Abstract
Dimeric basic leucine zipper (bZIP) proteins are conserved transcriptional enhancers found in all eukaryotes. A recently reported and novel function for bZIPs is association of these proteins with secondary metabolite production in filamentous fungi. In particular a Yap-like bZIP termed RsmA (restorer of secondary metabolism A) was identified in Aspergillus nidulans that positively regulates the carcinogen sterigmatocystin. To assess for conserved function for RsmA, we examined a role of this protein in secondary metabolism in the pathogen A. fumigatus. RsmA was found to positively regulate gliotoxin where overexpression (OE) of rsmA led to 2–100 fold increases of twelve gli cluster metabolites in culture medium including the newly identified gli metabolite cyclo(L-Phe-L-Ser). Lungs from both wild type and OErsmA infected mice contained gliotoxin (2.3 fold higher in OErsmA treatment) as well as the gliotoxin precursor cyclo(L-Phe-L-Ser) (3.2 fold higher in OErsmA treatment). The data here presents a conserved role for RsmA in secondary metabolite cluster activation and suggests cyclo(L-Phe-L-Ser) may serve as an alternative marker for diagnosis of invasive aspergillosis.
Introduction
Aspergillus fumigatus is an opportunistic pathogen of immunosuppressed individuals. In immunosuppressed hosts, invasive aspergillosis (IA) represents a major cause of morbidity and mortality, with mortality rates ranging from 40% to 90% [1], [2]. Part of the virulence of A. fumigatus is associated with its ability to produce a variety of toxic natural products. The natural product pathways in fungi result in the production of structurally diverse and low-molecular-weight compounds referred to as secondary metabolites (SMs) [3]. Although the functions of many fungal SMs remain to be determined, the known SMs of the genus Aspergillus have a tremendous impact on society. Compounds such as lovastatin (antihypercholesterolemic agent) and penicillin (antibiotic) are of clinical importance whereas the carcinogen, aflatoxin, causes both short term toxicities and long term liver cancer when humans eat contaminated crops [3].
Biosynthetic genes for individual SMs are typically clustered, and regulation of these clustered genes is often dependent on pathway-specific transcription factors. For example, the aflatoxin and related sterigmatocystin gene clusters contain one gene, aflR, that encodes a transcription factor regulating expression of the remaining biosynthetic genes [4], [5]; similarly gliZ encodes the transcription factor required for expression of the gliotoxin (gli) cluster genes in A. fumigatus [6]. In addition, SM cluster activation is also regulated at a higher hierarchic level by the nuclear protein LaeA [7]. Overexpression of laeA results in increase in the production of a multitude of SMs while deletion of laeA yields opposite results in many fungi [7]–[10]. Studies of the human pathogen, A. fumigatus, showed SMs such as gliotoxin are upregulated when laeA is overexpressed, and that deletion of laeA results in decreased virulence in both neutropenic and non-neutropenic murine models [6], [11].
The well-known A. fumigatus SM gliotoxin, a mycotoxin readily detected in IA, has immunosuppressive and apoptotic activities in the host [12]–[14]. Deletant strains of either gliZ or gliP (encoding a non-ribosomal peptide synthetase, NRPS, required for gliotoxin synthesis), are less damaging to host tissue during in vitro studies and insect model studies [15]–[17]. Additionally, these mutants show reduced virulence in non-neutropenic murine models but not neutropenic models [6], [18]–[20]. This toxin and/or its derivatives are also considered for use for diagnosis of IA [21], [22]. Aside from gliZ, no other transcription factors have been described to regulate the gli cluster.
Recently, several bZIP transcription factors have been found to regulate secondary metabolism in several fungal genera [23]–[27]. For example, a suppressor mutagenesis in A. nidulans identified rsmA, encoding a bZIP transcriptional enhancer, that when overexpressed could partially restore secondary metabolism in ΔlaeA strains [28]. Overexpression of rsmA (OErsmA) increased sterigmatocystin production in A. nidulans more than 40 fold through transcriptional activation of aflR [29]. Here we find that overexpression of the A. fumigatus rsmA ortholog, also called rsmA, increases gliotoxin synthesis to a similar fold as sterigmatocystin in OErsmA strains of A. nidulans. This increase is mediated via transcriptional activation of gliZ. In addition to gliotoxin, the gli cluster derivative cyclo(L-Phe-L-Ser) was found to accumulate in murine lungs from both OErsmA and wild type infected mice and may represent an alternative marker for diagnosis of IA. Furthermore, supporting previous research data that indicates gliotoxin negatively impacts neutrophil migration [30] we found that supernatants of A. fumigatus OErsmA but not those of OErsmAΔgliZ or wild type inhibit human neutrophil chemotaxis.
Materials and Methods
Ethics Statement
Neutrophils were obtained from whole blood of self-reportedly healthy donors, from which we obtained informed and written consent at the time of the blood draw with approval of the University of Wisconsin-Madison Center for Health Sciences Human Subjects committee. The Keller animal protocol number is M02468 as approved by the University of Wisconsin-Madison Research Animal Resources Center.
Fungal Strains and Culture Conditions
The strains used in this study are listed in Table 1 and plasmids are listed in Table 2. All fungal strains were maintained as glycerol stocks, and cultured at 25°C or 37°C on glucose minimal medium (GMM) [31] with appropriate supplements.
Table 1. Strains of Aspergillus fumigatus used in this study.
| Strains | Genotype | Source |
| AF293 | Wild-type | [51] |
| AF293.1 | pyrG1 | [51] |
| AF293.6 | pyrG1, argB1 | [51] |
| TDWC5.6 | pyrG1, ΔgliZ::pyrG | [43] |
| TJMP19.2 | pyrG1, argB1, ΔlaeA::argB | This study |
| TJMP36.2 | pyrG1, argB1, ΔlaeA::argB, pyrG | This study |
| TJMP54.16 | pyrG1, gpdA(p)::rsmA::pyrG | This study |
| TJMP52.2 | pyrG1, argB1, ΔlaeA::argB, gpdA(p)::rsmA::pyrG | This study |
| TJMP56.6 | pyrG1, argB1, gpdA(p)::rsmA::pyrG | This study |
| TRSR40.4 | pyrG1, argB1, gpdA(p)::rsmA::pyrG, ΔgliZ::argB | This study |
| TRSR1.2 | pyrG1, ΔrsmA::pyrG | This study |
Table 2. Plasmids used in this study.
Nucleic Acid Analysis and Molecular Genetic Manipulations
Extraction of fungal DNA and RNA, restriction enzyme digestion, gel electrophoresis, Southern and northern blotting, hybridization and probe preparation were performed using standard methods [32]. Primers for PCR and probes are listed in Table 3.
