colletotrichum species [33]. While C. gloeosporioides s.l. is established as a pathogen of a variety of important crop species worldwide [8], [14], [15], [31], [34]–[38], this study represents a unique perspective into the evolution of the species complex by investigating the genetic diversity of C. gloeosporioides s.l. in a single crop species and its surrounding habitat within the host’s native range. The specific objectives of this study are to determine: (1) whether there are multiple sympatric lineages within the species complex that infect cranberry, 2) whether host preference/specificity is evident among sympatric lineages within the species complex, 3) whether host organ or nature of the fungus-host association is predictive of phylogenetic structure, and 4) whether we can identify lineages with broad geographical and/or host associations suitable for fine-scale landscape genetic analysis. We sampled horizontally across five sympatric host species in wild and commercial cranberry bogs in order to address questions related to host specificity, and sampled vertically among different plant organs within V. macrocarpon to target questions related to organ specialization (Table 1). Utilizing the recent development of molecular phylogenetic markers useful for distinguishing lineages within the species complex [12]–[15] and isolates from five major cranberry agricultural areas, wild cranberry bogs, fruit and stem of V. macrocarpon, and from five sympatric host species, we assess lineage diversity among isolates from cranberry and surrounding habitats.
Table 1. Colletotrichum strains included in the four marker (D4G) phylogenetic analysis.
| Strain | Locality | Host | Organ | Species |
| Coll57 | New Jersey | Vaccinium macrocarpon | Stem | C. aff. acutatum |
| Coll60 | New Jersey | V. macrocarpon | Fruit | C. aff. acutatum |
| CBS124 | New Jersey | V. macrocarpon | NA | C. fructivorum |
| Coll1002 = CBS133128 | British Columbia | V. macrocarpon | Fruit | C. fructivorum |
| Coll1004 | British Columbia | V. macrocarpon | Fruit | C. fructivorum |
| Coll1062 | Massachussets | V. macrocarpon | Fruit | C. fructivorum |
| Coll1081 = CBS133121 | Massachussets | V. macrocarpon | Fruit | C. fructivorum |
| Coll1092* = CBS133135 | New Jersey | V. macrocarpon | Stem | C. fructivorum |
| Coll116 | Massachussets | V. macrocarpon | Fruit | C. fructivorum |
| Coll1164 = CBS133126 | Wisconsin | V. macrocarpon | Fruit | C. fructivorum |
| Coll1178 | Wisconsin | V. macrocarpon | Fruit | C. fructivorum |
| Coll1190 | Wisconsin | V. macrocarpon | Fruit | C. fructivorum |
| Coll1216 | Wisconsin | V. macrocarpon | Fruit | C. fructivorum |
| Coll1414 = CBS133125 | New Jersey | V. macrocarpon | Fruit | C. fructivorum |
| Coll445 = CBS133127 | New Jersey | V. macrocarpon | Fruit | C. fructivorum |
| Coll21 = CBS133133 | New Jersey | V. macrocarpon | Fruit | C. fructivorum |
| Coll864 = CBS133130 | New Jersey | Rhexia virginica | Stem | C. fructivorum |
| Coll873 | Delaware | V. macrocarpon | Fruit | C. fructivorum |
| Coll886 = CBS133124 | Pennsylvania | Vaccinium oxycoccos | Fruit | C. fructivorum |
| Coll1026 = CBS133134 | Delaware | Rhexia virginica | Stem | C. rhexiae |
| Coll1034 = CBS133136 | Delaware | V. macrocarpon | Fruit | C. rhexiae |
| Coll1041 | Delaware | V. macrocarpon | Fruit | C. rhexiae |
| Coll877* = CBS133132 | Delaware | V. macrocarpon | Fruit | C. rhexiae |
| Coll1038 | Delaware | R. virginica | Stem | C. rhexiae |
| Coll1306 | New Jersey | R. virginica | Stem | C. rhexiae |
| Coll952 = CBS133131 | New Jersey | R. virginica | Stem | C. rhexiae |
| Coll1470 = CBS133129 | Maryland | R. virginica | Leaf | C. rhexiae |
| GJS08214 | Cameroon | Coffea arabica | Fruit | C. kahawae |
| GJS08216 | Cameroon | C. arabica | Fruit | C. kahawae |
| GJS08211 | Cameroon | C. arabica | Fruit | C. kahawae |
| IMI319418 | Kenya | C. arabica | Fruit | C. kahawae |
| Coll1103* = CBS133120 | New Jersey | V. macrocarpon | Stem | C. temperatum |
| Coll883 = CBS133122 | New York | V. macrocarpon | Fruit | C. temperatum |
| Coll11* | Florida | Persea americana | Leaf | C. tropicale |
| Coll918 | Puerto Rico | Terpsichore taxifolia | Sporangium | C. tropicale |
| GJS0842 | Panama | Annona muricata | Leaf | C. tropicale |
| 4861 | Panama | T. cacao | Leaf | C. tropicale |
| 5101 * | Panama | T. cacao | Leaf | C. tropicale |
| 8401* | Panama | T. cacao | Leaf | C. tropicale |
| E1164* | Panama | Trichilia tuberculata | Leaf | C. tropicale |
| E2303 | Panama | Virola surinamensis | Leaf | C. tropicale |
| E406 | Panama | Pentagonia macrophylla | Leaf | C. tropicale |
| Q633* | Panama | Cordia alliodora | Leaf | C. tropicale |
| 7423* | Panama | Theobroma cacao | Leaf | C. tropicale |
| Coll126 = CBS133123 | New Jersey | V. macrocarpon | Stem | C. melanocaulon |
| Coll131 = CBS133251 | New Jersey | V. macrocarpon | Stem | C. melanocaulon |
| Coll6 | Florida | Mangifera indica | Leaf | C. siamense |
| 1092 | Panama | Theobroma cacao | Leaf | C. siamense |
| CollNC67 | North Carolina | NA | NA | C. siamense |
| Coll54 | New Jersey | Prunus persica | Fruit | C. sp. indet. B |
| CollNC60 | North Carolina | NA | ? | C. sp. indet. B |
| GJS0852 | Panama | T. cacao | Leaf | C. sp. indet. A |
| 7767 | Panama | T. cacao | Leaf | C. sp. indet. A |
| GJS0857 | Panama | P. americana | Fruit | ND |
| Coll38 | Florida | M. indica | Fruit | C. asianum |
| GJS08144 | Panama | M. indica | Fruit | C. asianum |
| GJS08147 | Panama | M. indica | Fruit | C. asianum |
| Coll1126* | New Jersey | V. macrocarpon | Stem | C. fructicola |
| Coll996 | New Jersey | R. virginica | Stem | C. fructicola |
| CollP1 | North Carolina | V. corymbosum | Leaf | C. fructicola |
| 1087* | Panama | T. cacao | Leaf | C. fructicola |
| 3589* | Panama | T. cacao | Leaf | C. fructicola |
| 3679* | Panama | T. cacao | Leaf | C. fructicola |
| 7574 | Panama | T. cacao | Leaf | C. fructicola |
| E886 | Panama | Tetragastris panamensis | Leaf | C. fructicola |
| Coll878 | New Jersey | Nuphar lutea | Leaf | C. nupharicola |
| cbs470 | Washington | N. lutea ssp. polysepala | Leaf | C. nupharicola |
| Coll922 | Washington | N. lutea | Leaf | C. nupharicola |
| cbs472 | Rhode Island | Nymphaea odorata | Leaf | C. nupharicola |
| Coll919 | Puerto Rico | T. taxifolia | Leaf seta | “C. ignotum 2″ |
| 8395 | Panama | T. cacao | Leaf | “C. ignotum 2″ |
| E183 | Panama | Genipa americana | Leaf | “C. ignotum 2″ |
| Coll940 | Oklahoma | Juglans nigra | Leaf | ND |
| Coll887 | W. Virginia | V. oxycoccos | Fruit | C. sp. indet. C |
| Coll920 | New Jersey | Chamaecyparis thyoides | Stem | C. sp. indet. C |
| Coll20* | Mexico | Solanum sp. | stem | C. gloeosporioides |
| IMI356878 | Italy | Citrus sinsensis | Fruit | C. gloeosporioides |
| Coll914 | Mexico | Solanum sp. | Leaf | C. gloeosporioides |
| GJS0848 | Panama | T. cacao | Fruit | C. theobromicola |
| GJS0843 | Panama | T. cacao | Leaf | C. theobromicola |
| GJS0850 | Panama | T. cacao | Leaf | C. theobromicola |
| 3386 | Panama | T. cacao | Leaf | C. sp. indet. D |
| 4801 | Panama | T. cacao | Leaf | C. sp. indet. D |
| 4766 | Panama | T. cacao | Leaf | C. sp. indet. D |
Accession numbers for cultures deposited at Centraalbureau voor Schimmelcultures (CBS) are included for relevant strains.
Endophyte isolates.
ND – not designated.
NA – not available.
Type strains are in bold-faced type.
Methods
Ethics Statement
All necessary permits were obtained for the described field studies from the Delaware Division of Parks and Recreation, the National Forest Service, and The Nature Conservancy. All other samples collected in this study were on private land and did not require permits. There were no endangered or protected species collected for this study.
Fungal Isolation and Culturing
Colletotrichum was isolated from symptomatic and asymptomatic tissue of several host species in North America with a focus on species sympatric with Vaccinium macrocarpon in wild and agricultural habitats. Sympatric host plant species from which C. gloeosporioides s.l. was isolated include Vaccinium oxycoccos, Rhexia virginica L., Chamaecyparis thyoides (L.) Britton, Sterns, & Poggenb., and Nuphar lutea (L.) Sm. (Table 1). Colletotrichum gloeosporioides s.l. was isolated from symptomatic and asymptomatic stem and fruit of V. macrocarpon (Table 1) in both wild and agricultural habitats. Both symptomatic and asymptomatic tissue was surface sterilized in 10% bleach (final concentration 0.6125% sodium hypochlorite) between 1.5 minutes and 5 minutes, depending on the permeability of the tissue, and plated on V8 juice agar, 2% malt extract agar (MEA: BD Diagnostics, Franklin Lakes, NJ, USA) or corn meal agar (CMA: BD Diagnostics), with the exception of Nuphar lutea. Nuphar lutea was incubated in a humid chamber under ambient light and conidia were transferred from anthracnose lesions to CMA and potato dextrose agar (PDA: BD Diagnostics) for purification. Isolates were characterized as endophytes if they were isolated from asymptomatic tissue after surface sterilization; all other isolates were considered pathogenic. Strains morphologically similar to Colletotrichum gloeosporioides s.l., based on an assortment of characters including growth rate, colony color, hyphal morphology and/or conidial shape and size, were isolated by transferring conidia or hyphal tips to sterile media and preserved on CMA slants stored at 6°C, stored in 1.5 mL microcentrifuge tubes at 6°C, and in 10% glycerol at −80°C. Types, epitypes, and representative cultures were deposited at the Centraalbureau voor Schimmelcultures (CBS) with corresponding dried cultures deposited as vouchers at the U.S. National Fungus Collections (BPI).
DNA Extraction and PCR Amplification
Isolates were grown on potato dextrose broth (Difco) for 5–7 days before mycelium was harvested, blotted dry on sterile paper towels and dehydrated for 6–10 hours in a vacuum centrifuge on low heat. Approximately 20 mg of dried tissue was used for DNA extraction using the DNeasy Plant MiniKit (Qiagen Inc., Valencia, CA, USA) or a standard phenol-chloroform extraction method after tissue homogenization in the FastPrep FP120 (MP Biomedicals, LLC., Solon, OH, USA).
PCR amplification reactions of nuclear ribosomal internal transcribed spacer (nrITS), beta-tubulin (tub2), DNA lyase (apn2), and an intergenic spacer between the 3′ end of the DNA lyase and the mating type locus MAT1-2 (apn2/matIGS) were performed in 25 µl reactions containing 9.3 µL of autoclaved ion-exchanged water, 2.5 µL of dNTP mixture (2.5 mM stock of each dNTP), 2.5 µL of bovine serum albumin (BSA; 0.25 µg µL−1 stock), 2.5 µL of buffer [200 mm Tris pH 8.8, 100 mM KCl, 100 mM (NH4)2SO4, 20 mm MgSO4·7H2O, 1% (v/v) Triton X-100, 50% (w/v) sucrose, 0.25% (w/v) cresol red], 5 µL betaine (1.2 M stock), 1 µL of each primer (0.67 µM final concentration), 0.2 µL of Taq polymerase (GenScript USA Inc., Piscataway, NJ, USA) and 1 µL (approximately 20–50 ng) of genomic DNA. The nrITS region was amplified and sequenced using primers ITS5 and ITS4 [39], tub2 with primers T1 and T224 [40], apn2 with CgDL_R1 and ColDL_F3 [14], and apn2mat/IGS using primers CgDL_F6 and CgMAT1_F2 [14].
PCR reactions were run on an Eppendorf Mastercycler pro S with the following cycling parameters: nrITS–initial denaturing for 2 min at 94°C, followed by 38 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 45 s, followed by a final extension at 72°C for 5 min; tub2– initial denaturing for 2 min at 94°C, followed by 30 cycles of 94°C for 35 s, 52°C for 55 s, and 72°C for 2 min, followed by a final extension at 72°C for 5 min; apn2/matIGS–initial denaturing for 5 min at 95°C, followed by 10 cycles of 95°C for 30 s, 62°C (decreasing by 1°C each cycle) for 30 s, and 72°C for 1 min, followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR products were purified and sequenced at the High Throughput Genomics Unit, Department of Genome Sciences, University of Washington using ABI 3730×l sequencers.
