Abstract
Porphyromonas gingivalis is a Gram-negative oral anaerobe which is strongly associated with periodontal disease. Environmental changes in the gingival sulcus trigger the growth of P. gingivalis and a concurrent shift from periodontal health to disease. Bacteria adjust their physiology in response to environmental changes and gene regulation by two-component phospho-relay systems is one mechanism by which such adjustments are effected. In P. gingivalis RprY is an orphan response regulator and previously we showed that the RprY regulon included genes associated with oxidative stress and sodium metabolism. The goals of the present study were to identify environmental signals that induce rprY and clarify the role of the regulator in the stress response. In Escherichia coli an RprY-LacZ fusion protein was induced in sodium- depleted medium and a P. gingivalis rprY mutant was unable to grow in similar medium. By several approaches we established that sodium depletion induced up-regulation of genes involved in oxidative stress. In addition, we demonstrated that RprY interacted directly with the promoters of several molecular chaperones. Further, both genetic and transcription data suggest that the regulator acts as a repressor. We conclude that RprY is one of the regulators that controls stress responses in P. gingivalis, possibly by acting as a repressor since an rprY mutant showed a superstress reponse in sodium-depleted medium which we propose inhibited growth.
Introduction
Periodontitis is a chronic inflammatory disease induced by polymicrobial infection of the periodontium. If untreated, the infection can lead to tooth destabilization due to destruction of supporting tissues. Porphyromonas gingivalis is a gram negative, commensal, black-pigmented, anaerobic oral bacterium and is widely considered as one of the causative agents of periodontitis due to its strong association with the disease [1]. Accordingly, this organism is studied extensively and several factors that contribute to its pathogenicity have been characterized [2]. P. gingivalis is part of the commensal microflora of the supra- and subgingival plaque biofilm [3] which can colonize gingival epithelium without apparent disease activity [4], [5]. The shift from periodontal health to a pathogenic state may partly depend on changes in the gingival microenvironment caused by early colonizing bacteria [6], [7], as well as host conditions including changes in ion concentration, exposure to blood, saliva, and host immunological factors.
Bacterial cells perceive and respond to environmental stimuli in order to ensure their survival. One type of response mechanism is mediated by two-component signal transduction in which a sensor, a transmembrane histidine kinase, perceives a change in the cell environment, and autoactivates by phosphorylation. The activation signal is relayed by phospho-transfer to activate the cognate response regulator, a DNA-binding protein that regulates transcription of response-specific genes. The RprY response regulator of P. gingivalis is the focus of the present study. RprY is an “orphan” in that the usual close genetic linkage to a candidate cognate kinase gene is absent, and hence, neither cognate kinase nor activating signals are yet known. However, in Bacteroidetes and other oral pathogens, the rprY homolog is found together with its cognate kinase gene rprX, and when the rprXY two component system (TCS) from B. fragilis was expressed in E. coli it conferred low level tetracycline resistance that correlated with a decrease in ompF expression suggesting that rprY may belong to EnvZ-OmpR family of transcriptional regulators [8].
Our previous investigation showed that in P. gingivalis several genes associated with oxidative stress and sodium translocation were part of the RprY regulon, suggesting a possible role in Na+ metabolism and the response to redox changes [9]. The goals of the present study were to determine the activation signal(s) for rprY and clarify its role in the oxidative stress response. We found that the regulator was essential for growth of P. gingivalis under sodium-limited growth conditions. While the parent strain adapted and was able to grow, albeit slowly, under these conditions, an rprY mutant strain could not, indicating that RprY was essential for the response to this stress and hence viability. By transcription profiling and metabolite analyses we determined that sodium limitation induced an oxidative stress response in both parent and rprY mutants strains. However, the response was highly amplified in the mutant and was accompanied by dysregulation of genes encoding protein chaperones. We found that RprY interacted directly with the promoters of several chaperone genes, and collectively our data indicate that the regulator acts as a repressor of their expression. We hypothesize that in the absence of RprY, P. gingivalis is unable control oxidative stress and protein chaperone responses compromising growth of the mutant under stress conditions.
Materials and Methods
Strains, Media and Growth Conditions
Escherichia coli strains used in this study were derivatives of E. coli K12 (Table 1). E. coli was grown and maintained in Luria-Bertani (LB) media supplemented with ampicillin (100 µg/l) as required. For RprY-LacZ expression assays cultures were grown at 37°C with aeration in modified LB broth. Briefly, the medium was reconstituted from individual components (tryptone and yeast extract). Sodium chloride (NaCl; 171 mM) was either included (LB) or omitted (LB0). To investigate the role of ion or stabilizers on RprY-LacZ expression, NaCl (171 mM), KCl (171 mM) or sorbitol (500 mM) were added to NaCl –depleted LB (LB0).