Table 3. Oligonucleotides used in this study.
| Primer Name | Sequence (5′-3′) | Purpose |
| JP Afumi argB For | GAACGCGGTCTGCATCCAAG | Create pJMP4/argB |
| JP Afumi argB Rev | GAAGGAGAGACCCATACATCC | Create pJMP4/argB |
| JP Apara pyrG Short For | GCTTGATATCGAATTCTCATGTTTG | Create pJMP7 |
| JP Apara pyrG Short Rev | TACGGAAGCTTATACGAACAGATGG | Create pJMP7 |
| JP gpdA(p) EcoRI For | TTTCGAATTCCATCCGGATGTCGAAGG | Create pJMP8 and pRS1 |
| JP gpdA(p) BamHI Rev | CTTGTGGATCCCGTCATCTTTGAAA | Create pJMP8 |
| MK Afumi Bzip Fusion Rev | ATTGGAATAAAATGAGTAGTCCATGGTGATGTCTGCTCAAGCGG | Create pRS1 |
| MK Afumi Bzip Fusion For | CCGCTTGAGCAGACATCACCATGGACTACTCATTTTATTCCAAT | Create pRS1 |
| MK Afumi Bzip rev (EcoRI) | GTTGTACTAGAAGAGAATTCGCAGTC | Create pRS1 |
| RS rsmA 5′F For | GTGAAGACGGCAATCTCTGAG | ΔrsmA 5′ flank |
| RS rsmA 5′ F+pyrg tail Rev | CCCTATAGTGAGTCGTATTACGATCTGTGCACTTCCCTCAAC | ΔrsmA 5′ flank |
| RS pyrG +3′F rsmA For | CGATGATAAGCTGTCAAACATGAGCCTTGGCAGGGCGAATTTGG | ΔrsmA 3′ Flank |
| RS rsmA 3′ F Rev | CAGCAAATCGTTCGTGCTCG | ΔrsmA 3′ Flank |
| RS rsmA screen For | GATCTACAACGCTTCGACGGAC | PCR screen rsmA |
| JP Af-laeA 5′ Fl For | CCTCGTTGAACTAATTGGCG | ΔlaeA 5′ Flank |
| JP Af-laeA 5′ Fl Rev | GGAAAATTTGTCTTGGATGCAGTCCGCGTTCAGGTCGAGGAGGTCCAATC | ΔlaeA 5′ Flank |
| JP Af-laeA 3′ Fl For | AGATCAAATGGATGTATGGGTCTCTCCTTCGCTTCAAACCTCTGAGATCG | ΔlaeA 3′ Flank |
| JP Af-laeA 3′ Fl Rev | CGACACACATATCATGACGG | ΔlaeA 3′ Flank |
| FY GZ 5′F FOR | GAGTTCTCGCTAGTGTCTGG | ΔgliZ 5′ Flank |
| RS ArgB tail +5′Rev | GTCTTGGATGCAGACCGCGTTCCGCAGTGCAACACGGGTGGC | ΔgliZ 5′ Flank |
| RS GliZ 3′ Fl For | CGCAGTGCAACACGGGTGGCGTCTTGGATGCAGACCGCGTTC | ΔgliZ 3′ Flank |
| FY GZ 3′F REV | CGAGTCAACATGATGACGG | ΔgliZ 3′ Flank |
| RS gliZ 5′ For | TGAGCCATACTGAAGTCTCCGTG | ΔgliZ nested |
| RS gliZ 3′ Rev | GACATCGACTGAACATGTCCACG | ΔgliZ nested |
| RS Af RsmA Int For | CAATCCTCAGTCACAGCAGC | rsmA probe |
| RS Af RsmA Int Rev | GGTCATATCAATTCATCGCGGC | rsmA probe |
| JP Modified T7 Promoter For | CGTAATACGACTCACTATAGGG | Amplify pyrG from pJMP7 |
| RS A.para pyrG Rev | CTCATGTTTGACAGCTTATCATCG | Amplify pyrG from pJMP7 |
| RS rsmA RT-PCR For | ACCGAGCTGCTCAGCGAGCG | rsmA RT-PCR |
| RS rsmA RT-PCR Rev | CTGGCCCTCCCTGAACGCCG | rsmA RT-PCR |
| RS actin RT-PCR For | GTCACCATGGTATCATGATTGG | actin RT-PCR |
| RS actin RT-PCR Rev | GGTAGTCCGTCAGATCACGG | actin RT-PCR |
| RS laeA RT-PCR For | CCAGTAGCAAGAATCCTGAC | laeA RT-PCR |
| RS laeA RT-PCR Rev | CCGGAGCCCATCTGCATGTG | laeA RT-PCR |
| RS gliZ int For | GCACCTACAGCTATTCCTCG | gliZ probe |
| RS gliZ int Rev | GTCACGGCCATGCTAATACTG | gliZ probe |
| RS actin int For | GTATGTCGGTGATGAGGCAC | Actin probe |
| RS actin int Rev | CCGTAGAGATCCTTACGGAC | Actin probe |
| GIINTF | TGTTGATCGAGACGCCGTTCTG | gliI probe |
| GIINTR | CAGAGCGGCTCGATTCTGGTG | gliI probe |
| RS gliT For | GCAAACTACTCTCCAACGGAG | gliT probe |
| RS gliT Rev | GCAAACTACTCTCCAACGGAG | gliT probe |
Construction of Plasmids
A 1.5-kb PCR fragment of the A. nidulans gpdA promoter was amplified from pTMH44.2 [33] using primers JP gpdA(p) EcoRI For and JP gpdA(p) BamHI Rev and subsequently cloned into pBluescript II SK- (Fermentas) to generate pJMP8. A plasmid containing the selectable marker gene for complementation of the argB1 allele in A. fumigatus was generated by PCR amplifying argB (Afu4g07190) from gDNA of AF293 with the primers JP Afumi argB For and JP Afumi argB Rev, the resulting PCR product was cloned into pCR-Blunt II-TOPO (Invitrogen) to create pJMP4. To construct a pyrG1 complementation plasmid, a 2.0-kb A. parasiticus pyrG PCR product was amplified from pJW24 [34] using primers JP Apara pyrG Short For and JP Apara pyrG Short Rev and subsequently cloned into pBluescript II SK- (Fermentas) to generate pJMP7. A plasmid to over-express rsmA was created by fusion PCR [35] of the gpdA promoter (JP gpdA(p) EcoRI For and MK Afumi Bzip Fusion Rev) and a 1.5-kb rsmA product (MK Afumi Bzip Fusion For and MK Afumi Bzip EcoRI Rev). The resulting 3.0-kb fusion PCR product was cloned into pJMP7 with EcoRI to generate pRS1.
Construction of Fungal Strains
Fungal transformation was done as previously described [36]. Gene replacement constructs for laeA, rsmA, and gliZ were made using double-joint PCR [35], [37]. Briefly, approximately 1-kb flanking regions were PCR amplified from gDNA of AF293 and fused to either the A. fumigatus argB gene from pJMP4 or the A. parasiticus pyrG gene from pJMP7 (primers are listed in Table 3). The laeA gene was disrupted with A. fumigatus argB in the AF293.6 (pyrG1, argB1) background to create TJMP19.2 (ΔlaeA, pyrG1) and TJMP19.2 was subsequently transformed with pJMP7 to create the prototrophic TJMP36.2 (ΔlaeA) strain. These strains were confirmed by Southern blot analysis (data not shown). Deletion of rsmA was achieved by replacement with the A. parasiticus pyrG gene in AF293.1 (pyrG1) to generate TRSR1.2 which was confirmed by Southern blot (Figure S1B). Over expression of rsmA (OErsmA) was achieved by transformation of pRS1 into AF293.1, TJMP19.2, and AF293.6, which created TJMP54.16 (OErsmA), TJMP52.2 (ΔlaeA, OErsmA), and TJMP56.6 (OErsmA, argB1) respectively (Figure S1A). To create a double OErsmAΔgliZ strain, gliZ was replaced with the A. fumigatus argB gene in TJMP56.6 resulting in TRSR40.4 (Figure S1C).
Gene Expression Analysis
107 fungal spores/ml were grown in liquid GMM at either 37°C, 250 rpm for 48 h or 25°C, 280 RPM for 72 h and total RNA was extracted via Isol-RNA Lysis Reagent (5 Prime) as previously described [36]. For semi-quantitative reverse transcriptase PCR, cDNA was generated using the iScript kit (Bio Rad) following manufacturers recommendations.