Contig Assembly, Sequence Editing, and Phylogenetic Inference
Sequences were automatically assembled into contigs, and edited manually in Sequencher version 4.9 (GeneCodes Corp., Ann Arbor, Michigan). Alignments were carried out with the online version of the sequence alignment program MAFFT version 6 [41], [42] using the iterative refinement option G-INS-i for each locus independently. GenBank accession numbers for sequences generated for this study are provided in Table S1.
Rojas et al. [14] sampled New World isolates of C. gloeosporioides s.l. representing several lineages within the species complex and demonstrated the utility of a combined analysis of nrITS, btub, apn2, and apn2/matIGS to resolve closely related lineages. Representative sequences from their study of all four markers were combined with sequences generated here to meet the aforementioned objectives. Independent gene trees were inferred under the parsimony (MP) optimality criterion in TNT [43] and maximum likelihood (ML) criterion in RAxML–HPC2 (7.2.8) implemented on the CIPRES Science Gateway portal [44]–[46]. Phylogenetic estimates of the concatenated matrix were inferred under parsimony in TNT, maximum likelihood in RAxML–HPC2 (implemented on the CIPRES cluster), and Bayesian inference in MrBayes v3.1.2 [47], [48]. Phylogenetic analyses in TNT were carried out with parsimony ratchet tree searches (one thousand random addition sequence replicates while holding 20 trees per replicate) and TBR branch swapping. Statistical support for the inferred nodes was determined with parsimony bootstrapping on 1000 pseudoreplicate datasets. Rapid bootstrapping in RAxML was carried out implementing the GTRCAT model and the ML tree search under the GTRGAMMA model (-m GTRCAT -x -f a). The best-fit models for Bayesian analyses were selected with MrModeltest 2.3 [49].
Independent analyses of nrITS and partial beta-tubulin were rooted in TNT and RAxML to Coll57 and Coll60, strains of Colletotrichum aff. acutatum Simmonds, a species complex related to Colletotrichum gloeosporioides s.l [50]–[52]. Strains Coll57 and Coll60 were identified based on morphological similarity and NCBI BLAST similarity searches of nrITS and partial beta-tubulin (nrITS: Glomerella fioriniae –GenBank accession JN121193, e-value 0.0; btub: Glomerella acutata – GenBank accession AB273716, e-value 0.0). Analyses of apn2 and apn2/matIGS were rooted in TNT and RAxML to strain 4766, which exploratory nrITS and tub2 analyses and a previous study of the group [14] indicated is a suitable outgroup for inferring phylogenetic relationships within C. gloeosporioides s.l. Statistical support for the inferred nodes was determined in RAxML–HPC2 by bootstrapping with the number of replicates for the independent gene trees determined by implementation of the extended majority rule (–N autoMRE) convergence criterion implemented in the CIPRES portal and the combined dataset with 1000 pseudoreplicates (–N 1000). Node frequencies for both parsimony and maximum likelihood bootstrap analyses were calculated with the SumTrees 3.1.0 program using the DendroPy Phylogenetic Computing Library version 3.7.1 [53].
The best-fit models implemented in MrBayes for each of nrITS, tub2, apn2 and apn2/matIGS respectively, were as follows: SYM+Γ, GTR+I, GTR+Γ, HKY+ Γ. Four parallel runs were conducted with one cold and three heated Markov chains per run for 10,000,000 generations sampling every 1,000 generations, with each of these models applied independently to each locus in MrBayes. Convergence of parameter estimates was monitored in Tracer v. 1.5 [54] and posterior probabilities calculated from the sampled trees after discarding the first 25% as burn-in. To assess convergence to a global optimum in both ML and Bayesian tree searches of the combined dataset, the log-likelihood scores (−lnL) for the best tree from the maximum likelihood and Bayesian tree searches were calculated in RAxML [45].
In order to estimate the primary concordance tree from the multilocus dataset we used Bayesian concordance analysis (BCA) implemented in BUCKy v. 1.4.0 [55], [56]. BUCKy integrates over gene tree uncertainty to estimate the proportion of sequenced markers that support each clade and constructs the tree that reflects the dominant vertical phylogenetic signal (primary concordance tree) [56]. We limited our analysis with BUCKy to a reduced dataset of 55 unique haplotypes (sequences with missing data were considered unique). The posterior distribution of single gene trees was inferred in MrBayes using the same model parameters for each partition as in the concatenated dataset. The primary concordance tree and concordance factors (CF) were estimated across a range of prior values for the discordance parameter (α) of 0.1, 1, 10, and 100. The discordance parameter (α) represents the prior probability distribution that all genes share the same tree, where α = 0 indicates all genes share the same tree while α = ∞ indicates all genes have a distinct set of trees [55]. All analyses were initially run with two Markov chain Monte Carlo search chains (MCMC) of 100,000 generations after a burn-in of 10,000 generations. All runs converged on the same primary concordance tree with identical concordance factors. The final analysis was run with α = 1 and two MCMC chains of 1,000,000 generations following a burn-in of 100,000 generations (-a 1 -c 2 -n 1000000).
Several species within the C. gloeosporioides species complex have been published since the epitypification of C. gloeosporioides [14], [17], [57], [58] and a comprehensive phylogenetic analysis of the species complex, including the description of new species and subspecies, was recently published [16]. Despite agreement regarding the necessity of multilocus genetic data to resolve taxonomic problems within the species complex and the recognition that the phylogenetic resolution provided by the most commonly used markers is significantly lacking, a common set of markers has not yet been adopted among independent research groups [14], [15], [17], [57]–[59]. Nonetheless, there is some overlap in the markers sequenced for recently described species. In order that isolates from this study could be placed in the broader context of the species complex and to evaluate the distinctiveness of the newly described species presented here (see ‘Species assignments’ below), we analyzed sequence data from representative isolates and type strains available in GenBank. In addition to the complete four-marker dataset described above, hereafter referred to as D4G, two additional datasets were constructed with the available sequence data from nrITS, partial tub2, and apn2/matIGS. The first dataset, hereafter referred to as D3G, included all isolates from the D4G dataset with the addition of sequence data from nine isolates, included in a recent study by Silva et al. (2012), representing three additional species and one subspecies. The combined data set included 2,134 nucleotides (nrITS: 557; tub2∶690; apn2mat/IGS: 887) for 91 isolates with no missing markers for any terminal. The second dataset, hereafter referred to as D3G+, included the addition of nrITS and partial tub2 sequence data for thirty additional strains, included in a comprehensive phylogenetic study of the species complex by Weir et al. (2012), representing sixteen species and one forma speciales with data for the apn2/matIGS marker coded as missing for these additional strains in an alignment of 2,148 nucleotides. Phylogenetic analyses of these concatenated matrices were conducted with RAxML–HPC2 and MrBayes v3.1.2 as previously described. The best-fit models determined in MrModeltest 2.3 for each locus in the expanded datasets were as follows: nrITS – GTR+Γ; tub2– GTR+Γ; apn2/matIGS – HKY+ Γ. GenBank accession numbers from previously published studies included in analyses presented here are provided in Table S2 and all multiple sequence alignments are available from the authors upon request. The presentation of results from the analysis of D3G+ is restricted to Text S1 for the sake of brevity and clarity.
Species Assignments
In order to delimit novel species, we applied the criteria of genealogical concordance phylogenetic species recognition (GCPSR) [60], [61] to phylogenetic estimates from the D4G dataset. Briefly, following Dettman et al. [60], novel species were recognized if they satisfied one of two criteria: genealogical concordance or genealogical non-discordance. Clades were genealogically concordant if they were present in three of the four single gene trees and genealogically non-discordant if they were strongly supported (MP≥75%; ML≥70%) in a single gene and not contradicted at or above this level of support in any other single gene tree. In addition, novel species were recognized if resolved with strong support (PP≥.95; ML≥70%; MP≥75%) in all analyses for the combined dataset of nrITS, tub2, apn2, and apn2/matIGS and were not nested within clades containing the type of any previously described species in any of the combined analyses. All isolates were assigned to a species with the exception of GJS0857 and Coll940, which remain singletons.
Quantitative Measures of Genealogical Sorting
The genealogical sorting index is used here to represent a quantitative assessment of the monophyly or genealogical exclusivity of a group of commonly labeled terminals in a set of trees. This quantitative measure enables the comparison of individual markers to assess their ability to differentiate lineages or species. In order to determine the degree of exclusive ancestry of each of the well–supported lineages inferred from the concatenated dataset of nrITS, btub, apn2, and apn2mat/IGS (D4G) across the independent gene trees, the bootstrap trees obtained from the parsimony bootstrap analyses for each independent gene and the concatenated dataset were used to calculate the genealogical sorting index (gsi) for each lineage [62]. The gsi was calculated for lineages of the C. gloeosporioides species aggregate using the R (2.13.0) package genealogicalSorting version 0.91 [63], [64] after removing outgroup strains (Coll57, Coll60, 4766, 4801, 4766). The genealogical sorting index (gsi) reaches a maximum when a commonly labeled group (species or terminal lineage) reaches monophyly in a tree or set of trees and a minimum when all nodes on the tree are required to unite a group. P-values represent the probability that the calculated gsi from the inferred tree or trees would be observed by chance. The ensemble genealogical sorting index (gsiT) is the sum of the gsi values for each topology weighted by the probability of a given topology based on the proportion of trees in the sample where it is represented. P-values are calculated by permutation of group labels on the terminals of a tree and determining the frequency distribution from the recalculated gsi value. P-values are estimated from 1,000 permutations of each dataset and represent the probability of observing gsi values ≥ to the reported gsi values by chance under the null hypothesis that labeled groups are of mixed ancestry.
Morphological Studies
Morphological observations were made from strains cultured on potato dextrose agar (PDA), corn meal agar (CMA), V8 agar, and Synthetischer nährstoffarmer agar (SNA) [65] incubated at 22–25°C. Growth rates were determined from cultures grown in darkness at 25°C on PDA. Cultures were first grown on CMA and 10 mm plugs were taken from the expanding margin and transferred to 20 mL of PDA in 100×15 mm Petri dishes. Each strain was plated to three replicate plates. Three radial measurements were taken from the edge of the plug to the margin of the colony every 24–48 hours over the course of 5 days by marking the bottom of the plate at the margin of the colony using a stereomicroscope equipped with stage lighting, resulting in 9 radial measurements per strain. Each plate was then photographed and the distance between markings was measured.
Microscopic observations of conidia, phialides, and ascospores were made from specimens mounted in water. Hyphal appressoria were observed using slide cultures; a 10 mm2 block of CMA was placed on a CMA plate, each of the four corners of the block was inoculated, covered with a sterile coverslip, and incubated at 25°C. Microscopic observation of perithecial development on cranberry fruit was made by surface sterilizing symptomatic field collected fruit, cutting in half transversely, placing face down on V8 agar, and incubating at room temperature (22°C) for approximately 3 weeks. The fruit was removed from the plate after perithecial development and fixed in FAA (3.7% formaldehyde, 5% glacial acetic acid, and 50% ethanol) before dehydration in an alcohol-xylene dehydration series and embedded in Paraplast X-TRA (Leica Microsystems, Buffalo Grove, IL, USA). The fruit was sectioned at 8 µm and stained with acid fuchsin and Cotton blue in lactic acid and mounted in Permount (Fisher Scientific, Pittsburgh, PA, USA).
Microscopic images were made with a Nikon DXM1200C digital camera attached to a Zeiss Axioplan compound microscope or with a Nikon SMZ1500 stereoscope equipped with a Nikon DXM1200F digital camera using Nikon ACT-1 software. All measurements were made using ImageJ 1.44p software [66], [67] and summary statistics calculated in R version 2.13.0. Growth rates are reported giving the minimum, average, maximum [(minimum-) average (-maximum)] and the standard deviation in millimeters per day. All other measurements additionally include the 1st quartile and the 3rd quartile with the following notation: (minimum-) 1st quartile – average –3rd quartile (-maximum).
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB. The online version of this work is archived and available from the following digital repositories: PubMed Central; LOCKSS.
Results
Individual Gene Trees
In order to test the hypothesis that multiple sympatric lineages within the C. gloeosporioides species complex infect cranberry in the field, North American isolates need to be placed in a broader phylogenetic context. Therefore, sequence data generated in this study were combined with an earlier study of isolates of C. gloeosporioides s.l. from the New World tropics [14]. Outgroup sampling was expanded from the aforementioned study to include two isolates of C. aff. acutatum. Individual locus data were analyzed separately to assess the topological congruence among datasets and the utility of each to resolve terminal lineages with robust statistical support for sister group relationships within the C. gloeosporioides species complex. Strain data are summarized in Table 1 and character and tree statistic data are summarized in Table 2.
Table 2. Tree statistics.
| Locus | # Taxa | # Chars | % PIC | #MPTs | MPT length | CI | RI | ML -ln L | BI -ln L |
| nrITS | 84 | 570 | 13.3 | 28 | 93 | 84 | 95 | −1324.92 | NA |
| tub2 | 84 | 900 | 31.2 | 12 | 403 | 87 | 97 | −3389.23 | NA |
| apn2 | 82 | 756 | 27.3 | 14 | 339 | 79 | 96 | −3024.04 | NA |
| apn2/MAT12IGS | 82 | 893 | 52.1 | 1 | 723 | 83 | 97 | −4531.11 | NA |
| Combined | 84 | 3119 | 33 | 1 | 1585 | 82 | 96 | −12981.97 | −12981.97 |
Taxa: number of taxa included in the analysis.