Table 1. Primers, strains and plasmids used in this study.
| Primers | Sequence 5'- 3' forward/reverse (amplicon size in bp) | Reference |
| rprY-lacZ cloning | ||
| rprY | ACCCGGGGATTTTTATTCCTGCCATAAC/GGGATCCTTACTATCGTCCTCGCAGAG (821) | This study |
| rprY cloning in pET28a | ||
| his-rprY | AAGCTTGCATGCCTGCTCAGACCTCTTTGATCAACTC/ | This Study |
| TATACATATGGAAGAAAAAACAAGAATCTTTCTCT | ||
| rprY cloning in pFD288 | ||
| rprYHis3 | TAGACGTCTCAGTGGTGGTGGTGGTGGTGGACCTCTTTGATCAACTCCTCA | This study |
| KAR6a | ACCCGGG GAT TTT TATT CCT GCC ATA AC | This study |
| OxyR deletion | ||
| 3-way-oxyRF1 | CGAGTGGCGATGAGCGTCAGGCC | This study |
| 3-way-oxyRR2 | TTCCGAGCTTTCTTCTTTTGCCGT | This study |
| 3-way-oxyR R1-EM | TGTAGATAAATTATTAGGTATACTACTGACAGCTTCGGGAAGGATCGTTTTGTATTAG | This study |
| 3-way-oxyR F1-EM | ACCGATGAGCAAAAAAGCAATAGCGGAAGCTATCGGACGAATCATATTTCCTATTTTTG | This study |
| RprY deletion | ||
| KK108 | GAGATAGAAGCATTAGAACTAATGGGCTACTAAAGGGCT | This study |
| KK109 | GGAGATAATTCGTTGTTTATCCAGCTGAGAACGGAATGAG | This study |
| KK111 | CTCATTCCGTTCTCAGCTGGATAAACAACGAATTATCTCC | This study |
| KK112 | AGCCCTTTAGTAGCCCATTAGTTCTAATGCTTCTATCTC | This study |
| KK107 | CTCGTGTTTATGGTTCCCAA | This study |
| KK110 | GTCAGGGGAGGCTCTGCTTC | This study |
| qPCR | ||
| 16S | TGTTACAATGGGAGGGACAAAGGG/TTACTAGCGAATCCAGCTTCACGG(118) | [42] |
| sod | AAAGAGCGAAGGCGGTATCT/CGAATGAGCCGAATTGTTTGTC(140) | [42] |
| ahpC | TCAAACTCAATGCCTATCACAATG/GATAGAAAACGACCAAAGACCACT(87) | [42] |
| rprY | CCATCGCGATCGATGATCAGGTAA/GGCATAGTTGCGTTCAAGGGTTTC(104) | [42] |
| PG0275 | ATCGGCAGATTGCCCATCGT/ATGCCGGTAAACTCACCGTCT(122) | This study |
| PG1286 | CAAGGCCGAAATGTGGTCTTCA/TCCATCATATCGTAGGCGTGCT(134) | This study |
| PG1134 | CTCATCATCGGTTCCGGACCT/TTCCACCTCGGTCGTAGTCGT(125) | This study |
| PG2008 | GGATCCGTTACAGGTACCGTAGT/ATTACCCGAAGGGATTCCCTTGA(141) | This study |
| PG0209 | GGTGAGCACCACAAGAATCAAG/GCCGGAGCATACATTGCTATC(144) | This study |
| PG1321 | GAGGTGGCTATGCACAGGTAC/GTCGCAAGTATTGCGGTTCTG(147) | This study |
| groES | CCTCTCAAGGGTGAAGTAATCGCT/ATTTGCCGTAGAGTACGGTGTCTC(91) | [42] |
| groEL | CGGCTACATCTCTCCCTACTTCGT/GAGGATCGGGAGCATCTCTTTCAG(121) | [42] |
| clpB | CTGCTCAAGGCCGTTATGGATCA/CTGGTTGGTCTCATGCGACAGA(160) | [42] |
| dnaK | CTGACCGGTGAGGTAAAGGATGTC/CTTCGTCGGGATAGTGGTATTGGC(120) | [42] |
| htpG | CGGTCGCTGACCGTGATCGT/TCTCCAATGCCGTGGATGCT(132) | [42] |
| EMSA probes | ||
| groES | CACCTTTTTTCCGACCGACT/TGTTGCTTGGTTTGTTATTGTTAGT(215) | This study |
| dnak | TGTACAGAGTCATTATTCGT/TCCCATAATTCTATATTCGT(199) | This study |
| htpG | AATAAAAATTTTCTCAGACCACGT/CATAGTGTATTATTATCTGTTAATGT(203) | This study |
| clpB | GTTCTGAGTTTCATAGTAGGGTA/GCTGTGTTTGTTGTTACCAGA(229) | This study |
| sod | GTTCGTGAGTCATAACGTCT/TTCGTCATGGAATGAATCGGT(164) | This study |
| ahpC | ACTGCGCCTTCTTAATGTTT/AGTATGAACGTTTGTTTCAGG(356) | [9] |
| Strains and plasmids | ||
| Name | Description or Genotype | Reference |
| P. gingivalis | ATCC33277(type strain) | Coykendall et al |
| oxyR mutant | ATCC33277ΔoxyR::ermF-ermAM | This study |
| rprY mutant | ATCC33277 rprY::ermF-ermAM | [9] |
| rprY tet deletion | ATCC33277 ΔrprY::tetQ | This study |
| E. coli DH5a | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Invitrogen |
| E. coli TB28 | rph-1 ilvG rfb-50 lacZYA <frt > | [43] |
| pRS414 | lacZ protein fusion vector, lacZYA lacks promoter, RBS and translational start, apr, colE1 origin | [44] |
| pFD972a | Expression vector, 9.1 kb; tetQ; (Apr) Tcr | [45] |
| pRprylacZ | pFD972a rprY-lacZ | This study |
| pFD288 | Bacteriodetes shuttle vector | [15] |
| pFDrprY | pFD288rprY | This study |
P. gingivalis ATCC 33277 and erythromycin-resistant (ErmR) rprY and ΔoxyR isogenic mutants were maintained on tryptic soy agar (TSA) supplemented with 5% sheep blood, menadione (1 µg/l), hemin (1 µg/l) and, when appropriate, erythromycin (5 µg/ml). Strains were grown in an anaerobic atmosphere containing 85% nitrogen, 5% hydrogen, 10% carbon dioxide either in sealed bags (Anaeropaks, Mitsubishi) or an anaerobic chamber (Coy). Trypticase Soy broth (TSB) was used for NaCl- depletion experiments with the medium constituted from individual components (pancreatic digest of casein, papaic digest of soybean, dextrose) with or without NaCl (85 mM), i.e. normal and NaCl-depleted TSB.