Physiological Studies
104 conidia from strains were point-inoculated onto solid glucose minimal media (GMM) in four replicates and incubated in continuous dark at 25°C for 12 days and at 37°C for 4 days. Radial growth was assayed by measuring the diameter of point-inoculated colonies after 4 days and 12 days incubation at 37°C and 25°C, respectively. Conidia were counted from 2 cm agar cores taken from the center of the point-inoculated cultures and enumerated using the hemocytometer.
Secondary Metabolite Analysis
Secondary metabolite production was assessed by thin-layer chromatography (TLC) from cultures grown in liquid glucose minimal medium (GMM) for 3 days at 25°C and at 280 rpm in triplicate. Secondary metabolites from the organic layer were extracted with chloroform and the air-dried extracts were resuspended in 100 µl of methanol and resolved in chloroform: acetone (7∶3) on UV infused TLC plates. Gliotoxin (Sigma) was spotted as standard. The TLC plates were then visualized at 366 nm and 254 nm.
HPLC-MS Profiling of Compounds 1–13 in Wild Type and OErsmA Cultures
(a) Liquid culture preparation: 107 fungal spores/ml were grown in triplicates in liquid GMM (50 mL) at 25°C, 280 rpm for 72 h. Three 50 mL liquid culture supernatant samples of both wild type and OErsmA were frozen and lyophilized. To each of the lyophilized samples, 50 mL of a mixture consisting of 80% acetonitrile, 15% ethyl acetate, and 5% H2O was added. After stirring for 2 h, the mixtures were filtered over extraction solvent-washed cotton. The extraction process was repeated twice more using 10 mL of the extraction solvent. The solvent was removed using rotary evaporation. (b) Liquid culture analysis: The evaporated samples were dissolved in 250 µL of 10% water in acetonitrile, and 2 µL of the resulting solution was injected into an Agilent 1100 series HPLC. The HPLC system, equipped with an Agilent Zorbax Eclipse XDB-C8 column (4.6×150 mm, 5 µm particle diameter), running a water-acetonitrile gradient (2 mL/min) starting with 5% acetonitrile for 5 min, followed by a linear increase to 36% acetonitrile over 25 min, followed by a rapid linear increase to 100% over 10 min, and ending with 10 min at 100% acetonitrile. The eluent of the HPLC was split (1∶10) into a Quattro II electrospray ionization (ESI) mass spectrometer operated in ESI positive (ESI+) ionization mode. Single ion monitoring (SIM) was used to increase sensitivity by monitoring the major ions produced by each analyte. Enriched samples of 1–13 served as standards to determine retention time and ionization/fragmentation for each analyte (enriched standards were obtained as described [38]). (c) Data processing: Ion chromatogram peak areas were measured for each analyte and used to assess the relative amounts in OErsmA compared to wild type.
Oxidative Stress Tolerance Assays
For studying the relative sensitivities of wild type and mutant strains to oxidative stress, 105 conidia in 5 µl were inoculated on GMM with 20, 30 and 40 µM menadione. Menadione was filter sterilized and added to agar medium at ∼50°C before solidification. Plates were incubated at 37°C for 48 h. GMM without menadione served as a control.
Neutrophil Recruitment
Neutrophil recruitment assays were performed using a microfluidic chemotaxis platform developed and described in more detail elsewhere [39]. In brief, the device is able to generate a linear gradient of soluble factors between a source channel containing the compounds to be tested and a sink channel containing the neutrophils. The gradient is generated in a thin cross-channel in which neutrophils from the sink channel can invade. The quantification was performed using software developed in-house (source code and software can be obtained upon request). The neutrophil purification was performed with a gradient centrifugation process using Polymorphprep solution, according to the manufacturer’s recommendations (Nycomed, Sheldon, UK). The neutrophils were re-suspended to a concentration of 4 million per mL in PBS, and kept no more than 2 h at 4 degrees Celsius. The microfluidic devices were prepared by filling them with modified HBSS (HBSS solution containing 2% HEPES buffer and 0.1% HAS), after which 4 µL of neutrophil suspension was added in each sink channel of each device, and finally 3 µL of the crude supernatants was inserted into each source channel of each device. The negative control was performed by adding blank GMM (the fungal growth medium) to the source channel, and the positive control was performed by adding a solution of 100 nM fMLP (F3506, Sigma Aldrich) in GMM.
Animal Model of Aspergillus Infection
(a) A. fumigatus wild type and OErsmA strains were assessed in a lung infection model of invasive aspergillosis. Briefly, female Swiss ICR mice (Harlan Sprague Dawley) weighing about 18–20 g were immunosuppressed via administration of cyclophosphamide by separate intraperitoneal injections, one at 4 days (200 mg/kg of body weight) and the other at 1 day (200 mg/kg) before infection. The treatment included administration of cortisone acetate at a dose of 250 mg/kg by separate intraperitoneal injection at 1 day before infection. Anesthetized mice (10 mice/fungal strain) were infected by nasal instillation of 50 µl of 1×107 conidia/ml (day 1) and monitored three times daily for 7 days postinfection. The control group was inoculated with saline (0.85% NaCl). All surviving mice were sacrificed at day 7. Health and comfort of animals was assessed twice a day by monitoring feeding and water intake. Animals were housed in the Microbial Science Building Vivarium. If animals were ill (decreased activity, difficulty moving, ruffled fur, decreased movement with stimulation) they were euthanized via CO2 inhalation. (b) Assessment of presence or absence of compounds 1–13 in the lungs of mice infected with wild type and OErsmA: Seven mice were infected per A. fumigatus strain as described above. Dissected lungs were lyophilized, and powdered using a metal spatula. Lung samples were extracted with 50 mL of acetonitrile and filtered over acetonitrile-washed cotton. Solvent was removed using rotary evaporation. Samples were redissolved in 20 µl acetonitrile, and 2 µL was injected into the HPLC-ESI+-SIMMS system described above. The chromatographic gradient consisted of water-acetonitrile starting with 5 min 5% acetonitrile, followed by a linear increase to 20% acetonitrile over 15 min, followed by a linear increase to 100% acetonitrile over 10 min, ending with 10 min at 100% acetonitrile. Enriched samples of 1–13 served as retention time and ionization standards.
Statistical Analysis
Statistical differences were analyzed using the JMP software package version 3.2.6 (SAS Institute, Inc., Cary, NC). Multiple comparisons of results for all strains were calculated for growth diameter and sporulation. Statistically significant mean values, indicated with different letters in the figures, are significant at P<0.0001.
Results
rsmA is a Low Expressed Gene
After confirming the creation of both the deletion of rsmA and gliZ and overexpression of rsmA in the appropriate backgrounds (Figure S1), northern blot analysis was carried out to assess rsmA expression in all strains. mRNA transcript level was barely detectable in wild type (WT, Figure S2A) and semi-quantitative RT-PCR was employed for confirmation of expression (Figure S2B). The OErsmA mutant showed multiple fold increase in mRNA transcript level, verifying the overexpression construct (Figure S2). The low expression of rsmA in wild type has also been characterized in A. nidulans [28].