Chars: number of characters per locus. %PIC: Percent of parsimony informative characters.
MPTs: number of most parsimonious trees. MPT: length of the most parsimonious trees. CI: ensemble consistency index. RI: ensemble retention index. ML –ln L: ML score of best tree calculated in RAxML under the GTRGAMMA model. BI –ln L: ML score of best tree from 4 runs of 10,000,000 generations inferred in MrBayes calculated in RAxML with GTRGAMMA model.
Sequence data from nrITS has been widely used in fungal phylogenetic studies and has been proposed as a barcode locus for fungi [68]. However, our analysis indicates it neither provides adequate resolution for reliable species assignment, nor does it reliably assess phylogenetic relationships within the C. gloeosporioides species complex, as has been reported in previous studies of Colletotrichum [14], [23], [69]. Despite the low phylogenetic resolution inferred from nrITS data, six nodes within the species complex were supported in more than 75% of the parsimony bootstrapped datasets (Figure S1) and eight nodes in more than 70% of the maximum likelihood bootstrapped datasets (Figure S5). In addition, C. sp. indet. D (4766, 3386, 4801) was determined to be closely related but peripheral to the C. gloeosporioides species complex, indicating this is a suitable outgroup, as previously suggested by Rojas et al. [14].
Phylogenetic analysis of partial tub2 sequence data largely supported the inferences made from nrITS data, but provided further resolution within the species complex, recovering 20 well-supported nodes in both MP and ML analyses (Figures S2 and S6). Isolates from cranberry (V. macrocarpon and V. oxycoccos) were distributed among 5 clades, including lineages originally described from tropical regions.
Phylogenetic analysis of the apn2 locus provided greater resolution for terminal lineages than that achieved with either tub2 or nrITS. Bootstrap analyses provided significant statistical support for 23 nodes and 20 nodes for MP (Figure S3) and ML (Figure S7) analyses, respectively, with six clades in both analyses containing isolates from V. macrocarpon and V. oxycoccos. While support for terminal lineages from apn2 sequence data was greater than that from partial tub2, support for sister group relationships was not as robust.
A similar topology was recovered from the analysis of nucleotide data from apn2/matIGS, the intergenic spacer bridging the apn2 and mating-type locus, with respect to the nrITS, tub2, and apn2 gene trees while providing greater resolution than the other three datasets. Bootstrap analyses of apn2/matIGS recovered strong branch support for 25 and 29 nodes for MP (Figure S4) and ML (Figure S8), respectively. In agreement with the apn2 analysis, six lineages include isolates from V. macrocarpon and V. oxycoccos. In addition to increased resolution of terminal clades within the species complex, compared with apn2, support for sister group relationships within the species complex is stronger than inferences based on partial tub2.
While statistical node support based on bootstrap resampling was somewhat inconsistent among ML and MP analyses, both sets of analyses were concordant, with differing levels of resolution. Similarly, the resolution of terminal clades among independent gene trees is consistent with a few exceptions. One strain, isolated as a leaf endophyte of Persea americana (Coll11), was relegated with strong support to a terminal clade that includes isolates designated as Colletotrichum siamense based on analysis of nrITS, but this isolate is strongly supported in both the partial tub2 and apn2/matIGS tree as belonging to a lineage that includes isolates of C. tropicale. The resolution of this isolate in the apn2 analysis was ambiguous. The phylogenetic placement of another strain, Coll940, is inconsistent among gene trees. While Coll940 is resolved as sister to a group that includes C. fructicola, “C. ignotum 2″, C. nupharicola and C. sp. indet. C in the tub2 (ML) and apn2 (MP) analyses, its placement is switched with C. sp. indet. C in the apn2/matIGS analyses.
Phylogenetic Analysis of the Combined Datasets
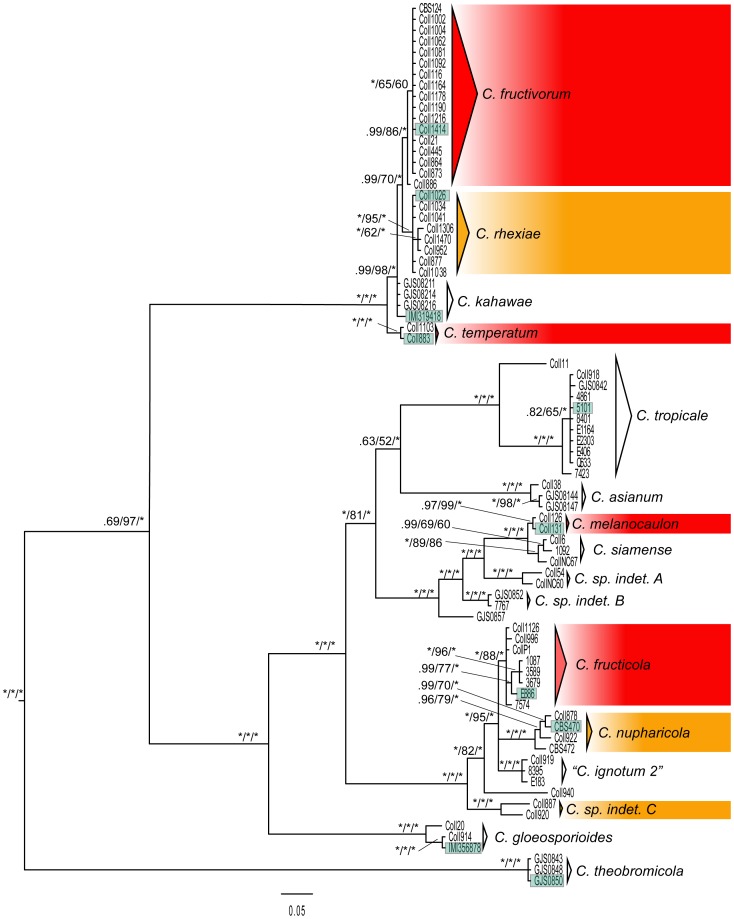
In order to infer organismal phylogenetic relationships and make species assignments, we relied on the combined dataset of 3,119 nucleotide characters from four nuclear markers and eighty-four terminals (D4G). The Bayesian consensus tree is presented in Figure 1 with node posterior probability and bootstrap proportion values from Bayesian, parsimony, and maximum likelihood analyses. The outgroup taxa, C. aff. acutatum and C. sp. indet. D have been trimmed from Figure 1 due to long branches between the species complex and the outgroup terminals. The relationships among clades in each of the three analyses converged on a topology, with varying levels of node support, identical to the Bayesian analysis presented in Figure 1. The log-likelihood of the tree with the best score (highest log-likelihood value) from the set of four Bayesian runs as calculated in RAxML under the GTRGAMMA model (−1298.97) was identical to the tree inferred under the maximum-likelihood criterion.
Figure 1. Bayesian majority rule consensus tree with support values (PP/ML-BS/MP-BS) for the combined analysis of nrITS, btub, apn2, and apn2mat/IGS.
Outgroup taxa (Coll57, Coll60, 3386, 4801, 4766) have been trimmed from the phylogram. Support values of 1.0 or 100% are represented with “*”. Terminals enclosed in shaded boxes represent sequence data from ex-type strains. Clades enclosed in red boxes contain isolates from Vaccinium macrocarpon in cultivated cranberry beds in North America. Clades enclosed in orange boxes contain isolates from host species sympatric with cultivated V. macrocarpon.
Thirty-six nodes were inferred with strong bootstrap and posterior probability support in the combined analysis of nrITS, tub2, apn2, and apn2mat/IGS. The multilocus analyses corroborated the majority of the single gene trees in recovering C. theobromicola as sister to two principal lineages within the species complex. The first lineage includes two previously described species, C. kahawae and C. rhexiae, and two newly described species, C. temperatum and C. fructivorum. The second lineage includes six previously described species, C. tropicale, C. asianum, C. siamense, C. fructicola, C. nupharicola, and C. gloeosporioides, one new species, C. melanocaulon and four undescribed sublineages. Added taxon sampling suggests the original circumscription of C. ignotum (recently placed in synonymy with C. fructicola by Weir et al. [16]) to include isolates in the lineage labeled “C. ignotum 2″ in Figure 1 was too broad, also discussed in Rojas et al. [14].
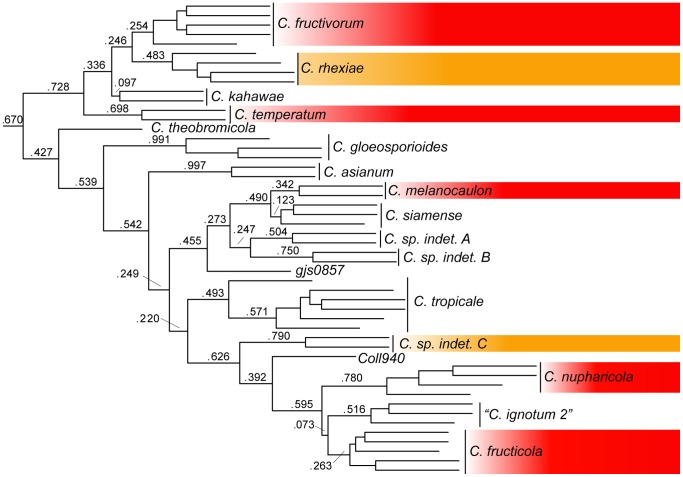
The primary concordance (PC) tree estimated with Bayesian concordance analysis from the four nuclear markers remained unchanged regardless of the value of the discordance parameter (α). The PC tree (Figure 2) is consistent with the assignment of individual isolates to terminal lineages on the basis of the concatenated analysis. Concordance factors are reported for all nodes above the species level. The low concordance factors (below 0.5) for several species, including C. fructicola, C. asianum, C. siamense, C. kahawae, C. rhexiae, and C. fructivorum appear to be due to the lack of resolution provided by individual markers rather than topological discordance. Similar to the analysis of the combined data, two principal lineages were resolved in the PC tree. However, relationships among a few species within these two lineages are distinctive from the combined analysis. For example, Colletotrichum theobromicola is placed within the principal lineage that includes C. gloeosporioides, and C. asianum is no longer sister to C. tropicale.
Figure 2. Primary concordance tree resulting from the Bayesian concordance analysis of the nrITS, btub, apn2, and apn2mat/IGS with concordance factors for nodes above the species level.
Clades enclosed in red boxes contain isolates from Vaccinium macrocarpon in cultivated cranberry beds in North America. Clades enclosed in orange boxes contain isolates from host species sympatric with cultivated V. macrocarpon.
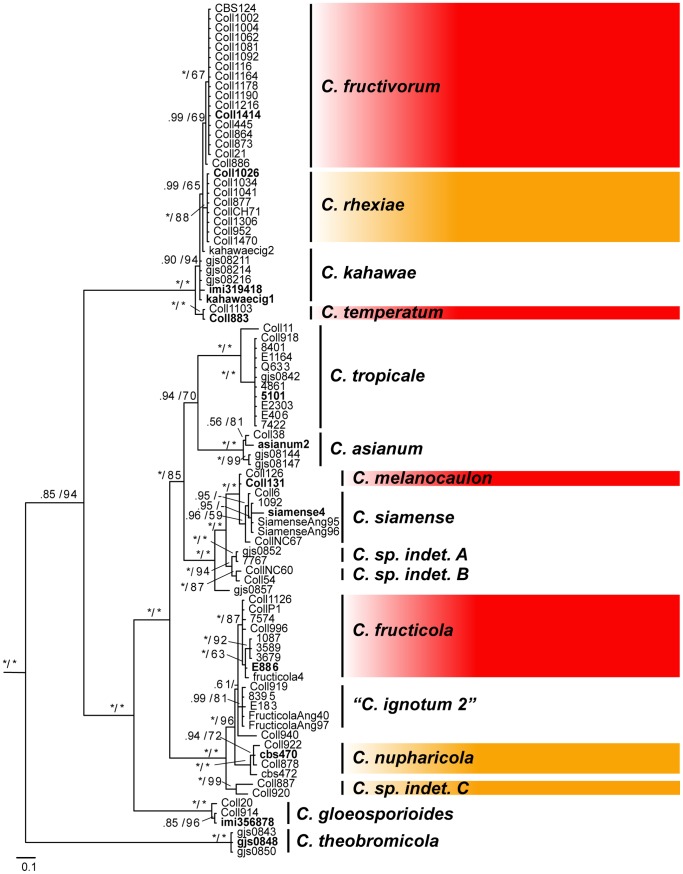
The monophyly of species delimited on the basis of the combined analysis presented in Figure 1 is largely supported by results from the analysis of D3G, which includes three additional species and one subspecies (Figure 3). Alternate sister group relationships are suggested by this analysis, however branch support values are generally lower than those in Figure 1 and the backbone of the tree topology remains unchanged. The addition of sequence data from ex-type strains of C. asianum and C. siamense allowed for the identification of isolates in clades that include these species. The assignment of isolates Coll38, G.J.S. 08-144, and G.J.S. 08-147 to C. asianum and isolates Coll6, 1092, and NC67 to C. siamense in Figure 1 and Figure 2 are based on the results presented in Figure 3. The inclusion of sequences from the ex-type of C. fructicola confirms that C. ignotum is a synonym as reported by Weir et al. [16]. In addition, the inclusion of two sequences of C. kahawae subsp. cigarro, including the ex-type, illustrates its close affinity to C. rhexiae and C. fructivorum. Weir et al. [16] also identified an isolate from cranberry (CBS124) as C. kahawae subsp. cigarro, however our analyses indicate this isolate belongs to a phylogenetically distinct lineage.