Plasmid Construction and RprY Recombinant Protein Purification
PCR primers used for strain constructions are listed in Table 1. Fusion plasmid pFDrprY- lacZ was constructed in a three- step procedure. The lacZ gene fragment from pRS414, an E. coli lacZ protein fusion vector, was PCR amplified and cloned into plasmid pJET (Fermentas). The fragment lacks a promoter and translational start codon which was provided by cloning a DNA fragment containing 776 bp DNA upstream and 41 bp downstream from the translational start of P. gingivalis rprY creating the protein fusion. The complete rprY-lacZ hybrid fragment was then cloned into pFD972a [10].
For recombinant RprY protein purification the rprY gene of P. gingivalis ATCC 33277 was PCR amplified using primers listed in table 1 and cloned into Nde1 and HindIII digested pET28a expression vector (Novagen). The resulting plasmid was transformed into E. coli BL21 DE3 (Novagen) for expression of the N-terminal 6X histidine tagged RprY recombinant protein which was purified using an Ni+2-NTA agarose affinity purification column. The eluate was dialysed and further concentrated by treating the membrane with polyethylene glycol (Sigma). The final protein concentration was determined using a Nanodrop 8000 (Thermo Scientific).
β-Galactosidase Assays
A Pierce yeast β -galactosidase assay kit was used according to the manufacturer’s instruction. Briefly, an overnight culture of the rprY-lacZ- containing E. coli strain was diluted 1∶50 in LB broth with and without added NaCl and grown for 2 hrs with shaking at 37°C. An aliquot of culture (350 µl) was added to a lysis and assay solution mix (350 µl), vortexed and incubated until sufficient color had developed. The assay was stopped by addition of 300 µl of assay stop solution. Cell debris was removed by centrifugation and the color was measured in spectrophotometer at OD420 nm. Miller units were calculated as described previously [11].
Construction of oxyR Deletion and rprY-complemented Mutant Strains
Three- way Soeing PCR [12] was performed to fuse 500 bp upstream and downstream sequences of the oxyR ORF to ermF-ermAM cassette [13]. The fused fragment was cloned into cloning vector pC2.1 TA (Invitrogen) which was electroporated into P. gingivalis strain ATCC 33277. Transformants were selected for erythromycin resistance generated by a double cross-over event which was confirmed by PCR.
Similarly, 552 bp upstream and 463 bp downstream sequence of rprY was fused with tetQ cassette [14] by Three-way Soeing PCR. The resultant PCR fragment was electroporated into P. gingivalis strain ATCC 33277 and transformants were selected for tetracycline resistance (2 µg/ml). A 1547 bp fragment containing the complete rprY ORF, 777 bp upstream sequence and restriction site linker was amplified from the ATCC 33277 genome. The fragment was digested with appropriate restriction enzymes and ligated to the similarly treated pFD288 plasmid vector [15]. The final plasmid, pFDrprY, was electroporated into ATCC 33277 ΔrprY(tetQ) strain and ErmR transformants were selected.
Porphyromonas gingivalis Growth and RNA Isolation for Microarray and qRT-PCR Analyses
Parent ATCC 33277 and rprY mutant strains were grown in tryptic soy broth to exponential growth phase (OD550 0.4–0.6). Cultures were split into two equal parts, centrifuged, and cells resuspended in pre-reduced modified TSBH broth with or without NaCl. Cultures were incubated further for 2 hr anaerobically, centrifuged and RNA either isolated immediately or the cell pellet was resuspended in RNA Later (Ambion) for storage. A control experiment was performed to rule out an oxidative stress response due to brief handling of cells outside the anaerobic chamber, i.e. the parent strain was grown in complete TSB medium and prepared as above. RNA was isolated immediately from one of the centrifuged cell pellets. The cells from the other pellet were resuspended in pre-warmed and pre-reduced complete TSB medium and incubated for 2 hr, 37°C in an anaerobic chamber, then centrifuged and RNA was isolated.
In all experiments RNA was isolated using an Epicenter kit according to the manufacturer’s instructions. Contaminating DNA was eliminated by treatment with Turbo DNase (Ambion). Isolated RNA was monitored for DNA contamination by PCR and RNA integrity was visually assessed after gel electrophoresis. The concentration and purity of RNA was determined using a Nanodrop1000 or 8000 Spectrophotometer (Fisher Scientific).