OErsmA Exhibits a Growth Defect at Low Growth Temperature
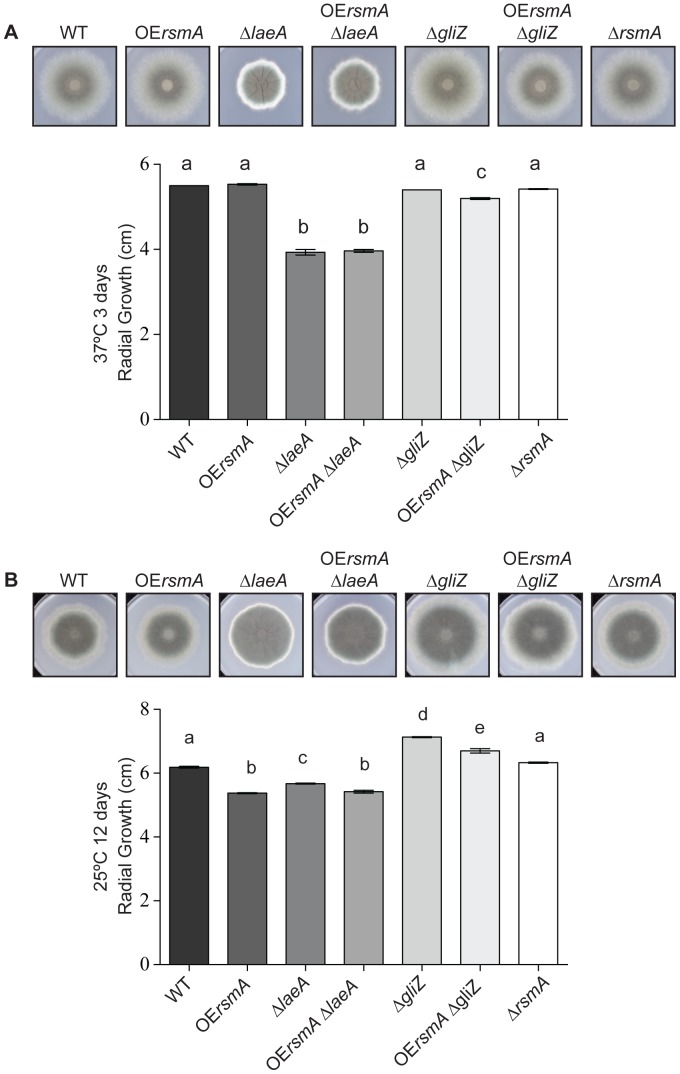
At 37°C, the radial growth of OErsmA strain is similar to that of wild type (Figure 1A). However, at 25°C, the radial growth of OErsmA is decreased compared to that of the wild-type strain (Figure 1B). Similar results were observed for the OErsmAΔgliZ and OErsmAΔlaeA strains, with a significant decrease in radial growth at 25°C compared to the ΔgliZ and ΔlaeA control strains. The radial growths of strains in the ΔlaeA background were decreased at both temperatures compared to wild type. No significant differences were observed in spore production between ΔrsmA, the OErsmA background strains and their respective controls (Figure S3).
Figure 1. Average radial growth of A. fumigatus strains.
104 conidia of each strain were point inoculated on GMM and grown at 37°C for 4 days and at 25°C for 12 days (Figure 1). Radial growth was measured at the end of each growth period. Means ± standard deviations are indicated for four replicates of each strain. Levels not connected by same letter are significantly different (P<0.0001) according to ANOVA analysis.
OErsmA Mutants are Resistant to Menadione
An examination of RsmA function in A. nidulans showed that RsmA binds a Yap1 DNA-binding site [29]. Although A. nidulans RsmA showed no response to oxidative stress [40], other fungal Yap-like bZIP proteins are often associated with the reactive oxygen species (ROS) response [41], [42]. To assess any possible role of A. fumigatus rsmA on oxidation stress, conidia of OErsmA, OErsmAΔlaeA, OErsmAΔgliZ and ΔrsmA mutants and their controls were inoculated on GMM with varying concentrations of menadione and incubated at 37°C for 48 h. All strains containing an OErsmA allele grew better than their respective controls on 20 and 30 µM menadione, regardless of the absence of either the laeA or gliZ allele, thus showing increased resistance of these strains to ROS induced by menadione (Figure 2).
Figure 2. OErsmA mutants are resistant to menadione.
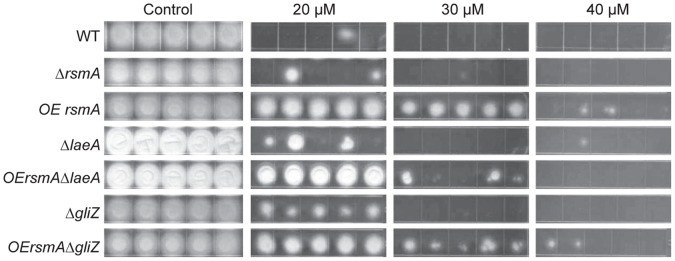
For each strain, 105 conidia in 5 µl were spotted on GMM plates with 20 µM, 30 µM and 40 µM of menadione, or on GMM only for a control. Each strain was replicated 5 times. The plates were incubated at 37°C for 48 h.
OErsmA Increases Production of Multiple gli Pathway Metabolites
In A. nidulans, overexpression of rsmA increased the production of sterigmatocystin 40–100 fold dependent on culture condition [28]. To examine any possible conserved role of RsmA in A. fumigatus, we examined SM production in several cultural conditions. First strains were grown in gliotoxin inducing conditions (liquid GMM for 3 days, shaking at 25°C). Here the OErsmA strain produced more gliotoxin than wild type, and no gliotoxin was visibly detected in ΔlaeA background strains by thin layer chromatography, nor was gliotoxin production rescued in the OErsmAΔlaeA double mutant (Figure S4). As expected the increase of gliotoxin was dependent on presence of the gliZ gene as observed by gliotoxin loss in the OErsmAΔgliZ mutant. By eye, ΔrsmA produced a similar TLC profile as wild type. Extracts from strains grown at 37°C on solid medium did not exhibit noticeable differences in SM profiles (data not shown).
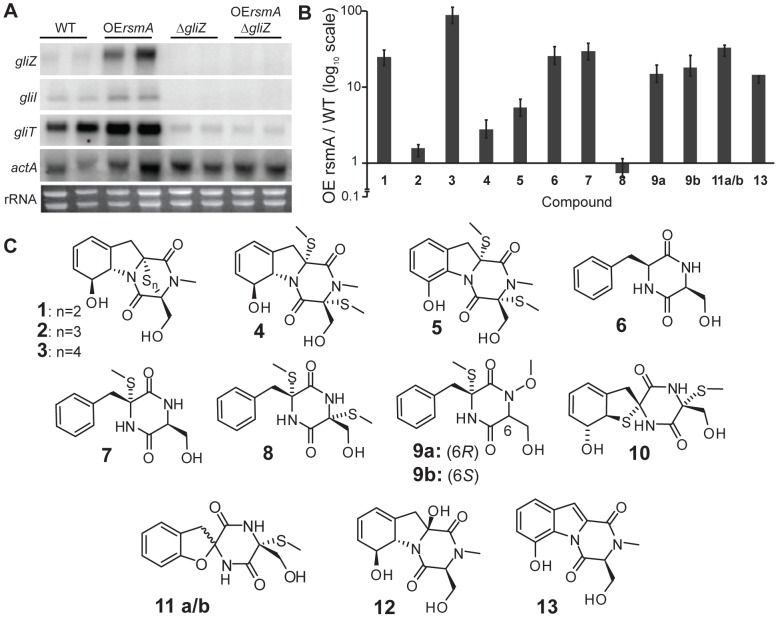
An examination of gliZ, gliI (a biosynthetic gene in the gli cluster) and gliT (encoding a gliotoxin oxidase providing endogenous protection against gliotoxin) expression at gliotoxin inducing conditions verified a positive impact of rsmA on gliotoxin gene expression where all three genes were upregulated in the OErsmA background (Figure 3A).