Figure 3. Bayesian majority rule consensus tree with support values (PP/ML-BS) for the combined analysis with expanded taxon sampling but a reduced dataset (D3G - nrITS, btub, and apn2mat/IGS).
Clades enclosed in red boxes contain isolates from Vaccinium macrocarpon in cultivated cranberry beds in North America. Clades enclosed in orange boxes contain isolates from host species sympatric with cultivated V. macrocarpon.
Genealogical Sorting Indices
The genealogical sorting index represents a quantitative assessment of the exclusive ancestry of each species in a set of bootstrap trees for each individual locus and in the combined dataset. Terminal labels for this analysis match the species assignments based on the combined four-marker analysis (D4G), as previously described. The results of these analyses are presented in Table 3 and the taxon assignments are presented in Table 4.
Table 3. Genealogical sorting indices of 1000 bootstrap trees calculated using genealogical Sorting R package with 1,000 permutations.
| Taxon | combined | nrITS | tub2 | apn2 | apn2/mat12IGS | |||||
| gsiT | p | gsiT | p | gsiT | p | gsiT | p | gsiT | p | |
| kahawae | 0.740 | *** | 0.155 | *** | 0.073 | NS | 0.110 | * | 0.572 | *** |
| fructivorum | 1.000 | *** | 0.370 | *** | 0.466 | *** | 0.703 | *** | 1.000 | *** |
| rhexiae | 1.000 | *** | 0.381 | *** | 0.179 | ** | 1.000 | *** | 1.000 | *** |
| temperatum | 1.000 | ** | 0.404 | ** | 1.000 | *** | 0.494 | ** | 0.901 | *** |
| tropicale | 1.000 | *** | 0.149 | *** | 1.000 | *** | 0.810 | *** | 1.000 | *** |
| CspindetA | 1.000 | *** | 1.000 | *** | 1.000 | ** | 1.000 | *** | 1.000 | *** |
| melanocaulon | 1.000 | ** | 0.022 | NS | 0.187 | * | 1.000 | *** | 0.241 | * |
| CspindetB | 0.952 | *** | 0.658 | *** | 0.220 | ** | 0.658 | *** | 0.487 | ** |
| CspindetC | 1.000 | *** | 0.022 | NS | 0.114 | * | 0.986 | *** | 1.000 | *** |
| CspindetD | 1.000 | ** | 0.022 | NS | 0.679 | *** | 1.000 | ** | 0.812 | *** |
| ignotum | 1.000 | *** | 0.221 | *** | 0.751 | *** | 0.757 | *** | 1.000 | *** |
| nupharicola | 1.000 | *** | 0.198 | *** | 1.000 | *** | 1.000 | *** | 1.000 | *** |
| ignotum2 | 1.000 | *** | 0.064 | * | 1.000 | *** | 1.000 | *** | 0.447 | *** |
| CspindetE | 1.000 | ** | 0.024 | NS | 1.000 | *** | 1.000 | *** | 0.786 | ** |
| gloeosporioides | 1.000 | *** | 1.000 | *** | 1.000 | *** | 1.000 | *** | 1.000 | *** |
| theobromicola | 1.000 | *** | 1.000 | *** | 1.000 | *** | 1.000 | *** | 1.000 | *** |
p-value <0.001.
p-value <0.01.
p-value <0.05.
NSp-value >0.05.
Table 4. Taxon assignments for the genealogical sorting index analysis.
| Strain | Assignment | Strain | Assignment |
| CBS124 | fructivorum | 3679 | fructicola |
| Coll1002 | fructivorum | 7574 | fructicola |
| Coll1004 | fructivorum | E886 | fructicola |
| Coll1062 | fructivorum | Coll919 | “ignotum 2″ |
| Coll1081 | fructivorum | 8395 | “ignotum 2″ |
| Coll1092 | fructivorum | E183 | “ignotum 2″ |
| Coll116 | fructivorum | GJS08211 | kahawae |
| Coll1164 | fructivorum | GJS08214 | kahawae |
| Coll1178 | fructivorum | GJS08216 | kahawae |
| Coll1190 | fructivorum | IMI319418 | kahawae |
| Coll1216 | fructivorum | Coll38 | asianum |
| Coll1414 | fructivorum | GJS08144 | asianum |
| Coll21 | fructivorum | GJS08147 | asianum |
| Coll445 | fructivorum | Coll878 | nupharicola |
| Coll864 | fructivorum | Coll922 | nupharicola |
| Coll873 | fructivorum | CBS470 | nupharicola |
| Coll886 | fructivorum | CBS472 | nupharicola |
| Coll1103 | temperatum | Coll1026 | rhexiae |
| Coll883 | temperatum | Coll1034 | rhexiae |
| Coll126 | melanocaulon | Coll1041 | rhexiae |
| Coll131 | melanocaulon | Coll1306 | rhexiae |
| Coll887 | C. sp. indet. C | Coll877 | rhexiae |
| Coll920 | C. sp. indet. C | Coll952 | rhexiae |
| Coll940 | Coll940 | Coll1038 | rhexiae |
| Coll6 | siamense | Coll1470 | rhexiae |
| CollNC67 | siamense | GJS0843 | theobromicola |
| 1092 | siamense | GJS0848 | theobromicola |
| Coll54 | C. sp. indet. B | GJS0850 | theobromicola |
| CollNC60 | C. sp. indet. B | Coll11 | tropicale |
| GJS0852 | C. sp. indet. A | Coll918 | tropicale |
| 7767 | C. sp. indet. A | GJS0842 | tropicale |
| GJS0857 | GJS0857 | 4861 | tropicale |
| Coll20 | gloeosporioides | 5101 | tropicale |
| Coll914 | gloeosporioides | 7423 | tropicale |
| IMI356878 | gloeosporioides | 8401 | tropicale |
| Coll1126 | fructicola | E1164 | tropicale |
| Coll996 | fructicola | E2303 | tropicale |
| CollP1 | fructicola | E406 | tropicale |
| 1087 | fructicola | Q633 | tropicale |
| 3589 | fructicola |
Colletotrichum gloeosporioides, C. asianum, and C. theobromicola reached the maximum ensemble gsi (gsiT) value of 1 when calculated across all bootstrapped trees from the nrITS data. Several species including C. kahawae, C. tropicale, C. fructicola, C. nupharicola, and “C. ignotum 2″ had low, but significant, gsiT values based on the nrITS trees ranging from 0.064 to 0.221. Four species, C. fructivorum, C. rhexiae, C. temperatum, and C. siamense had moderate values of gsiT ranging from 0.370 to 0.658. The remaining species, C. melanocaulon, C. sp. indet. C, C. sp. indet. B, and C. sp. indet. A had non-significant gsiT values across the nrITS bootstrapped trees.
Unlike the gsiT values of the nrITS trees, all values calculated from the tub2 trees were significant with the exception of C. kahawae. Colletotrichum temperatum, C. tropicale, C. asianum, C. nupharicola, “C. ignotum 2″, C. sp. indet. C, C. gloeosporioides, and C. theobromicola reached the maximum gsiT value of 1 when calculated across all bootstrapped trees. Colletotrichum rhexiae, C. melanocaulon, C. siamense, and C. sp. indet. B, had low but significant gsiT values ranging from 0.114 to 0.220. Colletotrichum fructivorum, C. sp. indet. A and C. fructicola had moderate to high gsiT values ranging from 0.466 to 0.751.
All gsiT values of the apn2 trees were significant. Colletotrichum kahawae had the lowest value of 0.11, while all others were moderate to high. Colletotrichum rhexiae, C. asianum, C. melanocaulon, C. sp. indet. A, C. nupharicola, “C. ignotum 2″, C. sp. indet. C, C. gloeosporioides, and C. theobromicola had the maximum gsiT value of 1 across all bootstrapped trees. Colletotrichum fructivorum, C. temperatum, C. tropicale, C. siamense, C. sp. indet. B, and C. fructicola had moderate to high gsiT values ranging from 0.494 to 0.986.
Similarly, all gsiT values of the apn2mat/IGS trees were significant. Colletotrichum melanocaulon had the lowest value of 0.241, while all others were moderate to high. Colletotrichum fructivorum, C. rhexiae, C. tropicale, C. asianum, C. sp. indet. B, C. fructicola, C. nupharicola, C. gloeosporioides, and C. theobromicola had gsiT values of 1 when calculated across all bootstrapped trees, while C. kahawae, C. temperatum, C. siamense, C. sp. indet. A, “C. ignotum 2″, and C. sp. indet. C had gsiT values ranging from 0.447 to 0.901.
All gsiT values of trees from the combined dataset were significant with high values ranging from 0.740 to 1. The only species that did not reach the maximum possible gsiT value of 1 were C. kahawae (0.740) and C. siamense (0.952). All values based on the combined four-marker dataset represent strong measures of genealogical divergence across all 1000 bootstrap replicates.
Taxonomy and Morphology
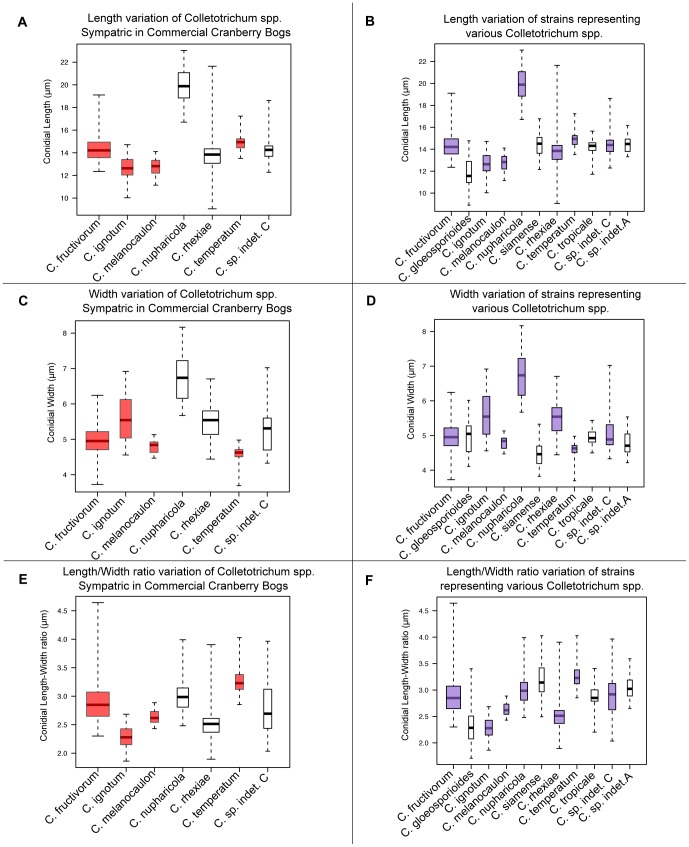
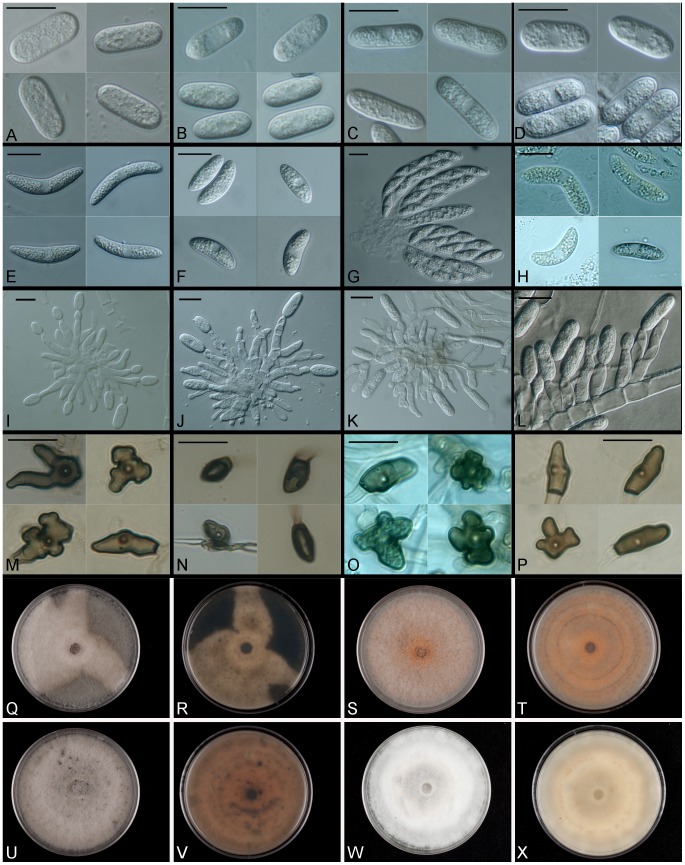
Multilocus phylogenetic analysis of Colletotrichum gloeosporioides s.l. strains isolated from V. macrocarpon and other sympatric host species revealed several distinct lineages within the species complex. Comparison of growth rates and conidial morphology indicates that there is significant morphological overlap between Colletotrichum species isolated from V. macrocarpon and sympatric host species as well as between strains representing additional species within the species complex. Colletotrichum nupharicola is exceptional, exhibiting a very slow growth rate, with significantly longer and wider conidia than all other species included in this study. Comparisons of growth rates and conidial dimensions are presented in Figure 4 and Figure 5 respectively. Three new species are described below and C. rhexiae is epitypified. Conidial, ascospore, and colony morphology are represented in Figure 6 for each new species and C. rhexiae. Seta morphology is depicted in Figure 7.