Microarray Analysis
Microarray slides were fabricated based on annotated open reading frames in the genome of strain W83, therefore gene numbers used in this work are based on strain W83 annotation in order to avoid confusion. The slides were provided by the J. Craig Venter Institute and their in-house protocol was used for cDNA synthesis and microarray processing. Briefly, 5–15 µg RNA was reverse transcribed using Superscript III (Invitrogen) or Smartscribe (Clonetech) reverse transcriptase, random hexamers (Invitrogen 3 µg/ml), RNase Out (Invitrogen) and 25 mM dNTP-aaUTP mix. Synthesis was carried out for 16–20 hr at 42°C and RNA was hydrolysed using 0.5 M EDTA and 1 M sodium hydroxide followed by incubation at 65°C,15 min, followed by addition of Tris (1 M, pH 7.4) to neutralize the pH. The cDNA was purified (Qiagen Qiaquick PCR purification kit) then precipitated by sodium acetate -ethanol treatment and resuspended in 4.5 µl of 0.1 M sodium carbonate buffer (pH 9.5) and 4.5 µl of Cy3 or Cy5 dyes (Amersham Biosciences). cDNA labeling reactions were carried out for at least 2 hr in the dark and quenched with 100 mM sodium acetate (pH 5.2). Uncoupled dye was removed (Qiaquick PCR purification kit) and labeled cDNAs were mixed together and precipitated by sodium acetate- ethanol treatment. Labeled cDNA was hybridized onto the microarray slides for 16 hr, 42°C. Washed slides were scanned in a Genepix 4000B (Axon) scanner and the spot intensities measured with Genepix 6.0 software. The data were analysed using Significant Analysis for Oral Pathogen Microarray Data (SAOPMD) tools available at the Bioinformatics Resource for Oral Pathogen (BROP) website (www.brop.org) hosted by The Forsyth Institute. The combined data was obtained by analysis of all repeats within and between arrays using LIMMA (Linear Models for Microarray package; http://bioinf.wehi.edu.au/limma/; [16], [17]).
Quantitatve RT- PCR Analysis
Quantitative RT-PCR was used to quantify relative-fold changes of differential expression and to validate microarray data. iQ SYBR Green super mix (Bio-Rad) was used according to the manufacturer’s instructions. RNA (1 µg) was reverse transcribed with a RevertAid First strand cDNA synthesis kit (Fermentas) according to the instructions provided. Real-time PCR quantitation of double-stranded DNA fragments was performed with an iCylcer (BioRad). A three-step protocol was used for PCR amplification: 95°C, 3 mins for initial denaturation, 50 cycles at 95°C for 15 s, 55°C for 15 s and 72°C for 15 s. A melt curve analysis was performed to determine the generation of a single amplicon under the following conditions: 95°C for 1 min, 55°C for 1 min, and 55°C to 95°C with a heating rate of 0.5°C per 10 s. Each experiment was performed in triplicate and mean values of at least two independent experiments is shown. Fold- changes in gene expression between the samples were determined as previously described [18].
Electrophoretic Mobility Shift Assay (EMSA)
The intergenic region upstream of the start codon of the gene of interest was PCR amplified using Takara polymerase (Clontech, Takara Bio) or platinum PCR supermix (Invitrogen) and primers listed in the table 1. The DNA fragment was resolved on a 1% agarose gel and purified using a QIAquick gel extraction kit (Qiagen). Digoxygenin (DIG) labeled probes were prepared using DIG labeling Kit (Roche) according to the manufactures instructions. Electrophoretic mobility assays using recombinant RprY were performed as previously described [9], with the addition of poly[d(I-C)] (0.01 ug/ul) to the binding buffer.
Metabolomic Analysis
Mid-logarithmic phase (OD550 0.4–0.6) cultures of P. gingivalis parent and rprY mutant strains were split into two and cells were pelleted by centrifugation. Pellets were resuspended in either complete or NaCl-depleted TSB. Media used for resuspension were pre-warmed and reduced in an anaerobic chamber overnight. Resuspended pellets were incubated anaerobically for 2 hr, 37°C, then centrifuged to pellet the differentially- treated cells. Cell pellets were flash frozen in an ethanol-dry ice bath and stored at −80°C. The samples were shipped to Metabolon Inc. (NC) on dry ice for metabolomic analysis.
Samples were analysed by GC/MS and LC/MS/M as previously described [19], [20]. Identification of metabolites was performed by automated comparison of the ion features with reference library of chemical standards developed at Metabolon Inc. Welch’s two-sample t-tests were performed for pair-wise comparison of NaCl-depleted and normal samples also between two different genotypes (parent vs rprY mutant). Two- way Anova was used to identify biochemicals exhibiting a significant genotype effect, treatment effect (NaCl presence or absence) and genotype-treatment interaction.
Results
Sodium Depletion Induces Expression of the P. gingivalis rprY Promoter-LacZ Fusion in E. coli
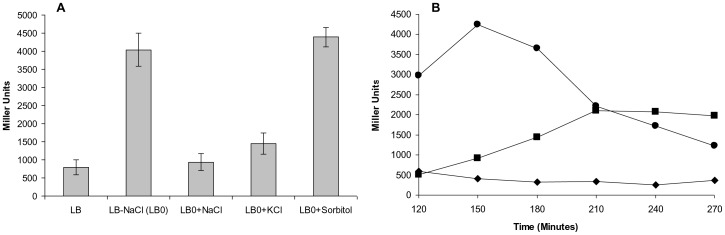
We constructed lacZ protein fusions to study expression of rprY in response to various stimuli. Although up-regulated expression of a response regulator is not necessarily required under inducing conditions, increased expression due to a stimulus may be an indicator of activation. In E. coli K12 (the intermediate cloning host) we did observe significant expression (>500 Miller units) of the rprY-lacZ fusion in normal LB, suggesting that the rprY promoter was active in E. coli (Fig. 1A). This view was also supported by an earlier study in which rprYX genes from B. fragilis conferred tetracycline resistance and decreased ompF expression in E. coli [8].
Figure 1. RprY-LacZ production in E.coli.