Figure 3. RsmA regulates production of gliotoxin and is dependent on gliZ.
A. gliZ, gliI and gliT gene expression in liquid shaking cultures. gliZ, gliI and gliT expression in A. fumigatus (AF293) wild type, OErsmA, ΔgliZ, and OErsmAΔgliZ. All strains were grown in 50 mL GMM liquid shaking culture at 25°C and 280 rpm for 72 h. Total RNA was extracted from mycelia and probed for gliZ, gliI and gliT. WT = wild type. B. Relative amounts of compounds 1–13 in OErsmA compared to WT. Compounds 10 and 12 were not detected in the OErsmA nor WT cultures. Compounds 11a/b and 13 were not detected in wild type, their values represent the measured S/N ratio as determined by their diagnostic mass ion chromatograms. Data was triplicated and the error bars represent 1 standard deviation. C. Collection of gliZ-dependent metabolites profiled in this study, structures and gliZ-dependency in A. fumigatus was previously established in [38].
The increased expression of gliZ in the OErsmA strain was of considerable interest as GliZ has been found to be required for the production of a multitude of gli cluster metabolites, including the family of diketopiperazines shown in Figure 4C (1–13) [38]. We found that compounds 6, 7, 9, 11, 13, and the gliotoxins 1 and 3 are at least 10-fold up-regulated in OErsmA, whereas the methylsulfanyl containing compounds 4, 5, and 8 are much less or not at all upregulated (Figure 4B). The methylsulfanyl groups in compounds 4, 5, and 8 likely arise from SAM-dependent methylation of free thiols. Their relatively lower abundance compared to the other compounds in OErsmA logically follows that of overexpression of gliT in OErsmA (Figure 3A), the gene encoding the oxidase responsible for disulfide formation from dithiol-containing metabolites. Overexpression of gliT would be expected to prevent the accumulation of large amounts of free thiols. We further found that production of the four sulfur atom-containing gliotoxin G (3) is dramatically increased in OErsmA whereas the three sulfur atom-containing gliotoxin E (2) is only modestly up-regulated, suggesting that the biosynthesis of different gliotoxin derivatives may be regulated independently.
Figure 4. Neutrophil chemotaxis is reduced when exposed to extracts from OErsmA.
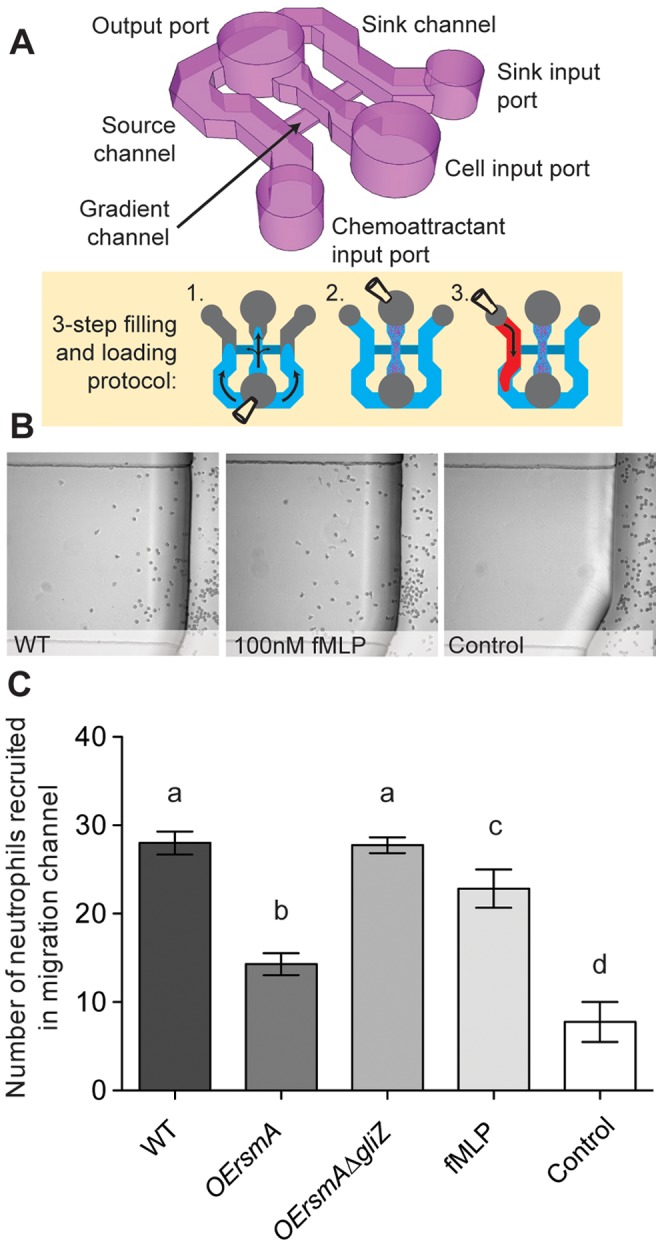
A. Schematic of the neutrophil migration platform used. The neutrophils are placed in the center channel; the compound to be tested is placed in one of the side channels, while the other channel acts as a negative control. The device is prepared in 3 steps: (1) filling, (2) adding neutrophils, and (3) adding the fungal culture supernatants. B. Representative microscopy images of the migration area after 1 h incubation. Supernatant of wild type A. fumigatus (AF WT) has chemoattractive properties on par with the known chemoattractant fMLP. C. Quantification of neutrophil recruitment to the crude supernatant of wild type, OErsmA, and OErsmAΔgliZ strains (significant differences at P<0.5 are indicated by different letters).
gli Cluster Metabolites Decrease Neutrophil Migration
As an earlier study had suggested gliotoxin inhibited neutrophil chemotaxis [30], the supernatants from wild type, OErsmA and OErsmAΔgliZ were assessed for the ability to alter neutrophil migration. Results show that neutrophils are strongly recruited to the supernatant of A. fumigatus wild type, in a magnitude on par with the recruitment towards a known chemotactic compound, fMLP at 100 nM (Figure 4). The supernatant of OErsmA, however, induces a significant decrease in neutrophil migration. The decrease is rescued in the OErsmAΔgliZ strain. These results suggest that gliotoxin, or a metabolite in the gliotoxin pathway, is the cause of the neutrophil migration defect observed in the over-expressing rsmA strain.
Detection of Cyclo(L-Phe-L-Ser) in Murine Infection
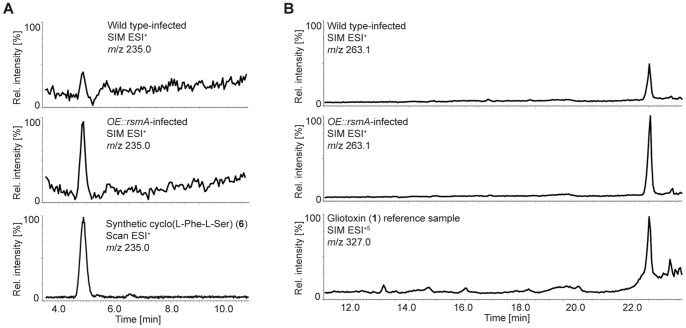
Gliotoxin has previously been isolated from the murine lung environment [43], however it is unknown if gliotoxin intermediates also accumulate in vivo. To test this hypothesis, we conducted a neutropenic IA mouse assay with wild type and OErsmA strains and assayed for the presence or absence of compounds 1–13 from murine lungs. There was no significant difference in virulence between wild type versus OErsmA in the neutropenic model of IA (Figure S5). Lungs from wild type and OErsmA-infected mice were homogenized and extracted and HPLC-ESI+-SIMMS analysis showed that gliotoxin (1) and cyclo(L-Phe-L-Ser) (6), were present in lung tissue from both wild type and OErsmA treated mice. Both compounds were present in higher amounts (2- and 3-fold, respectively) in OErsmA lungs (Figure 5A and 5B). Compound 6 has not previously been observed in A. fumigatus-lung infection models. The absence of the remaining gliZ-dependent compounds, 2–5 and 7–13 in the wild type and OErsmA-infected lung tissue suggests that these compounds are not produced in amounts large enough for detection or that they were not detected due to decomposition, as the methylsulfanyl groups in 4, 5, and 7–11 are easily substituted by other nucleophiles, including water [38].