Figure 4. Box plots of daily growth rates representing various species in the Colletotrichum gloeosporioides species complex.
A) Box plot of species isolated from Vaccinium macrocarpon and other sympatric host species in commercial cranberry beds. Red boxes represent species where strains have been isolated from V. macrocarpon in commercial beds only. Non-colored boxes represent species isolated only from sympatric host species. B) Box plot of several strains representing species within the C. gloeosporioides species complex. Purple boxes represent species that contain strains isolated from host plants found in wild and commercial cranberry bogs in North America. Non-colored boxes represent species not found in these habitats.
Figure 5. Box plots of conidial sizes representing various species in the Colletotrichum gloeosporioides species complex.
A, C, E) Box plot of species isolated from Vaccinium macrocarpon and other sympatric host species in commercial cranberry beds. Red boxes represent species where strains have been isolated from V. macrocarpon in commercial beds only. The measurements for C. sp. indet. C were taken from the only strain to have been isolated from commercial cranberry beds. Non-colored boxes represent species isolated only from sympatric host species. B, D, F) Box plot of several strains representing species within the C. gloeosporioides species complex. Purple boxes represent species that contain strains isolated from host plants found in wild and commercial cranberry bogs in North America. The measurements for Colletotrichum sp. indet. C were taken from both known strains. Non-colored boxes represent species not found in these habitats.
Figure 6. New and epitypified species of Colletotrichum from North American cranberry bogs.
A-D. Conidia. A) C. rhexiae on CMA. B) C. melanocaulon on CMA. C) C. temperatum on CMA. D) C. fructivorum on CMA. E-H. Asci and ascospores. E) C. rhexiae on PDA. F) C. temperatum on CMA. G) C. temperatum on CMA. H) C. fructivorum on V8 agar. I-L. Phialides. I) C. rhexiae on CMA. J) C. melanocaulon on SNA. K) C. temperatum on CMA. L) C. fructivorum on CMA. M-P. Hyphal appressoria. M) C. rhexiae. N) C. melanocaulon. O) C. temperatum. P) C. fructivorum. Q-X. Colonies on PDA after 10 days at 25°C in the dark (100×15 mm plates). Q) C. rhexiae obverse. R) C. rhexiae reverse. S) C. melanocaulon obverse. T) C. melanocaulon reverse. U) C. temperatum obverse. V) C. temperatum reverse. W) C. fructivorum obverse. X) C. fructivorum reverse. All scale bars = 10 µm.
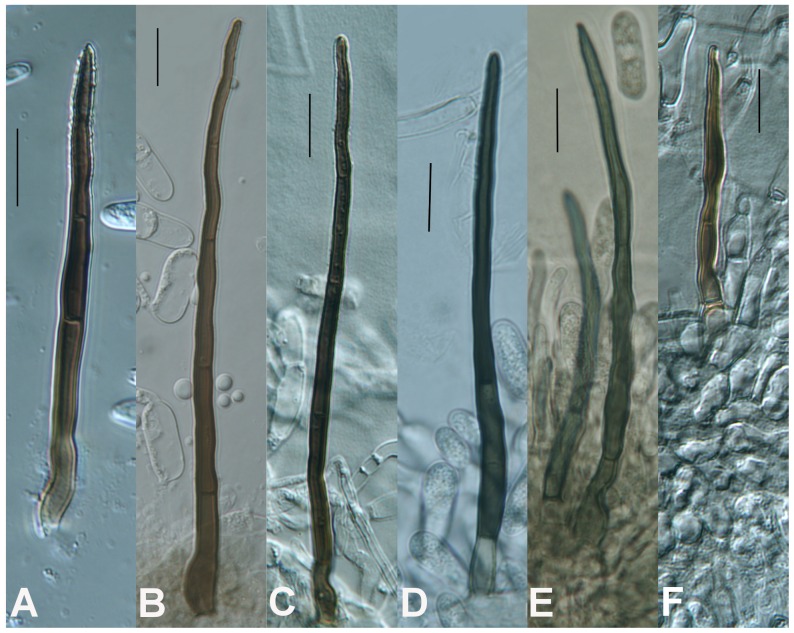
Figure 7. Setae.
A) Colletotrichum melanocaulon on SNA. B) Colletotrichum rhexiae on CMA. C) Colletotrichum rhexiae on SNA. D) Colletotrichum fructivorum on CMA. E) Colletotrichum temperatum on CMA. F) Colletotrichum sp. indet. B on SNA. Scale bars = 10 µm.
Colletotrichum fructivorum V. Doyle, P.V. Oudem. & S.A. Rehner, sp. nov.
Index Fungorum LSID: urn:lsid:indexfungorum.org:names:801462.
MycoBank MB 801462 (Figure 6, Figure 7).
Similar to Colletotrichum rhexiae Ellis & Everh. but setae less abundant, the interquartile range of the ascospore length to width ratio smaller (3.2–3.9 um) and the interquartile range of the conidial length to width ratio larger on CMA. Common fruit-rot pathogen of cranberry (Vaccinium macrocarpon Aiton) in commercial production.
Growth rate (3.0–) 5.4 (–6.8) mm per day with standard deviation of 1.1 mm on PDA at 25°C [n = 54]; aerial mycelium floccose, white to greyish white, medium grey and brownish grey in some strains; sectoring common. Perithecia developing and maturing on V8 agar, clustered or solitary, dark brown to black, globose to obpyriform to papillate; ascospores allantoid, olive yellow, (15.7–) 16.6–18.2–18.5 (–26.9)×(3.8–) 4.5–5.1–5.6 (–7.1) µm with length/width ratio (2.6–) 3.2–3.7–3.9 (–5.6) µm [n = 30]. On CMA and SNA perithecial fundaments with melanized hyphae radiating from the base but mature ascospores not observed. On SNA aerial mycelium flocculose; conidial masses flesh to light orange, (12.4–) 13.2–14.3–15.2 (–18.6)×(3.8) 4.3–4.9–5.6 (–6.2) µm with length/width ratio (2.0–) 2.4–3.0–3.5 (–4.3). On CMA mycelium barely visible; conidial masses usually abundant, embedded in the medium and on the surface, flesh to light orange; phialides tapering towards the tip, monoblastic, (9.0–) 10.5–12.9–14.2 (–18.1)×(2.9–) 3.7–3.9–4.1 (–4.9) µm [n = 30] at the widest point; conidia subcylindrical to slightly tapered with obtuse apices, (12.4–) 13.6–14.3–14.9 (–19.1)×(3.7–) 4.7–5.0–5.2 (–6.2) µm with length/width ratio (2.3–) 2.7–2.9–3.1 (–4.6) µm [n = 240]; hyphal appressoria melanized, clavate or irregular, shallowly lobed, terminal, (3.9–) 8.0–9.4–10.8 (–15.2)×(3.1–) 3.9–4.5–4.7 (–7.7) µm with length/width ratio (0.6–) 1.8–2.2–2.6 (–3.5) µm [n = 30].
Habitat and Distribution
Commonly isolated as a fruit-rot pathogen and from asymptomatic tissue of Vaccinium macrocarpon in commercial cranberry beds throughout North America. Also isolated from Rhexia virginica from asymptomatic infections growing in commercial cranberry beds and symptomatic fruit of V. oxycoccos in a wild cranberry bog in Pennsylvania.
Etymology
The specific epithet, “fructivorum”, refers to the propensity of the species to be associated with fruit-rot of cranberry. From the Latin fructus, fruit, and –vorous, eating.
Holotype
USA. New Jersey: isolated as a fruit-rot pathogen of Vaccinium macrocarpon, Burlington County, 39.8128 N, 74.6399 W, Oct 2010, V. Doyle, P.V. Oudemans, C. Constantelos Coll1414 (BPI 884103, CBS 133125).
Additional specimens examined
USA. New Jersey: isolated as a stem endophyte of V. macrocarpon, Burlington County, 39.9518 N, 74.5005 W, Oct 2010, V. Doyle Coll1092 (BPI 884114, CBS 133135); isolated from Rhexia virginica, Burlington County, 39.7348 N, 74.5120 W, Sept 2009, V. Doyle Coll864 (BPI 884108, CBS 133130); isolated as a fruit-rot pathogen of V. macrocarpon, Burlington County, 39.9518 N, 74.5005 W, Oct 2009, V. Doyle, P.V. Oudemans Coll445 (BPI 884105, CBS 133127); isolated as a fruit-rot pathogen of V. macrocarpon, Burlington County, Fall 2007, P.V. Oudemans GLO7-17 (BPI 884111, culture V. Doyle Coll21 = CBS 133133); Collected by C.L. Shear, deposited to CBS in Apr 1922 (CBS124.22). Massachusetts: isolated as a fruit-rot pathogen of V. macrocarpon, Plymouth County, Nov 2010, V. Doyle Coll1062; isolated as a fruit-rot pathogen of V. macrocarpon, Plymouth County, Nov 2010, V. Doyle Coll1081 (BPI 884099, CBS 133121); isolated as a fruit-rot pathogen of V. macrocarpon, P.V. Oudemans (culture V.Doyle Coll116). Wisconsin: isolated as a fruit-rot pathogen of V. macrocarpon, Monroe County, 44.0685 N, 90.4015 W, Sept 2010, P.V. Oudemans, C. Constantelos, E. Zeldin WI-1 (BPI 884104, culture V. Doyle Coll1164 = CBS 133126); isolated as a fruit-rot pathogen of V. macrocarpon, Wood County, 44.3854 N, 90.0151 W, Sept 2010, P.V. Oudemans, C. Constantelos, E. Zeldin WI-53 (culture V. Doyle Coll1216); isolated as a fruit-rot pathogen of V. macrocarpon, Juneau County, 44.2072 N, 90.0995 W, Sept 2010, P.V. Oudemans, C. Constantelos, E. Zeldin WI-27 (culture V. Doyle Coll1190); Delaware: isolated as a fruit-rot pathogen of V. macrocarpon, Kent County, 39.3130 N, 79.5555 W, Fall 2009, P.V. Oudemans, C. Constantelos GLO9-1 (culture V. Doyle Coll873). Pennsylvania: isolated as a fruit-rot pathogen of V. oxycoccos, Monroe County, Tannersville Cranberry Bog Preserve, 41.0376 N, 75.2648 N, V. Doyle Coll886 (BPI 884102, CBS 133124). CANADA. British Columbia: isolated as a fruit-rot pathogen of V. macrocarpon, Oct 2010, V. Doyle, P.V. Oudemans, B. Mouza Coll1002 (BPI 884106, CBS 133128); isolated as a fruit-rot pathogen of V. macrocarpon, Oct 2010, V. Doyle, P.V. Oudemans, B. Mouza Coll1004;
Notes
Glomerella rufomaculans var. vaccinii was described by C.L. Shear in 1907 [70] from leaves of Vaccinium macrocarpon in New Jersey. The type material was a slide (slide no. 1447A C.L.S.) of a single ascospore isolate made from this collection deposited in the “pathological collection of the Department of Agriculture”, now housed in the collections at the Systematic Mycology and Microbiology Laboratory in Beltsville, Maryland (BPI). While other slides designated by C.L. Shear as type material for new species published in the same protologue were located at BPI, slide no. 1447A C.L.S. was not located and is thought to be lost. However, a culture deposited by Shear in April, 1922 to Centraalbureau voor Schimmelcultures in the Netherlands was obtained, sequenced and found to be conspecific with other isolates described here. The nomenclature used by CBS in designating this isolate as Glomerella rufomaculans-vaccinii, however, seems to be based on a typographical error as C.L. Shear clearly intended recognition of this taxon at the varietal level and there is no indication in the literature that it has been formally elevated to species. Furthermore, Glomerella rufomaculans was subsumed into Glomerella cingulata (anamorph: Colletotrichum gloeosporioides) with the revisionary work of von Arx in 1957 [11], necessitating the designation of the new specific epithet, Colletotrichum fructivorum. We have chosen a recently isolated strain as the ex-type strain due to the lack of sporulation observed in the culture deposited by Shear and the fact the holotype designated here is a single ascospore isolate for which we have morphological data for both the anamorph and the teleomorph (Figure 6).
Colletotrichum melanocaulon V. Doyle, P.V. Oudem. & S.A. Rehner sp. nov.
Index Fungorum LSID: urn:lsid:indexfungorum.org:names:801464.
MycoBank MB 801464 (Figure 6, Figure 7).
Similar to Colletotrichum gloeosporioides (Penz.) Sacc. but associated with stem canker of Vaccinium macrocarpon Aiton.