A. E. coli strain MG1655 (ΔlacZ) carrying the rprY-lacZ fusion was subcultured in LB broth with or without NaCl (LB0). β-Galactosidase was assayed after 2 hr growth at 37°C. LB0 was supplemented with 171 mM NaCl, 171 mM KCl or 350 mM sorbitol to measure their effects on rprY-lacZ production. The results are the mean of three independent experiments. B. The RprY-lacZ fusion strain was diluted in LB or LB0 and incubated for 2 hrs, followed by addition of LiCl (500 mM) to the culture grown in LB medium. β-Galactosidase production was measured over time from cells cultured in LB (control): black diamonds; LB0: black circles; and LB+LiCl (500 mM): black squares. The experiment was performed twice and a representative experiment is shown.
Since RprY belongs to EnvZ-OmpR family of regulators (18) and our previous results indicated that RprY regulates sodium-dependent channels [9], we determined the effect of NaCl on RprY expression. In this experiment, normal LB medium (LB0+ NaCl) contains separately added 171 mM NaCl. As shown in Fig. 1A, omission of NaCl (LB0) caused a three- to six-fold increase in expression of the LacZ fusion, although there was an approximate three-fold decrease in growth of the strain (not shown). However, the increase in LacZ expression still occurred when sorbitol at equivalent osmolarity (500 mM) was substituted for NaCl (i.e., LB0+ sorbitol) which did not compromise growth (not shown), indicating that growth rate did not affect expression of the lacZ fusion. On the other hand, substitution with KCl (LB0+KCl) conferred lower expression similar to that in normal medium. (Not shown is that as much as 400 mM additional NaCl to normal medium had no effect).
The E.coli rprY- lacZ fusion strain was grown in complete and NaCl-depleted LB and in complete LB containing lithium chloride (LiCl). As a substrate analogue of Na, Li uses many of the same permeases for uptake, therefore when Li+ is present in excess bacterial cells use it instead of Na+, leading to disruption of the normal sodium cycle [21]. RprY- lacZ fusion expression was assayed at different times during growth in three media: normal LB with and without Na+, and normal LB with LiCl (500 mM). Expression of rprY-lacZ was elevated for up to 150 mins in Na+ -depleted broth before declining (Fig. 1B). The data indicated that the rprY promoter responded immediately to Na+ depletion and also suggested that after cells adapted to the condition rprY expression is no longer required. Addition of LiCl resulted in growth retardation of the strain (data not shown), however the expression of the rprY-lacZ fusion increased up to 90 mins after addition of LiCl suggesting that the uptake of Li+ caused Na+ depletion (Fig. 1B). In summary, these experiments indicated that depletion of Na+ activated expression of the rprY promoter in E. coli and expression of rprY was affected by monovalent cation concentration and not osmotic stress.
Sodium Depletion Prevents Growth of a P. gingivalis rprY Mutant
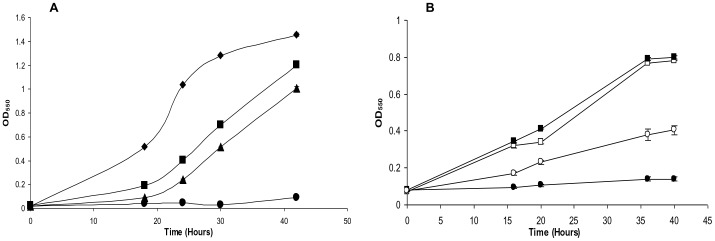
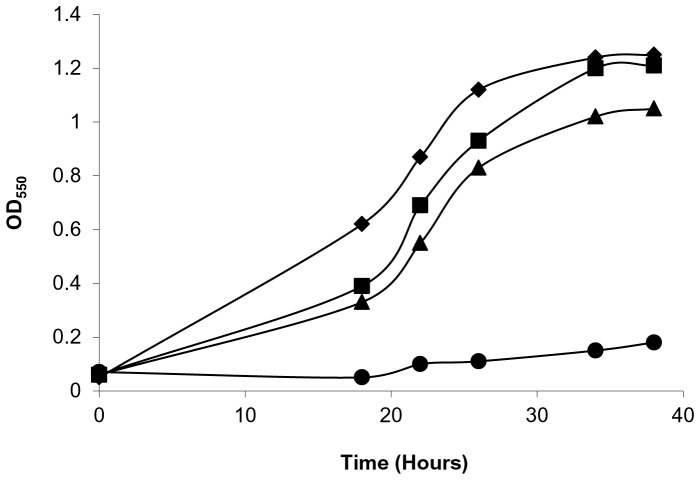
It was clear from our experiments with E. coli that RprY responds to Na+ depletion and thus may be involved in sodium-dependent metabolism. Next, we determined whether RprY played a role in the response of P. gingivalis to Na+ depletion, starting with the effect on growth. Overnight cultures of P. gingivalis ATCC 33277 parent and rprY mutant strains were grown in normal TSB medium, then diluted 1∶40 into (pre-reduced) normal or Na+-depleted TSB and incubated under anaerobic conditions. The rprY mutant grew slower than the parent even in complete medium, but in the absence of Na+ did not grow for up to 42 hours (Fig. 2A). These data indicated that the rprY mutant was hypersensitive to Na+ depletion and was unable to adapt to this growth condition. Complementation of the rprY mutation with the wild type gene on a plasmid partially restored growth of the mutant in the absence of Na+ (Fig. 2 B), indicating that RprY function was responsive to sodium.