Figure 5. Infected mouse lung extracts (IMLE) HPLC- single ion monitoring (SIM)MS ion chromatograms.
HPLC-SIMMS analysis of crude mouse lung extracts corresponding to mice infected with wild type, or OErsmA. (A) Ion chromatogram showing compound 6 is approximately two to three times as abundant in the OErsmA-IMLE relative to WT-IMLE. (B) Similarly, gliotoxin is about two times as abundant in OErsmA-IMLE than WT-IMLE. Reference chromatograms (bottom panels) show diagnostic ions of cyclo(L-Phe-L-Ser) (6) and gliotoxin (1). Lung extract chromatograms are scaled to the peaks measured in the OErsmA-IMLE sample (bottom panels of standards are not to scale).
Discussion
Our interest in exploring RsmA in impacting secondary metabolism in A. fumigatus arose from studies using the model genetic system, A. nidulans. RsmA (restorer of secondary metabolism A) was identified in a mutagenesis screen of an A. nidulans ΔlaeA strain where overexpression of rsmA (OErsmA) partially rescued secondary metabolism in a ΔlaeA genetic background and greatly over produced the carcinogen sterigmatocystin in a laeA wild type background [28]. LaeA, a conserved protein in filamentous fungi, governs the production of multiple SMs that contribute to virulence in all pathogenic fungi examined to date including A. fumigatus [6], [8], [10], [44]. Here we found that although OErsmA did not restore gliotoxin synthesis in the ΔlaeA background in A. fumigatus, rsmA overexpression did have a profound effect on gli cluster gene expression and concomitant metabolite production, mediated by GliZ, the gli gene cluster transcription factor.
In A. nidulans, RsmA binds to the promoter and activates expression of aflR, encoding the transcription factor required for sterigmatocystin and aflatoxin synthesis [29]. The A. fumigatus RsmA ortholog shares overall 58% identity to that of A. nidulans and 100% identity in the DNA binding region suggesting that both proteins bind to the same DNA sites, several of which are found in the gli gene cluster and possibly involved in the activation observed in this study. Several bZIP proteins have recently been found to regulate fungal SM clusters – both positively and negatively - including the bZIPs AtfB and ApYap1 regulating aflatoxin in A. parasiticus [24], [25], AoYap1 regulating ochratoxin in A. ochraceus [23], MeaB regulating bikaverin in Fusarium fujikuroi [26] and BcAft1 regulating botrydial, botryendial and botcinin A in Botrytis cinerea [27]. A distinguishing characteristic of RsmA, in both A. nidulans and A. fumigatus, is the extreme up-regulation of a primary SM in each species. In A. nidulans OErsmA led to 40-fold induction of sterigmatocystin, an anti-predation metabolite protecting this species from fungivory [29]. Here OErsmA increased production of a multitude of GliZ dependent metabolites 10 to near 100 fold over wild type. Gliotoxin is a potent anti-fungal thought to help establish a niche for A. fumigatus in natural settings [45]; it may be that RsmA represents a bZIP pathway hardwired for defensive SM production in these species [29].
One difference between OErsmA in A. nidulans and A. fumigatus was the role of this protein in the stress response of the two species. bZIP factors are associated with the stress response in fungi, such as NapA in A. nidulans [41] and Afyap1 in A. fumigatus [42], [46]. Despite the lack of a stress response in the A. nidulans rsmA mutants [40], the A. fumigatus strains containing an OErsmA allele showed increased resistance to menadione, an indicator of resistance to reactive oxidative species (ROS), than the control strains. This was irrespective of presence of gliZ or laeA (Figure 2), the latter result in contrast to the non-affect of OErsmA on secondary metabolism in the ΔlaeA background. These observations suggest that RsmA directly or indirectly activates genes that are involved in oxidative stress response of A. fumigatus. In the animal host, the fungus faces a variety of toxic environmental challenges, which are aimed at eliminating the fungus. For example, neutrophils produce high levels of reactive oxygen species (ROS) during oxidative burst. However, pathogenic fungi possess oxidative-stress response genes that help to evade the otherwise lethal elevation of ROS. However, because the OErsmA strain showed no difference compared to the WT in the murine model, this factor of enhanced resistance to ROS likely has small impact on virulence of OErsmA. This is reminiscent of the A. fumigatus Afyap1 mutant which while Afyap1 is important for resistance to ROS, the deletant strain did not exhibit a pathogenicity defect [42]. Possibly these two proteins play a redundant role in A. fumigatus ROS resistance.
The inhibition of neutrophil chemotaxis with OErsmA extracts supports earlier studies where gliotoxin or A. fumigatus extracts were found to diminish PMN chemotaxis [30], [47]. Currently the mechanism of this impact of chemotaxis is not known and could simply be attributable to the apoptotic properties of gliotoxin and, perhaps, other gli cluster metabolites. The great increase in gli cluster metabolites as observed in culture grown OErsmA was tempered in the animal model (ca. 10 fold less increase in murine lung). This may explain differences in the results from the in vivo (no difference in pathogenicity in the murine model) and in vitro (inhibition of chemotaxis) results of the OErsmA strain in this study.
In terms of gli cluster regulation, the greatly enhanced production of numerous gli cluster metabolites in OErsmA was striking and supported and extended our earlier studies showing that gliotoxin is not the only metabolite produced by the gli gene cluster [38]. Here we show that in addition to gliotoxin, cyclo(L-Phe-L-Ser) (6) accumulates in lungs of IA mice. Two pertinent observations from this study include further support for GliT as an oxidase responsible for disulfide formation from dithiol-containing metabolites. Notably, the lesser accumulation of the methylsulfanyl containing compounds (4, 5, and 8) would be expected in the OErsmA as gliT expression was particularly upregulated in this strain. GliT is also responsible for A. fumigatus self-resistance to gliotoxin, and likely other gli metabolites [48], [49] and, along with bis(dethio)bis(methylthio)gliotoxin (4) [22], has been proposed as an antigen for diagnosis of IA [50]. Our work here reveals an additional metabolite, cyclo(L-Phe-L-Ser) (6), that could serve as a marker for IA diagnosis. This compound, along with gliotoxin, is produced in high amounts by the fungus both in vivo and in vitro and, at least in vitro, is chemically more stable than gliotoxin or bis(dethio)bis(methylthio)gliotoxin (4).