Growth rate (4.9–) 6.4 (–7.3) mm per day with standard deviation of 0.8 mm on PDA at 25°C [n = 18]; aerial mycelium floccose, white to light grey, sectoring observed; fertile perithecia not observed; conidiomata forming abundantly in concentric rings in type strain, conidial masses orange. On SNA aerial mycelium flocculose; conidiomata abundant, conidial masses light orange; phialides tapering towards the tip, monoblastic, (8.6–) 11.5–13.6–15.3 (–19.0)×(2.1–) 2.8–3.3–3.9 (–4.8) µm [n = 34] at the widest point; conidia subcylindrical to slightly tapered with obtuse apices, (11.3–) 12.6–13.1–13.4 (–14.8)×(4.4–) 5.1–5.5–5.9 (–6.3) µm with length/width ratio (2.1–) 2.3–2.4–2.5 (–3.0) µm [n = 60]. On CMA mycelium barely visible, conidial masses pale orange to melon; phialides tapering toward the tip, monoblastic, (11.7–) 13.5–14.1–14.4 (–17.0)×(2.8–) 3.2–3.4–3.7 (–3.9) µm [n = 10] at the widest point; conidia subcylindrical to slightly tapered with obtuse apices, (11.1–) 12.2–12.7–13.3 (–14.1)×(4.5–) 4.7–4.8–4.9 (–5.1) µm with length/width ratio (2.4–) 2.6–2.6–2.7 (–2.9) µm [n = 30]; hyphal appressoria melanized, subglobose to clavate, not lobed, terminal, (4.4–) 5.8–6.3–7.2 (–8.2)×(3.3–) 4.1–4.5–4.9 (–5.9) µm with length/width ratio (1.0–) 1.1–1.4–1.6 (–1.9) µm [n = 10].
Habitat and Distribution
Isolated from stem canker lesions of Vaccinium macrocarpon in commercial cranberry beds in New Jersey.
Etymology
The specific epithet, “melanocaulon”, refers to the brown or black stems from which the species was isolated. From the Greek, melano-, black or very dark, and caulon, stem, in agreement with the neuter generic name Colletotrichum.
Holotype
USA. New Jersey: isolated from cankered stems of Vaccinium macrocarpon, Burlington County, 39.7539 N, 74.5387 W, Aug 2008, V. Doyle Coll131 (BPI 884113, CBS 133251).
Additional specimens examined
USA. New Jersey: isolated from cankered stems of Vaccinium macrocarpon, Burlington County, 39.7538 N, 74.5400 W, Aug 2008, V. Doyle Coll126 (BPI 884101, CBS 133123).
Colletotrichum temperatum V. Doyle, P.V. Oudem. & S.A. Rehner, sp. nov.
Index Fungorum LSID: urn:lsid:indexfungorum.org:names:801463.
MycoBank MB 801463 (Figure 6, Figure 7).
Similar to Colletotrichum rhexiae Ellis & Everh. and C. fructivorum but ascospore length smaller, and ascospore length to width ratio smaller.
Growth rate (6.3–) 6.6 (–7.2) mm per day with standard deviation of 0.2 mm on PDA at 25°C [n = 18]; aerial mycelium floccose, white to greyish white, sectoring observed. Mature ascospores not observed on PDA. On CMA aerial mycelium flocculose to barely visible; perithecia solitary to clustered, dark brown to black, subglobose to obpyriform, ascospores hemisphaeroid to reniform, olive brown, (13.5–) 14.3–14.7–15.3 (–17.1)×(4.6–) 5.3–5.5–5.7 (–5.8) µm with length/width ratio (2.3–) 2.6–2.7–2.8 (–3.3) µm [n = 22]; conidial masses abundant on surface and embedded in agar, orange yellow; phialides tapering toward the tip, monoblastic, (7.7–) 11.3–13.9–15.7 (–22.6)×(2.2–) 3.3–3.4–3.7 (–4.2) µm [n = 30] at the widest point; conidia subcylindrical to slightly tapered, rarely medially constricted, with obtuse apices, (13.5–) 14.5–14.9–15.2 (–17.2)×(3.7–) 4.5–4.6–4.7 (–4.9) µm with length/width ratio (2.9–) 3.1–3.3–3.4 (–4.0) µm [n = 30]; hyphal appressoria melanized, irregular, shallowly to deeply lobed, terminal, (7.7–) 9.3–10.7–11.7 (–13.2)×(4.9–) 6.0–7.4–8.6 (–11.5) µm with length/width ratio (1.0–) 1.2–1.5–1.8 (–2.3) µm [n = 50].
Habitat and Distribution
Isolated from rotten fruit of an ornamental cultivar, Vaccinium macrocarpon ‘Hamilton’, growing at The New York Botanical Garden and as a stem endophyte of V. macrocarpon in a commercial cranberry bog in New Jersey.
Etymology
The specific epithet, “temperatum”, refers to the known distribution of the species. From the Latin, temperatum, temperate.
Holotype
USA. New York: isolated from rotten fruit of Vaccinium macrocarpon, Bronx County, The New York Botanical Garden, 40.8674 N, 73.8780 W, Nov 2009, V. Doyle & C. Mozzicato Coll883 (BPI 884100, CBS 133122).
Additional specimens examined
USA. New Jersey: isolated as a stem endophyte of Vaccinium macrocarpon, Burlington County, 39.9514 N, 74.5008 W, Oct 2010, V. Doyle Coll1103 (BPI 884098, CBS 133120).
Colletotrichum rhexiae Ellis & Everh. Proceedings of the Academy of Natural Sciences of Philadelphia 46∶372. 1894.
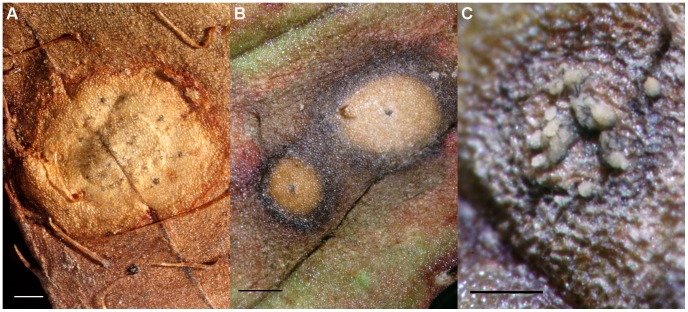
MycoBank MB 178511 (Figure 6, Figure 7, Figure 8).
Figure 8. Acervuli of Colletotrichum rhexiae on Rhexia virginica.
A) Acervulus of the holotype, A. Commons 2534, on leaf. Scale bar = 500 µm. B) Acervuli on leaves of R. virginica. Scale bar = 250 µm. C) Acervulus with setae and conidial masses from which V. Doyle Coll1470 was isolated. Scale bar = 250 µm.
Growth rate (4.9–) 6.2 (–7.1) mm per day with standard deviation of 0.6 mm on PDA at 25°C [n = 36]; aerial mycelium floccose, white to light grey to greyish brown, sectoring common; perithecia solitary to clustered, dark brown to black, ascospores fusiform to lunate, greyish yellow (18.1–) 21.7–23.2–24.4 (–27.5)×(3.5–) 3.9–4.1–4.3 (–4.7) µm with length/width ratio (4.5–) 5.2–5.8–6.4 (–7.6) µm [n = 60]; On SNA aerial mycelium flocculose; conidial masses flesh to light orange, phialides tapering toward the tip, monoblastic, (9.5–) 13.5–14.7–16.2 (–17.6)×(3.0–) 3.6–3.7–4.0 (–4.4) µm [n = 15] at the widest point; conidia subcylindrical with obtuse apices, (11.7–) 13.4–14.0–14.3 (–27.9)×(3.9–) 5.4–5.8–6.4 (–7.4) µm with length/width ratio (1.8–) 2.1–2.5–2.6 (–4.4) µm [n = 60]. On CMA mycelium barely visible; conidial masses abundant on the surface, flesh to light orange, phialides tapering towards the tip, monoblastic, (5.9–) 10.9–13.4–15.0 (–28.9)×(2.6–) 3.3–3.7–4.2 (–4.7) µm [n = 30] at the widest point; conidia subcylindrical with obtuse apices, (9.1–) 13.1–13.7–14.4 (–21.7)×(4.4–) 5.1–5.5–5.8 (–6.7) µm with length/width ratio (1.9–) 2.4–2.5–2.6 (–3.9) µm [n = 120]; hyphal appressoria melanized, clavate, obclavate to fusiform and irregular, shallowly to deeply lobed, terminal, (7.0–) 9.4–10.9–12.8 (–15.2)×(3.9–) 5.0.–6.1–7.1 (–9.7) µm with length/width ratio (1.0–) 1.3–1.9–2.2 (–3.4) µm [n = 30].
Holotype
USA. Delaware: acervuli on several leaves of Rhexia virginica, Kimensi, Delaware, A. Commons 2534.
Epitype
isolated from lesioned stem tissue of Rhexia virginica, Sussex County, Delaware, Cape Henlopen State Park, 38.7778 N, 75.1062 W, Nov 2010, V. Doyle Coll1026 (BPI 884112, CBS 133134).
Additional Specimens Examined. USA. Delaware: isolated from lesioned stem tissue of Rhexia virginica, Sussex County, Cape Henlopen State Park, 38.7772 N, 75.1075 W, Nov 2010, V. Doyle Coll1038; isolated from healthy fruit of Vaccinium macrocarpon, Sussex County, Cape Henlopen State Park, 38.7772 N, 75.1072 W, Nov 2009, V. Doyle, P.V. Oudemans, C. Constantelos Coll877 (BPI 884110, CBS 133132); isolated from rotten fruit of Vaccinium macrocarpon, Sussex County, Cape Henlopen State Park, 38.7770 N, 75.1075 W, Nov 2010, V. Doyle Coll1034 (BPI 884115, CBS 133136); isolated from rotten fruit of Vaccinium macrocarpon, Sussex County, Cape Henlopen State Park, 38.7772 N, 75.1075 W, Nov 2010, V. Doyle Coll1041. New Jersey: isolated from stem tissue of R. virginica in commercial cranberry bog, Burlington County, 39.7546 N, 74.5388 W, Sept 2010, V. Doyle Coll952 (BPI 884109, CBS 133131); isolated from stem tissue of R. virginica in commercial cranberry bog, Burlington County, 39.7309 N, 74.5160 W, July 2010, V. Doyle Coll1306. Maryland: isolated from leaf anthracnose lesions of R. virginica, Prince George’s County, 39.0308 N, 76.7886 W, Sept 2011, V. Doyle & S.A. Rehner Coll1470 (BPI 884107, CBS 133129).
Notes
Colletotrichum rhexiae was described from leaves of Rhexia virginica collected in “Kimensi”, Delaware in 1894. We attempted to find R. virginica in an area thought to be the type locality but were unable to find any host plants. The type material, consisting of R. virginica leaves with circular lesions, is morphologically consistent in vitro with material collected in Cape Henlopen State Park in Delaware on R. virginica and is conspecific with material isolated from circular lesions on the same host in Maryland (Figure 8). We have decided to epitypify C. rhexiae with a living culture based on the necessity of molecular data for species delimitation in Colletotrichum and the need to evaluate morphological characters from a living culture.
Discussion
Sympatric Lineages and Host Distribution in North American Cranberry Bogs
Several phylogenetic studies have confirmed the ability of multiple lineages within Colletotrichum gloeosporioides s.l. to colonize the same host family, genus, or species (e.g. Coffea arabica L. [58]; Jasminum sambac (L.) Aiton [71]; Hemerocallis spp. [72]; Amaryllidaceae [73]). Seven well-supported lineages of C. gloeosporioides s.l. were isolated from North American cranberry bogs, either from V. macrocarpon, V. oxycoccos, or other sympatric host plant species (Table 1 and Figure 1). Similar to the results from previous studies focused on a single host or group of closely related hosts, multiple lineages within the species complex can be isolated from Vaccinium spp., including five species (C. fructivorum, C. rhexiae, C. temperatum, C. melanocaulon, and C. fructicola) as endophytes and/or pathogens of V. macrocarpon, two lineages (C. fructivorum and C. sp. indet. C) from V. oxycoccos, and one species (C. fructicola) from V. corymbosum. Likewise, three species have been found to colonize Rhexia virginica L., an herbaceous perennial inhabiting well-drained soils that are seasonally inundated; often found sympatric with V. macrocarpon in wild and agricultural habitats in eastern North America.
In contrast, a single lineage has been isolated from Nuphar and Nymphaea within and beyond North American cranberry bogs. Colletotrichum nupharicola was previously described from Nuphar lutea subsp. polysepala (Engelm.) E.O. Beal in Washington and Idaho and Nymphaea odorata Aiton in Rhode Island. Our study indicates that this species is also present in irrigation reservoirs in agricultural cranberry beds in New Jersey, growing on Nuphar lutea (L.) Sm. The morphological distinctiveness of C. nupharicola makes it one of the few species within the species complex that can be reliably identified on the basis of conidial and cultural morphology and thus can be more easily tracked. Colletotrichum nupharicola was frequently encountered on Nuphar during the course of this study; however, other lineages were not isolated from either Nuphar or Nymphaea.
Chamaecyparis thyoides (L.) Britton, Sterns & Poggenb. is a common and dominant tree species adjacent to cranberry bogs in parts of eastern North America and can be found as a weed in commercial cranberry beds. Bills and Polishook [25] reported C. gloeosporioides s.l. from Chamaecyparis thyoides collected near cranberry bogs in eastern North America, however they reported just two isolates in a survey of endophytes of the host in New Jersey. A single isolate of C. gloeosporioides s.l. was isolated from C. thyoides in this study and found to be sister to Coll887, isolated from a diseased fruit of V. oxycoccos in West Virginia. These isolates form a phylogenetically distinct but undescribed lineage (C. sp. indet. C). Given the apparent ecological distinctiveness, geographical separation, and relative sequence dissimilarity of these isolates, this lineage will remain undescribed until additional, closely related isolates are collected.