Figure 2. Growth of P. gingivalis strains in the absence of NaCl.
Overnight cultures of P. gingivalis parent, rprY mutant (A), and complemented rprY mutant strains (B) were diluted 1∶40 (OD550 ∼0.02) in pre-reduced TSB media with or without NaCl and grown anerobically at 37°C. Parent strain with NaCl: black diamonds; rprY mutant with NaCl: black squares; parent without NaCl: black triangles; rprY mutant without NaCl: black circles; complemented rprY mutant with NaCl: open squares; complemented mutant without NaCl: open circles. The average of two independent experiments is shown. The experiment was repeated at least three times.
Sodium Depletion Induces an Oxidative Stress Response in P.gingivalis
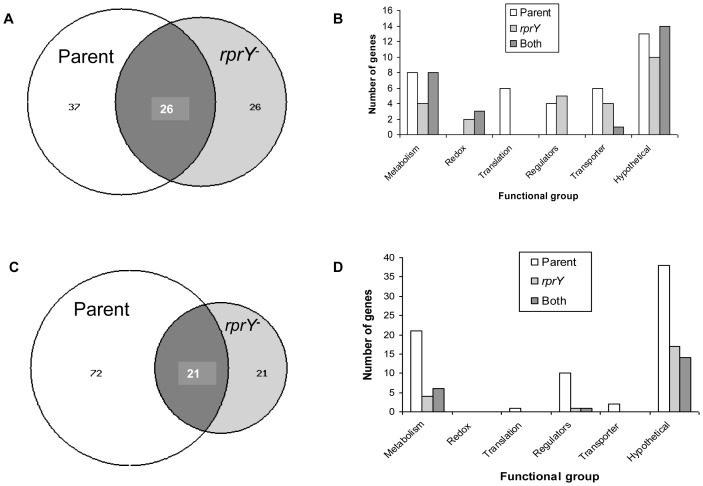
We used comparative gene expression profiling to analyze further the role of RprY in the physiology of P. gingivalis. Because the rprY mutant cannot grow in Na+-depleted TSB we compared expression profiles after parent and mutant strains were grown to mid-logarithimic phase in complete TSB, then centrifuged and resuspended in modified TSB with or without NaCl, and incubated for 2 hrs anaerobically. Induction of rprY-lacZ in E. coli was rapid; therefore, to capture adequately the RprY response in slower-growing P. gingivalis we previously established by QRT-PCR that RprY was expressed in the parent strain after 2 hrs in the absence Na+ (data not shown). Using a conservative cut-off of 1.9-fold expression in either parent or mutant strains, a total of sixty-three genes were up-regulated in response to Na+ depletion (Na−/Na+) in the parent strain (Fig. 3A). The majority of these genes coded for hypothetical proteins, as shown in Fig. 3B. The next highest class of up-regulated genes were those associated with metabolism. A total of fifty-two genes were up-regulated in the rprY mutant upon Na+ depletion, again with the highest number coding for hypotheticals, followed by those involved in gene regulation or transport functions. Parent and mutant strains had twenty-six up-regulated genes in common. Of genes that responded to redox, at least two were uniquely up-regulated in the mutant while the rest were also upregulated in the parent. Regulators induced by Na+ depletion were either parent- or mutant-specific. Many more genes were down-regulated in the parent in response to Na+ depletion, i.e. ninety-three compared to forty-two in the mutant, and of these twenty-one genes were common to both strains (Fig. 3C and D). Again, the largest numbers of genes down-regulated in response to Na+ stress were hypotheticals and those involved in metabolic functions. Interestingly, few genes associated with redox, translation, and transport functions were down-regulated in either strain, but expression of more regulators was repressed in the parent including PG0020 (MarR family transcriptional regulator), PG0245 (universal stress protein family), PG0928 (response regulator), and PG0985 (ECF, RNA polymerase sigma-70 factor). The complete microarray data can be viewed at the Bioinformatics Resource for Oral Pathogens (www.brop.org).
Figure 3. Gene expression analysis of parent and rprY mutant strains under Na+ depleted conditions.
A and C. Venn diagrams showing functional categories and numbers of genes up-(A) or down-regulated (C), respectively, in the absence of Na+. B and D. Functional categories and numbers of genes up-(B) or down-regulated (D), respectively, in the absence of Na+. Open columns represent the number of genes up-regulated in the parent only, grey columns show the number up-regulated in the mutant only, and black columns the number of up-related genes in common.
In both parent and mutant strains Na+ depletion lead to significant upregulation of genes previously identified as responsive to oxidative stress in P. gingivalis [22], [23]. As shown in Table 2, these included formate-tetrahydrofolate ligase (PG1321), formate-nitrite transporter (PG0209), thiol peroxidase (PG1729), ferritin (PG1286) and thioredoxin (PG0034). In the rprY mutant upregulation of these genes was more robust and included the induction of additional oxidative stress response genes e.g. Fe-Mn superoxide dismutase (PG1545), thioredoxin family protein (PG0275), and tetrapyrrole methylase family protein (PG0433), and to lesser levels alkyl peroxidase reductase C (ahpC, PG0618), nitrite reductase-related protein (PG2213), a putative membrane protein (PG1868), and ATP:cobalamine adenosyltransferase (PG1124).