Supporting Information
Overexpression and deletion of rsmA , and deletion of a transcription factor of gliotoxin biosynthesis, gliZ. A. Southern blot analysis of the OErsmAΔlaeA, OErsmA and wild type (WT) strains. Genomic DNA was digested with KpnI. Expected hybridization band patterns: 3.5 kb and 5.6 kb for mutants, and 3.5 kb for wild type strain. B. Southern blot analysis of the ΔrsmA mutant strains and wild type (WT) strain. Genomic DNA was digested with BglI and PvuII. Expected hybridization band patterns: 5.572 kb (BglI), 4.571 kb and 2.457 kb (PvuII) for mutants; 1.633 kb and 3.149 kb (BglI), 6.038 kb (PvuII) for wild type strain. C. Southern blot analysis of the ΔgliZ and OErsmAΔgliZ mutant strains and wild type(WT) strain. Genomic DNA was digested with SpeI and EagI. Expected hybridization band patterns: 5.17 kb (SpeI) and 6.21 kb (EagI) for mutants; 4.03 kb (SpeI), 1.13 kb and 3.96 kb (EagI) for wild type strain. The arrowheads denote the correct mutants.
(TIF)
Detection of rsmA by Northern blot and reverse transcriptase- PCR (RT-PCR). A. Northern blot analysis of A. fumigatus wild type (WT), ΔlaeA, OErsmAΔlaeA and OErsmA strains. 107 spores/ml were inoculated in triplicates into liquid GMM and incubated at 37°C shaking for 48 h. Mycelia were collected, frozen in liquid nitrogen and lyophilized overnight. Total RNA was isolated. An internal fragment of the rsmA open reading frame was used as a probe. As a control, RNA blots were also hybridized with an internal fragment of the gpdA gene. B. Reverse transcriptase-PCR of wild type (WT), ΔlaeA, OErsmAΔlaeA and OErsmA strains. RNA of each strain was extracted from freeze-dried mycelia, treated with DNase1, reverse transcribed and the resultant cDNA was amplified with primers specific for rsmA, laeA and actin (serves as a control).
(TIF)
Average sporulation of A. fumigatus strains. 104 conidia of each strain were point inoculated on GMM and grown under dark conditions at 25°C for 12 days (A) and at 37°C for 4 days (B). Agar plugs were extracted from the center of the colonies, homogenized in water and spores counted using a haemocytometer. Means ± standard errors are indicated for four replicates of each strain. Levels not connected by same letter are significantly different (P>0.15) according to the student’s t test.
(TIF)
Thin-layer chromatography profiles of secondary metabolites produced by A. fumigatus wild type (WT), OE rsmA , ΔlaeA , OE rsmAΔlaeA, ΔgliZ, OE rsmAΔgliZ and ΔrsmA strains. Secondary metabolites were extracted by chloroform from cultures grown in liquid GMM at 25°C, 280 rpm for 3 days. Dried extracts were resuspended in 100 µl of methanol, and 10 µl was used for separation on TLC plate. All strains were triplicated. The solvent condition was chloroform:acetone (7∶3), and the plates were visualized at 254 nm (top) and 366 nm (bottom). G, gliotoxin standard.
(TIF)
Virulence of OErsmA in a murine lung infection model. 10 female Swiss ICR mice immunosuppressed by intraperitoneal injection of cyclophosphamide (200 mg/kg) and cortisone acetate (250 mg/kg) were inoculated intranasally with 50 µl of 1×107 conidia/ml of A. fumigatus wild type (AF293) and OErsmA and control (saline). Pairwise comparisons indicated no significant differences from WT vs OErsmA (P = 0.1924).
(TIF)
Funding Statement
This research was supported in part by National Institutes of Health (NIH) (R01 Al065728-01 to NPK and GM008500 to RRF), a DuPont Young Professor Award (to FCS) and a grant from the American Asthma Foundation to NPK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dagenais TRT, Keller NP (2009) Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22: 447–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Latgé JP (2001) The pathobiology of Aspergillus fumigatus . Trends Microbiol 9: 382–389. [DOI] [PubMed] [Google Scholar]
- 3. Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24: 393–416. [DOI] [PubMed] [Google Scholar]
- 4. Woloshuk CP, Foutz KR, Brewer JF, Bhatnagar D, Cleveland TE, et al. (1994) Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol 60: 2408–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandes M, Keller NP, Adams TH (1998) Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol 28: 1355–1365. [DOI] [PubMed] [Google Scholar]
- 6. Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, et al. (2005) LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell 4: 1574–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bok JW, Keller NP (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amaike S, Keller NP (2009) Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus . Eukaryot Cell 8: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butchko RAE, Brown DW, Busman M, Tudzynski B, Wiemann P (2012) Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides . Fungal Genet Biol 49: 602–612. [DOI] [PubMed] [Google Scholar]
- 10. Wu D, Oide S, Zhang N, Choi MY, Turgeon BG (2012) ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus . PLoS Pathog 8: e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sugui JA, Pardo J, Chang YC, Müllbacher A, Zarember KA, et al. (2007) Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus . Eukaryot Cell 6: 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewis RE, Wiederhold NP, Lionakis MS, Prince RA, Kontoyiannis DP (2005) Frequency and species distribution of gliotoxin-producing Aspergillus isolates recovered from patients at a tertiary-care cancer center. J Clin Microbiol 43: 6120–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pahl HL, Krauss B, Schulze-Osthoff K, Decker T, Traenckner EB, et al. (1996) The immunosuppressive fungal metabolite gliotoxin specifically inhibits transcription factor NF-kappaB. J Exp Med 183: 1829–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanzani M, Orciuolo E, Lewis RE, Kontoyiannis DP, Martins SL, et al. (2005) Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood 105: 2258–2265. [DOI] [PubMed] [Google Scholar]
- 15. Ramirez-Ortiz ZG, Lee CK, Wang JP, Boon L, Specht CA, et al. (2011) A nonredundant role for plasmacytoid dendritic cells in host defense against the human fungal pathogen Aspergillus fumigatus . Cell Host Microbe 9: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Speth C, Kupfahl C, Pfaller K, Hagleitner M, Deutinger M, et al. (2011) Gliotoxin as putative virulence factor and immunotherapeutic target in a cell culture model of cerebral aspergillosis. Mol Immunol 48: 2122–2129. [DOI] [PubMed] [Google Scholar]
- 17. Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, et al. (2008) Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis 197: 479–486. [DOI] [PubMed] [Google Scholar]
- 18. Kupfahl C, Heinekamp T, Geginat G, Ruppert T, Hartl A, et al. (2006) Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol Microbiol 62: 292–302. [DOI] [PubMed] [Google Scholar]
- 19. Cramer RA, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, et al. (2006) Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell 5: 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, et al. (2007) Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot Cell 6: 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Puri A, Ahmad A, Panda BP (2010) Development of an HPTLC-based diagnostic method for invasive aspergillosis. Biomed Chromatogr 24: 887–892. [DOI] [PubMed] [Google Scholar]
- 22. Domingo MP, Colmenarejo C, Martínez-Lostao L, Müllbacher A, Jarne C, et al. (2012) Bis(methyl)gliotoxin proves to be a more stable and reliable marker for invasive aspergillosis than gliotoxin and suitable for use in diagnosis. Diagn Microbiol Infect Dis 73: 57–64. [DOI] [PubMed] [Google Scholar]
- 23. Reverberi M, Gazzetti K, Punelli F, Scarpari M, Zjalic S, et al. (2012) Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus . Appl Microbiol Biotechnol 95: 1293–1304. [DOI] [PubMed] [Google Scholar]
- 24. Reverberi M, Zjalic S, Punelli F, Ricelli A, Fabbri AA, et al. (2007) Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit Contam 24: 1070–1075. [DOI] [PubMed] [Google Scholar]