Host Preference, Host Organ Preference, and Habitat Distribution
Species of C. gloeosporioides s.l. are largely considered to be host generalists with few exceptions (such as C. salsolae [16]). There are other species that have been isolated from single host species or genera, but these have been sampled primarily from cultivated plants where isolates may be transferred with plant material or from a narrow geographical range in native habitats (e.g. C. ti [16]; C. psidii [16]). Similarly, strains isolated from V. macrocarpon are distributed throughout the C. gloeosporioides species complex, with little indication of host specificity for most lineages. Two of those lineages, C. temperatum and C. melanocaulon, originate solely from V. macrocarpon, however these were isolated only from cultivated habitats and given further sampling may be found on additional hosts. Colletotrichum temperatum has been isolated as a stem endophyte of V. macrocarpon and from rotten fruit in a horticultural variety (V. macrocarpon cv. Hamilton: not grown for agricultural production). The other species, C. melanocaulon, is associated with an emerging stem canker disease on cranberry where it has been isolated from affected stem tissue collected from field populations in commercial cranberry beds. This study shows, however, that a composite of host, habitat, and host organ origin can be a useful indicator of lineage identity in some cases.
Isolates of C. fructivorum and C. rhexiae are not host specific; C. fructivorum has been isolated from V. macrocarpon, V. oxycoccos and R. virginica, while C. rhexiae has been isolated from V. macrocarpon and R. virginica. Nevertheless, the fact that all isolates originating from V. macrocarpon in wild habitats are conspecific with C. rhexiae, and all isolates from diseased fruit in cranberry agricultural production areas are conspecific with C. fructivorum, suggests that host, host organ, and habitat can be useful indicators of lineage identity. Gonzalez et al. [33] noted a correlation between organ-specific pathogenicity and genotype in isolates from the C. gloeosporioides complex. Given their findings, it appears selection for pathogenicity to fruit may be enabling C. fructivorum to colonize fruit to the exclusion of all other lineages within the species complex that can be found in commercial cranberry beds.
Episodic selection has been proposed to be a common force driving divergence among fungal species [74]–[76]. Host shifts are prominent among the factors responsible for episodic selection and speciation, and has recently been proposed for C. kahawae [59]. In contrast, habitat transformation has been implicated as a factor contributing to population divergence in Colletotrichum cereale, a broadly distributed turfgrass pathogen [77]. The broad distribution of C. fructivorum in commercial cranberry beds and restriction to cultivated habitats suggests that the shift to agricultural production of cranberry may have led to its divergence from C. rhexiae. However, this conclusion is preliminary and would better be addressed with additional sampling and more variable molecular markers.
The only North American lineage of C. gloeosporioides s.l. sympatric with cranberry with some indication of host preference over a broad geographical range is C. nupharicola. While C. nupharicola has been isolated from two aquatic host genera, Nuphar and Nymphaea, both of these genera are in the same family, Nymphaeaceae, indicating host specificity at the family level. This species is broadly distributed on Nymphaeaceae from Alaska through the Rocky Mountains to eastern North America (Dennis A. Johnson, pers. communication, and V. Doyle, unpublished data).
Geographic and Host Distribution of Colletotrichum fructivorum
This and other studies of Colletotrichum gloeosporioides s.l. have demonstrated the utility of delimiting species using multi-locus DNA sequence data (e.g. [14], [16]). However, given the cryptic nature of many species within the complex and the apparently broad diversity of species at small spatial scales it is difficult to design an appropriate sampling scheme to capture the species diversity of Colletotrichum within a given host, habitat or geographic range. To develop a better understanding of scenarios that promote species divergence, it is necessary to understand the factors that lead to population substructure within a focal species. One of the objectives of this study was to identify one or more broadly distributed lineages within the C. gloeosporioides species complex that occur on cranberry to serve as a system for investigation of the factors driving the divergence between populations. Given the widespread occurrence of Colletotrichum gloeosporioides s.l. as the causal agent of fruit-rot in cranberry agricultural areas, the economic importance of cranberry production in North America, and the ease at which C. gloeosporioides s.l. can be isolated from diseased fruit in commercial cranberry production areas, we sampled diseased fruit across several of the major cranberry production areas in North America and Canada (Table 1). All strains of C. gloeosporioides s.l. isolated from fruit in commercial cranberry production areas were found to belong to C. fructivorum regardless of geographic origin. This reveals that C. fructivorum is broadly distributed in North America from Delaware to Massachusetts and west to Washington and British Columbia. In addition, there is evidence that strains of C. fructivorum are not organ specific (fruit and stems) and not host specific (V. macrocarpon, V. oxycoccos, and R. virginica). The combination of economic importance, broad geographic distribution, and diverse organ and host association makes this a suitable species for examining biotic and abiotic factors that influence microevolutionary processes in C. gloeosporioides s.l.
Diversity of North American Lineages
Fungal strains included in this study have been placed in a broader phylogenetic context to better understand the relationships among species within the Colletotrichum gloeosporioides species complex. Despite the fact that there is a significant body of research on North American isolates thought to be members of the species complex (e.g. [8], [23], [24], [27], [33], [78]–[83]) there is a paucity of modern systematic research addressing the phylogenetic and geographic distribution of North American lineages of C. gloeosporioides s.l. However, a recent study by Weir et al. [16] included several North American isolates.
With the inclusion of C. rhexiae, C. temperatum, C. melanocaulon, and C. asianum, our research indicates there is a broader diversity of species in North America than has previously been reported. The species that have been isolated from native North American plant species include C. aeschonymenes, C. asianum, C. clidemiae, C. fructicola, C. fructivorum, C. melanocaulon, C. nupharicola, C. rhexiae, C. temperatum, C. theobromicola, and C. “f. sp. camelliae”, while C. gloeosporioides, C. musae, and C. siamense have been reported from introduced host species. However, there has been very little work addressing the geographic distribution of the species complex in North America. The only species that have been isolated from more than a very localized range are C. fructivorum, C. rhexiae, and C. nupharicola. Our understanding of the importance of the C. gloeosporioides species complex in North America will continue to improve as more North American mycologists and plant pathologists begin to place their isolates in the appropriate phylogenetic context.
Colletotrichum rhexiae and C. fructivorum each present a unique opportunity to investigate the role of host and geography in shaping the microevolutionary patterns that may ultimately lead to species divergence. Colletotrichum rhexiae was described by Ellis and Everhart in the late 1800’s [18] from Delaware while C. fructivorum has been known from the same region since at least 1907 with its formal recognition by Shear [19], [70] as Glomerella rufomaculans var. vaccinii and likely earlier as implicated by the work of Byron Halsted [18]. These species have apparently been sympatric for at least 105 years. While we do not have DNA evidence that C. rhexiae has been present for this extent of time, we have examined the original type material and it is not apparently distinct from our collections of C. rhexiae in New Jersey, Delaware and Maryland. In contrast, we do have a culture deposited to CBS in 1922 by Shear of what he considered to be G. rufomaculans var. vaccinii and it is identical in sequence across 4 loci to isolates collected from cranberry fruit in 5 US states and BC, from cranberry stem in New Jersey, and from Rhexia virginica growing as a weed in agricultural cranberry beds. The presence of sexual reproductive structures in both species suggests the potential for genetic exchange among conspecific individuals (unless they are obligately homothallic). However, despite their sympatry and ability to reproduce sexually, strong statistical support at the bifurcation between C. rhexiae and C. fructivorum indicates the presence of other biological factors reinforcing species limits. However, an objective test of species boundaries is better approached with more rapidly evolving markers variable within species, such as microsatellites, and using more appropriate non-phylogenetic algorithms to determine if there is any evidence of introgression. Further investigation using more rapidly evolving genetic markers could also lend the potential for inferring the reproductive strategy of these species and better understand the factors influencing species divergence within Colletotrichum. In addition, with the advancement of next-generation sequencing technology, it is possible to investigate these sister species at the genomic level to understand how genomic modifications (gene divergence, content/turnover or synteny) may influence pathogenicity on different hosts or in distinct organs.
Conclusions
Colletotrichum gloeosporioides s.l. has long been implicated as a pathogen of cranberry in commercial production areas, but a more refined understanding of the species responsible for fruit-rot of cranberry has not been possible due to the difficulty of species delimitation using morphological features. This study resolves this issue, identifying C. fructivorum as the species associated with fruit-rot in cranberry. This study also lays the groundwork for future studies regarding the natural history and ecology of members of C. gloeosporioides s.l. and should provide useful tools for plant breeders and plant pathologists, in their efforts to develop resistant cultivars for an industry that utilizes one of North America’s few native crop species, V. macrocarpon. We have also determined that C. fructivorum is broadly distributed across North America and Canada in areas of commercial cranberry production and is capable of infecting alternate host species as well. This makes this species an appropriate model for addressing questions of population structure and dispersal at broad geographical and landscape level spatial scales. By virtue of a horizontal (among sympatric species) and vertical (among organs within a species) sampling scheme, we were able to uncover greater diversity of C. gloeosporioides s.l. in wild and commercial cranberry beds than has previously been suggested, revealing seven distinct lineages associated with V. macrocarpon and sympatric host species. Likewise, this level of sampling allowed us to determine that host specificity is not strongly implicated in determining macroevolutionary patterns among these species in temperate regions, with the exception of C. nupharicola. Similarly, despite the generalist nature of the species recovered in this study with respect to organ specificity, C. fructivorum does have an affinity for fruit of V. macrocarpon in commercial cranberry beds, to the exclusion of all other related species.
Supporting Information
Nelsen consensus from maximum parsimony analysis of nrITS gene from D4G. Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of partial beta-tubulin gene from D4G. Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of apn2 gene from D4G . Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of apn2/matIGS gene from D4G . Bootstrap support values shown above branches.
(TIF)
Maximum-likelihood majority-rule consensus tree of nrITS gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of partial beta-tubulin gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of apn2 gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of apn2/matIGS gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Bayesian majority-rule consensus tree with support values (PP/ML-BS) resulting from the combined analysis of D3G+.
(TIF)
GenBank accession numbers for sequence data generated in phylogenetic study of C. gloeosporioides s.l.
(DOCX)
GenBank accession numbers for sequence data generated in previous studies of C. gloeosporioides s.l.
(XLSX)
Discussion of the inferences drawn from the analysis of D3G+.
(DOCX)
Acknowledgments
There are many people to thank for assistance with this work. Discussion with the following people helped to improve the manuscript: Peter Smouse, Larry Kelly, Drew Minnis, and Gary Samuels. The following people provided technical assistance: Natalia Pabon-Mora, Dennis Stevenson, Luiza Maciejak, and Josh Simpson. Chris Constantelos, Jen Vaicunas, Lindsay Wells, Donna Larsen, Patty McManus, Frank Caruso, Rob Hiller, Chris Bennett, Kent Karriker, Christine Worthington, Brian Mouza, Kim Patten, Eric Zeldin, Roger Spots, and Christina Mozzicato assisted with collections. We also thank two anonymous reviewers for their assistance in improving the manuscript.
Funding Statement
Financial support from the National Science Foundation [www.nsf.gov] (DBI 0749751 and DEB 1011120 - Doctoral Dissertation Improvement Grant), the Torrey Botanical Society Graduate Research Fellowship [www.torreybotanical.org], and The New York Botanical Garden [www.nybg.org] helped to fund this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Morris CE, Bardin M, Kinkel LL, Moury B, Nicot PC, et al. (2009) Expanding the Paradigms of Plant Pathogen Life History and Evolution of Parasitic Fitness beyond Agricultural Boundaries. PLoS Pathogens 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey JA, Jeger MJ, editors (1992) Colletotrichum: Biology, Pathology and Control: Wallingford, UK: CAB International.
- 3. Latunde-Dada AO (2001) Colletotrichum: tales of forcible entry, stealth, transient confinement and breakout. Molecular Plant Pathology 2: 187–198. [DOI] [PubMed] [Google Scholar]
- 4. Guozhong L, Cannon PF, Reid A, Simmons CM (2004) Diversity and molecular relationships of endophytic Colletotrichum isolates from the Iwokrama Forest Reserve, Guyana. Mycological Research 108: 53–63. [DOI] [PubMed] [Google Scholar]
- 5. Hyde KD, Cai L, McKenzie EHC, Yang YL, Zhang JZ, et al. (2009) Colletotrichum: a catalogue of confusion. Fungal Diversity 39: 1–17. [Google Scholar]
- 6. Hyde KD, Cai L, Cannon PF, Crouch JA, Crous PW, et al. (2009) Colletotrichum - names in current use. Fungal Diversity 39: 147–182. [Google Scholar]
- 7.Sutton BC (1992) The genus Glomerella and its anamorph Colletotrichum. In: Bailey JA, Jeger MJ, editors. Colletotrichum: Biology, Pathology and Control. Surrey, UK: CAB International. 1–26.
- 8. Oudemans PV, Caruso FL, Stretch AW (1998) Cranberry fruit rot in the Northeast: A complex disease. Plant Disease 82: 1176–1184. [DOI] [PubMed] [Google Scholar]
- 9.Adaskaveg JE, Forster H (2000) Occurrence and Management of Anthracnose Epidemics Caused by Colletotrichum Species on Tree Fruit Crops in California. In: Prusky D, Freeman S, Dickman MB, editors. Colletotrichum: Host Specificity, Pathology and Host-Pathogen Interaction. St. Paul, Minnesota: The American Phytopathogical Society Press. 317–336.
- 10.Legard DE (2000) Colletotrichum diseases of Strawberry in Florida. In: Prusky D, Freeman S, Dickman MB, editors. Colletotrichum: Host Specificity, Pathology, and Host-Pathogen Interaction. St. Paul, Minnesota: American Phytopathological Society. 292–299.