Table 2. Genes involved in the oxidative stress response that are differentially expressed under Na+ depleted conditions.
| Parent ATCC 33277−/+Na+ | rprY mutant −/+Na+ | ||||||
| Locusa | Common name | Fold expb | P valc | Repeats | Fold exp | P val | Repeats |
| PG0034 | Thioredoxin | 2.95 | 0.0004 | 12 | 3.25 | 0.0001 | 12 |
| PG0046 | Phosphatidate cytidylyltransferase | 2.21 | 0.0028 | 12 | 2.25 | 0.0001 | 12 |
| PG0047 | Cell division protein FtsH, putative | 2.04 | 0.0026 | 12 | 2.12 | 0.0016 | 12 |
| PG0095 | DNA mismatch repair protein MutS | 0.65 | 0.0806 | 8 | 1.91 | 0.0041 | 8 |
| PG0209 | Formate-nitrite transporter | 3.71 | 0.0004 | 12 | 3.39 | 0.0241 | 11 |
| PG0258 | ABC transporter, ATP-binding protein | 1.78 | 0.0028 | 12 | 3.94 | 0.0000 | 12 |
| PG0275 | Thioredoxin family protein | 1.71 | 0.0049 | 12 | 4.09 | 0.0000 | 12 |
| PG0432 | NOL1-NOP2-sun family protein | 2.82 | 0.0517 | 12 | 3.58 | 0.0001 | 12 |
| PG0433 | Tetrapyrrole methylase family protein | 2.88 | 0.0165 | 12 | 7.43 | 0.0001 | 8 |
| PG0618 | Alkyl hydroperoxide reductase, C subunit | 1.17 | 0.1644 | 12 | 1.85 | 0.0119 | 12 |
| PG0889 | Peptidase, M24 family | 2.35 | 0.0004 | 12 | 2.94 | 0.0002 | 12 |
| PG0890 | Alkaline phosphatase, putative | 2.37 | 0.0002 | 12 | 3.46 | 0.0000 | 12 |
| PG1043 | Ferrous iron transport protein B | 0.96 | 0.7002 | 7 | 2.01 | 0.0501 | 9 |
| PG1044 | Iron dependent repressor, putative | 1.50 | 0.2211 | 12 | 3.27 | 0.0067 | 8 |
| PG1088 | Acetyltransferase, GNAT family | 2.14 | 0.0001 | 12 | 0.94 | 0.5204 | 12 |
| PG1089 | DNA-binding response regulator RprY | 1.58 | 0.0001 | 12 | 2.05d | 0.0000 | 12 |
| PG1124 | ATP:cob(I)alamin adenosyltransferase, putative | 1.12 | 0.4240 | 12 | 2.02 | 0.0000 | 12 |
| PG1134 | Thioredoxin reductase | 1.27 | 0.0248 | 12 | 1.93 | 0.0075 | 12 |
| PG1190 | Glycerate dehydrogenase | 1.56 | 0.1968 | 12 | 4.20 | 0.0000 | 10 |
| PG1286 | Ferritin | 2.03 | 0.0005 | 12 | 4.00 | 0.0000 | 12 |
| PG1321 | Formate–tetrahydrofolate ligase | 4.79 | 0.0001 | 12 | 6.78 | 0.0000 | 12 |
| PG1545 | Superoxide dismutase, Fe-Mn | 1.96 | 0.0069 | 12 | 4.44 | 0.0001 | 12 |
| PG1640 | DNA-damage-inducible protein F | 1.60 | 0.0011 | 12 | 2.16 | 0.0001 | 10 |
| PG1642 | Cation-transporting ATPase, authentic frameshift | 1.88 | 0.0000 | 12 | 3.50 | 0.0000 | 12 |
| PG1729 | Thiol peroxidase | 3.03 | 0.0000 | 12 | 3.68 | 0.0000 | 12 |
| PG1841 | Conserved hypothetical protein | 3.20 | 0.0006 | 12 | 4.02 | 0.0005 | 12 |
| PG1868 | Membrane protein, putative | 1.50 | 0.4660 | 10 | 3.83 | 0.0013 | 12 |
| PG2008 | TonB-dependent receptor, putative | 1.03 | 0.8227 | 5 | 3.11 | 0.0620 | 8 |
| PG2213 | Nitrite reductase-related protein | 1.54 | 0.0004 | 12 | 2.26 | 0.0005 | 12 |
Loci numbers are those from the annotation of the genome of strain W83 and the microarrays.
Fold expression.
P values were calculated using LIMMA.
The microarray PCR amplicon for rprY is upstream from the erythromycin resistance cassette that was used to generate the rprY mutation, thus an expression signal was still detectable.
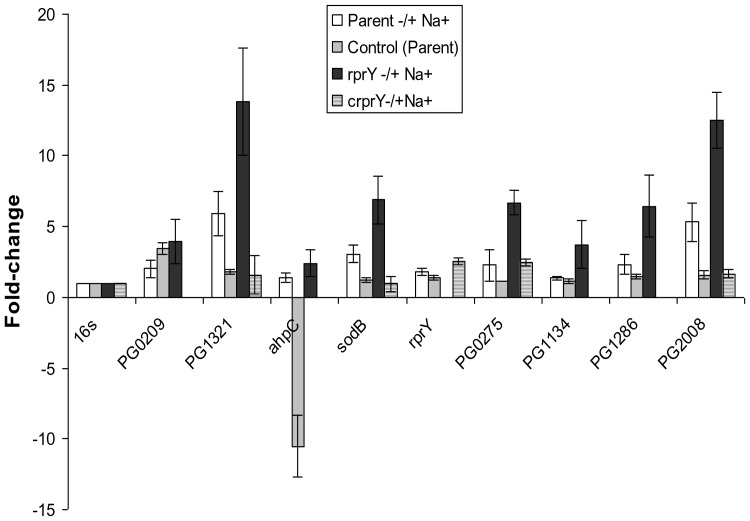
Preliminary QRT-PCR control experiments ruled out the possibility that brief handling of cultures outside the anaerobic chamber induced this response. Differential regulation of several of these genes was confirmed by qRT-PCR (Fig. 4) in which the fold- changes of the genes tested was higher than seen by microarray, however, the trends of differential regulation were consistent with the microarray data. In addition, for the majority of transcripts assayed, complementation of the rprY mutant with the wild type gene restored expression to parental levels.