- 25. Roze LV, Chanda A, Wee J, Awad D, Linz JE (2011) Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J Biol Chem 286: 35137–35148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wagner D, Schmeinck A, Mos M, Morozov IY, Caddick MX, et al. (2010) The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specific loci. Eukaryot Cell 9: 1588–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Temme N, Oeser B, Massaroli M, Heller J, Simon A, et al. (2012) BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea . Molecular Plant Pathology 13: 704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaaban MI, Bok JW, Lauer C, Keller NP (2010) Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell 9: 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin W-B, Amaike S, Wohlbach DJ, Gasch AP, Chiang Y-M, et al. (2012) An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR . Mol Microbiol 83: 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shah DT, Jackman S, Engle J, Larsen B (1998) Effect of gliotoxin on human polymorphonuclear neutrophils. Infect Dis Obstet Gynecol 6: 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimizu K, Keller NP (2001) Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans . Genetics 157: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 33. McDonald T, Brown DW, Keller NP, Hammond TM (2005) RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact 18: 539–545. [DOI] [PubMed] [Google Scholar]
- 34. Calvo AM, Bok JW, Brooks W, Keller NP (2004) veA is required for toxin and sclerotial production in Aspergillus parasiticus . Appl Environ Microbiol 70: 4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. [DOI] [PubMed] [Google Scholar]
- 36. Palmer JM, Perrin RM, Dagenais TR, Keller NP (2008) H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot Cell 7: 2052–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, et al. (2006) Fusion PCR and gene targeting in Aspergillus nidulans . Nat Protoc 1: 3111–3120. [DOI] [PubMed] [Google Scholar]
- 38. Forseth RR, Fox EM, Chung D, Howlett BJ, Keller NP, et al. (2011) Identification of cryptic products of the gliotoxin gene cluster using NMR-based comparative metabolomics and a model for gliotoxin biosynthesis. J Am Chem Soc 133: 9678–9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D (2010) An arrayed high-content chemotaxis assay for patient diagnosis. Integr Biol 2: 630–638. [DOI] [PubMed] [Google Scholar]
- 40. Yin W-B, Reinke AW, Szilágyi M, Emri T, Chiang Y-M, et al. (2013) bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans . Microbiology 159: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Asano Y, Hagiwara D, Yamashino T, Mizuno T (2007) Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans . Biosci Biotechnol Biochem 71: 1800–1803. [DOI] [PubMed] [Google Scholar]
- 42. Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, et al. (2007) The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell 6: 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bok JW, Chung D, Balajee SA, Marr KA, Andes D, et al. (2006) GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun 74: 6761–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, et al. (2010) FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol 77: 972–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carberry S, Molloy E, Hammel S, O’Keeffe G, Jones GW, et al. (2012) Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet Biol 49: 302–312. [DOI] [PubMed] [Google Scholar]
- 46. Qiao J, Kontoyiannis DP, Calderone R, Li D, Ma Y, et al. (2008) Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclophosphamide and triamcinolone. Med Mycol 46: 773–782. [DOI] [PubMed] [Google Scholar]
- 47. Murayama T, Amitani R, Ikegami Y, Nawada R, Lee WJ, et al. (1996) Suppressive effects of Aspergillus fumigatus culture filtrates on human alveolar macrophages and polymorphonuclear leucocytes. Eur Respir J 9: 293–300. [DOI] [PubMed] [Google Scholar]
- 48. Scharf DH, Remme N, Heinekamp T, Hortschansky P, Brakhage AA, et al. (2010) Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus . J Am Chem Soc 132: 10136–10141. [DOI] [PubMed] [Google Scholar]
- 49. Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, et al. (2010) Self-protection against gliotoxin–a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog 6: e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shi L-n, Li F-q, Huang M, Lu J-f, Kong X-X, et al. (2012) Immunoproteomics based identification of thioredoxin reductase GliT and novel Aspergillus fumigatus antigens for serologic diagnosis of invasive aspergillosis. BMC Microbiol 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue T, Nguyen CK, Romans A, Kontoyiannis DP, May GS (2004) Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch Microbiol 182: 346–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression and deletion of rsmA , and deletion of a transcription factor of gliotoxin biosynthesis, gliZ. A. Southern blot analysis of the OErsmAΔlaeA, OErsmA and wild type (WT) strains. Genomic DNA was digested with KpnI. Expected hybridization band patterns: 3.5 kb and 5.6 kb for mutants, and 3.5 kb for wild type strain. B. Southern blot analysis of the ΔrsmA mutant strains and wild type (WT) strain. Genomic DNA was digested with BglI and PvuII. Expected hybridization band patterns: 5.572 kb (BglI), 4.571 kb and 2.457 kb (PvuII) for mutants; 1.633 kb and 3.149 kb (BglI), 6.038 kb (PvuII) for wild type strain. C. Southern blot analysis of the ΔgliZ and OErsmAΔgliZ mutant strains and wild type(WT) strain. Genomic DNA was digested with SpeI and EagI. Expected hybridization band patterns: 5.17 kb (SpeI) and 6.21 kb (EagI) for mutants; 4.03 kb (SpeI), 1.13 kb and 3.96 kb (EagI) for wild type strain. The arrowheads denote the correct mutants.
(TIF)
Detection of rsmA by Northern blot and reverse transcriptase- PCR (RT-PCR). A. Northern blot analysis of A. fumigatus wild type (WT), ΔlaeA, OErsmAΔlaeA and OErsmA strains. 107 spores/ml were inoculated in triplicates into liquid GMM and incubated at 37°C shaking for 48 h. Mycelia were collected, frozen in liquid nitrogen and lyophilized overnight. Total RNA was isolated. An internal fragment of the rsmA open reading frame was used as a probe. As a control, RNA blots were also hybridized with an internal fragment of the gpdA gene. B. Reverse transcriptase-PCR of wild type (WT), ΔlaeA, OErsmAΔlaeA and OErsmA strains. RNA of each strain was extracted from freeze-dried mycelia, treated with DNase1, reverse transcribed and the resultant cDNA was amplified with primers specific for rsmA, laeA and actin (serves as a control).
(TIF)
Average sporulation of A. fumigatus strains. 104 conidia of each strain were point inoculated on GMM and grown under dark conditions at 25°C for 12 days (A) and at 37°C for 4 days (B). Agar plugs were extracted from the center of the colonies, homogenized in water and spores counted using a haemocytometer. Means ± standard errors are indicated for four replicates of each strain. Levels not connected by same letter are significantly different (P>0.15) according to the student’s t test.
(TIF)
Thin-layer chromatography profiles of secondary metabolites produced by A. fumigatus wild type (WT), OE rsmA , ΔlaeA , OE rsmAΔlaeA, ΔgliZ, OE rsmAΔgliZ and ΔrsmA strains. Secondary metabolites were extracted by chloroform from cultures grown in liquid GMM at 25°C, 280 rpm for 3 days. Dried extracts were resuspended in 100 µl of methanol, and 10 µl was used for separation on TLC plate. All strains were triplicated. The solvent condition was chloroform:acetone (7∶3), and the plates were visualized at 254 nm (top) and 366 nm (bottom). G, gliotoxin standard.
(TIF)
Virulence of OErsmA in a murine lung infection model. 10 female Swiss ICR mice immunosuppressed by intraperitoneal injection of cyclophosphamide (200 mg/kg) and cortisone acetate (250 mg/kg) were inoculated intranasally with 50 µl of 1×107 conidia/ml of A. fumigatus wild type (AF293) and OErsmA and control (saline). Pairwise comparisons indicated no significant differences from WT vs OErsmA (P = 0.1924).
(TIF)