- 11. von Arx JA (1957) Die Arten der Gattung Colletotrichum Corda. Phytopathologische Zeitschrift 29: 414–468. [Google Scholar]
- 12. Du M, Schardl CL, Nuckles EM, Vaillancourt LJ (2005) Using mating-type gene sequences for improved phylogenetic resolution of Colletotrichum species complexes. Mycologia 97: 641–658. [DOI] [PubMed] [Google Scholar]
- 13. Crouch JA, Clarke BB, Hillman BI (2006) Unraveling the evolutionary relationships among the divergent lineages of Colletotrichum causing anthracnose disease in turfgrass and corn. Phytopathology 96: 46–60. [DOI] [PubMed] [Google Scholar]
- 14. Rojas EI, Rehner SA, Samuels GJ, Van Bael SA, Herre EA, et al. (2010) Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panamá: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102: 1318–1338. [DOI] [PubMed] [Google Scholar]
- 15. Silva DN, Talhinas P, Várzea V, Cai L, Paulo OS, et al. (2012) Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.). Mycologia 104: 396–409. [DOI] [PubMed] [Google Scholar]
- 16. Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cannon PF, Buddie AG, Bridge PD (2008) The typification of Colletotrichum gloeosporioides . Mycotaxon 104: 189–204. [Google Scholar]
- 18.Halsted B (1889) Some fungus diseases of the cranberry. New Jersey Agricultural College Experiment Station. 1–40.
- 19.Shear CL (1907) Cranberry Diseases. In: USDA, editor: Government Printing Office.
- 20. Stiles CM, Oudemans PV (1997) Cranberry fruit rot pathogens isolated from various cranberry plant tissues over two growing seasons. Phytopathology 87: S94. [Google Scholar]
- 21.Zhu P (1999) Genetic diversity and systematics of Colletotrichum species on cranberry (Vaccinium macrocarpon) [Ph.D.]. New Brunswick: Rutgers University. 143 p.
- 22. Olatinwo RO, Hanson EJ, Schilder AMC (2003) A First Assessment of the Cranberry Fruit Rot Complex in Michigan. Plant Disease 87: 550–556. [DOI] [PubMed] [Google Scholar]
- 23. Polashock JJ, Caruso FL, Oudemans PV, McManus PS, Crouch JA (2009) The North American cranberry fruit rot fungal community: a systematic overview using morphological and phylogenetic affinities. Plant Pathology 58: 1116–1127. [Google Scholar]
- 24. Ellis JB, Everhart BM (1894) New Species of Fungi from Various Localities Proceedings of the Academy of Natural Sciences of Philadelphia. 46: 322–386. [Google Scholar]
- 25. Bills G, Polishook JD (1992) Recovery of endophytic fungi from Chamaecyparis thyoides . Sydowia 44: 1–12. [Google Scholar]
- 26. Johnson DA, Carris LM, Rogers JD (1997) Morphological and molecular characterization of Colletotrichum nymphaeae and C. nupharicola sp. nov. on water-lilies (Nymphaea and Nuphar). Mycological Research 101: 641–649. [Google Scholar]
- 27. Stiles CM, Oudemans PV (1999) Distribution of cranberry fruit-rotting fungi in New Jersey and evidence for nonspecific host resistance. Phytopathology 89: 218–225. [DOI] [PubMed] [Google Scholar]
- 28. Rodriguez-Saona C, Vorsa N, Singh AP, Johnson-Cicalese J, Szendrei Z, et al. (2011) Tracing the history of plant traits under domestication in cranberries: potential consequences on anti-herbivore defences. Journal of Experimental Botany 62: 2633–2644. [DOI] [PubMed] [Google Scholar]
- 29. Vorsa N, Polashock JJ (2005) Alteration of Anthocyanin Glycosylation in Cranberry Through Interspecific Hybridization. Journal of the American Society for Horticultural Science 130: 711–715. [Google Scholar]
- 30.Vorsa N (1994) Breeding the American Cranberry. In: Roper TR, editor. Wisconsin State Cranberry Growers Association.
- 31. Freeman S, Katan T, Shabi E (1996) Characterization of Colletotrichum gloeosporioides isolates from avocado and almond fruits with molecular and pathogenicity tests. Applied and Environmental Microbiology 62: 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Photita W, Taylor PWJ, Ford R, Hyde KD (2005) Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand. Fungal Diversity 18: 117–133. [Google Scholar]
- 33. González E, Sutton TB, Correll JC (2006) Clarification of the etiology of Glomerella Leaf Spot and Bitter Rot of apple caused by Colletotrichum spp. based on morphology and genetic, molecular, and pathogenicity tests. Phytopathology 96: 982–992. [DOI] [PubMed] [Google Scholar]
- 34. Agostini JP, Timmer LW (1994) Population dynamics and survival of strains of Colletotrichum gloeosporioides on Citrus in Florida. Phytopathology 84: 420–425. [Google Scholar]
- 35. Chakraborty S, Fernandes CD, Charchar MJdA, Thomas MR (2002) Pathogenic variation in Colletotrichum gloeosporioides infecting Stylosanthes spp. in a center of diversity in Brazil. Phytopathology 92: 553–562. [DOI] [PubMed] [Google Scholar]
- 36. Colon-Garay J, Rivera-Vargas L, McGovern RJ, Rodriquez RdP (2002) Hypovirulent isolates of Colletotrichum gloeosporioides induce resistance to anthracnose in detached mango fruits and seedlings. J Agric Univ Puerto Rico 84: 56–64. [Google Scholar]
- 37. Kaufmann PJ, Weidemann GJ (1996) Isozyme analysis of Colletotrichum gloeosporioides from five host genera. Plant Disease 80: 1289–1293. [Google Scholar]
- 38.MacKenzie SJ (2005) Population structure and pathogenicity of Colletotrichum gloeosporioides from strawberry and noncultivated hosts in Florida [Ph.D.]. United States – Florida: University of Florida.
- 39.White TJ, Bruns T, Lee JS, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. New York: Academic Press, Inc. 315–322.
- 40. O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- 41. Katoh K, Kuma K-I, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298. [DOI] [PubMed] [Google Scholar]
- 43. Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- 44.Miller MA, Pfeifer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees; New Orleans, LA. 1–8.
- 45. Stamatakis A (2006) RAxML-VI-HPC: Maximum Likelihood-based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 46. Stamatakis A, Hoover P, Rougemont J (2008) A Fast Bootstrapping Algorithm for the RAxML Web-Servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- 47. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 48. Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 49.Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 50. Than PP, Shivas RG, Jeewon R, Pongsupasamit S, Marney TS, et al. (2008) Epitypification and phylogeny of Colletotrichum acutatum J.H. Simmonds. Fungal Diversity 28: 97–108. [Google Scholar]
- 51. Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, et al. (2010) Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity 44: 33–43. [Google Scholar]
- 52. Simmonds JH (1965) A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Queensland Journal of Agricultural and Animal Science 25: 437–459. [Google Scholar]
- 53. Sukumaran J, Holder MT (2010) DendroPy: A Python library for phylogenetic computing. Bioinformatics 26: 1569–1571. [DOI] [PubMed] [Google Scholar]
- 54.Rambaut A, Drummond AJ (2007) Tracer v.1.4. Beast website. Available: http://beast.bio.ed.ac.uk/Tracer. Accessed March 25, 2013.
- 55. Ané C, Larget B, Baum DA, Smith SD, Rokas A (2007) Bayesian estimation of concordance among gene trees. Molecular Biology and Evolution 24: 412–426. [DOI] [PubMed] [Google Scholar]
- 56. Larget BR, Kotha SK, Dewey CN, Ané C (2010) BUCKy: Gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26: 2910–2911. [DOI] [PubMed] [Google Scholar]
- 57. Phoulivong S, Cai L, Parinn N, Chen H, Abd-Elsalam KA, et al. (2010) A new species of Colletotrichum from Cordyline fruticosa and Eugenia javanica causing anthracnose disease. Mycotaxon 114: 247–257. [Google Scholar]
- 58. Prihastuti H, Cai L, Chen H, McKenzie EHC, Hyde KD (2009) Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity 39: 89–109. [Google Scholar]
- 59. Silva DN, Talhinas P, Cai L, Manuel L, Gichuru EK, et al. (2012) Host-jump drives rapid and recent ecological speciation of the emergent fungal pathogen Colletotrichum kahawae . Molecular Ecology 21: 2655–2670. [DOI] [PubMed] [Google Scholar]
- 60. Dettman JR, Jacobson DJ, Taylor JW (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora . Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- 61. Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. (2000) Phylogenetic Species Recognition and Species Concepts in Fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- 62. Cummings MP, Neel MC, Shaw KL (2008) A genealogical approach to quantifying lineage divergence. Evolution 62: 2411–2422. [DOI] [PubMed] [Google Scholar]
- 63.Bazinet AL, Myers DS, Khatavkar P (2009) genealogicalSorting v.0.91. Genealogical sorting index website. Available: http://www.genealogicalsorting.org/. Accessed March 31, 2013.
- 64.R-Development-Core-Team (2011) R: A Language and Environment for Statistical Computing.
- 65. Nirenberg H (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land-und Forstwirtschaft 169: 1–117. [Google Scholar]
- 66.Rasband WS (1997–2008) ImageJ. Bethesda, Maryland, USA: National Institutes of Health.
- 67. Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- 68. Seifert KA (2009) Progress towards DNA barcoding of fungi. Molecular Ecology Resources 9: 83–89. [DOI] [PubMed] [Google Scholar]
- 69. Crouch JA, Clarke BB, Hillman BI (2009) What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate spored graminicolous Colletotrichum group. Mycologia 101: 648–656. [DOI] [PubMed] [Google Scholar]
- 70. Shear CL (1907) New species of fungi. The Bulletin of the Torrey Botanical Club 34: 305–317. [Google Scholar]
- 71. Wikee S, Cai L, Pairin N, McKenzie EHC, Su YY, et al. (2011) Colletotrichum species from Jasmine (Jasminum sambac). Fungal Diversity 46: 171–182. [Google Scholar]
- 72. Yang YL, Liu ZY, Cai L, Hyde KD (2012) New species and notes of Colletotrichum on daylilies (Hemerocallis spp.). Tropical Plant Pathology 37: 165–174. [Google Scholar]
- 73. Yang YL, Liu ZY, Cai L, Hyde KD, Yu ZN, et al. (2009) Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity 39: 123–146. [Google Scholar]
- 74. Brasier CM (1995) Episodic selection as a force in fungal microevolution, with special reference to clonal speciation and hybrid introgression. Canadian Journal of Botany 73: S1213–S1221. [Google Scholar]
- 75. Giraud T, Gladieux P, Gavrilets S (2010) Linking the emergence of fungal plant diseases with ecological speciation. Trends in Ecology and Evolution 25: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gladieux P, Guerin F, Giraud T, Caffier V, Lemaire C, et al. (2011) Emergence of fungal pathogens by ecological speciation: importance of the reduced viability of immigrants. Molecular Ecology 20: 4521–4532. [DOI] [PubMed] [Google Scholar]
- 77. Crouch JA, Tredway LP, Clarke BP, Hillman BI (2009) Phylogenetic and population genetic divergence correspond with habitat for the pathogen Colletotrichum cereale and allied taxa across diverse grass communities. Molecular Ecology 18: 123–135. [DOI] [PubMed] [Google Scholar]
- 78. Chilton SJP, Lucas GB, Edgerton CW (1945) Genetics of Glomerella III: Crosses with a conidial strain. American Journal of Botany 32: 549–554. [Google Scholar]
- 79. Edgerton CW (1914) Plus and minus strains in the Genus Glomerella . American Journal of Botany 1: 244–254. [Google Scholar]
- 80. Edgerton CW, Chilton SJP, Lucas GB (1945) Genetics of Glomerella II: Fertilization between strains. American Journal of Botany 32: 115–118. [Google Scholar]
- 81. MacKenzie SJ, Seijo TE, Legard DE, Timmer LW, Peres NA (2007) Selection for pathogenicity to strawberry in populations of Colletotrichum gloeosporioides from native plants. Phytopathology 97: 1130–1140. [DOI] [PubMed] [Google Scholar]
- 82. Shear CL, Wood A (1907) Ascogenous forms of Gloeosporium and Colletotrichum . Botanical Gazette 43: 259–266. [Google Scholar]
- 83. Wheeler HE, McGahen JW (1952) Genetics of Glomerella X: Genes affecting sexual reproduction. American Journal of Botany 39: 110–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nelsen consensus from maximum parsimony analysis of nrITS gene from D4G. Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of partial beta-tubulin gene from D4G. Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of apn2 gene from D4G . Bootstrap support values shown above branches.
(TIF)
Nelsen consensus from maximum parsimony analysis of apn2/matIGS gene from D4G . Bootstrap support values shown above branches.
(TIF)
Maximum-likelihood majority-rule consensus tree of nrITS gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of partial beta-tubulin gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of apn2 gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Maximum-likelihood majority-rule consensus tree of apn2/matIGS gene from D4G . Bootstrap support values shown above branches. Outgroups (C. aff. acutatum and strains 4766, 3386, and 4801) have been trimmed from the tree. Branch lengths represent the mean across samples.
(TIF)
Bayesian majority-rule consensus tree with support values (PP/ML-BS) resulting from the combined analysis of D3G+.
(TIF)
GenBank accession numbers for sequence data generated in phylogenetic study of C. gloeosporioides s.l.
(DOCX)
GenBank accession numbers for sequence data generated in previous studies of C. gloeosporioides s.l.
(XLSX)
Discussion of the inferences drawn from the analysis of D3G+.
(DOCX)