Figure 4. Confirmation of oxidative stress gene expression by QRT-PCR.
The expression of several oxidative stress genes that were differentially regulated in Na+ depleted versus Na+ replete conditions was quantified by QRT-PCR. Positive fold values refer to increased, and negative values refer to decreased expression under Na+ depleted conditions. To verify that culture handling did not induce an oxidative stress response, control fold-expression changes (grey columns) were calculated by comparing gene expression in parent cultures processed immediately after growth with that in cultures after centrifugation and resuspension in pre-reduced normal TSB and anaerobic incubation (2 hrs). PG209: formate nitrate transporter; PG1321:formate tetrahydrofolate reductase; PG0275: thioredoxin family protein; PG1134: thioredoxin reductase; PG1286: ferritin; PG2008: putative TonB-dependent receptor.
Overall, these results indicate that Na+ depletion induced an oxidative stress response. The “super” oxidative stress response in the rprY mutant may result from increased superoxide production in the mutant upon Na+ depletion. On the other hand, the data are also consistent with a model in which RprY directly or indirectly represses expression of several stress response genes.
To further verify that Na+ depletion induces an oxidative stress response of P. gingivalis, we carried out comparative analyses of small metabolites formed in parent and rprY mutant cells grown (2 hr) in the presence and absence of Na+. The data was analyzed in several ways including differences between parent and mutants strains in normal medium; in Na+-depleted medium; and metabolites that responded to Na+ stress in parent or mutant. The p-values obtained from Two-Way ANOVA statistical analyses indicated whether differences were due to the effect of the rprY mutation or to Na+ stress or both. Accordingly, as seen in Table 3, certain significant effects appeared to relate to the sodium status, e.g., in Na+ stress, higher methionine sulfoxide, lower ketosphingosine, and more of certain dipeptides. Other effects appeared more related to RprY function than to Na+, e.g., in the mutant, higher undecanoate and lower thiamine. While it is difficult to make close connections of these changes with physiology (see Discussion), we here observe only that this initial analysis of a large number of metabolites from biosynthesis, catabolism and central metabolism reveals perturbations in relatively few of them in the four situations examined.
Table 3. Small metabolite analysis.
| Class | Compound | Relative Values | Two-Way ANOVA | ||||
| Parent (−) Na+ Parent (+) Na+ | rprY (+) Na+ Parent (+) Na+ | rprY (−) Na+ rprY (+) Na+ | rprY (−) Na+ Parent (−) Na+ | Genotype Main Effecta | Treatment Main Effecta | ||
| p-Value | p-Value | ||||||
| Lysine metabolism | Diaminopimelate | 0.90 | 3.88 | 0.32 | 1.37 | 0.0022 | 0.0112 |
| Methionine metabolism | Methionine sulfoxide | 2.98 | 0.91 | 3.12 | 0.96 | 0.7805 | <0.001 |
| Dipeptides | Alanylglutamate | 3.93 | 0.75 | 3.79 | 0.72 | 0.0538 | <0.001 |
| Alanylleucine | 1.99 | 0.62 | 3.04 | 0.94 | 0.5429 | 0.0141 | |
| Monoacylglycerol | 1-Myristoylglycerol | 0.40 | 1.50 | 0.13 | 0.50 | 0.9050 | 0.0078 |
| 1-Palmitoylglycerol | 0.75 | 4.44 | 0.11 | 0.65 | 0.1367 | 0.0061 | |
| Medium chain fatty acids | Undecanoate (11∶0) | 1.61 | 10.64 | 0.94 | 6.20 | <0.001 | 0.3274 |
| Sphingolipids | 3-Ketosphinganine | 0.18 | 6.04 | 0.15 | 5.09 | 0.0020 | 0.0050 |
| Thiamine metabolism | Thiamin (Vitamin B1) | 1.06 | 0.18 | 1.40 | 0.24 | 0.0971 | 0.6976 |
Specific effects, gene or Na+ stress are labeled in bold.
Finally, we compared growth of a P. gingivalis oxyR mutant with that of the parent strain in Na+-depleted and normal media. As observed previously, the parent strain was able to grow in the Na+-depleted medium while the oxyR mutant could not (Fig. 5), similar to the rprY mutant (Fig. 2). Since OxyR is a master regulator of genes involved in oxidative stress in Gram-negative bacteria [18], we infer that Na+ depletion causes oxidative stress in the P. gingivalis cells.
Figure 5. Growth of a P. gingivalis oxyR mutant in the absence of NaCl.
Overnight cultures of P. gingivalis parent and oxyR mutant strains were subcultured in medium with and without NaCl and grown anerobically at 37°C. Parent strain with NaCl: black diamonds; oxyR mutant with NaCl: black squares; parent without NaCl: black triangles; oxyR mutant without NaCl: black circles. A representative of at least three independent experiments is shown.
RprY Directly Regulates Heat Shock Chaperones
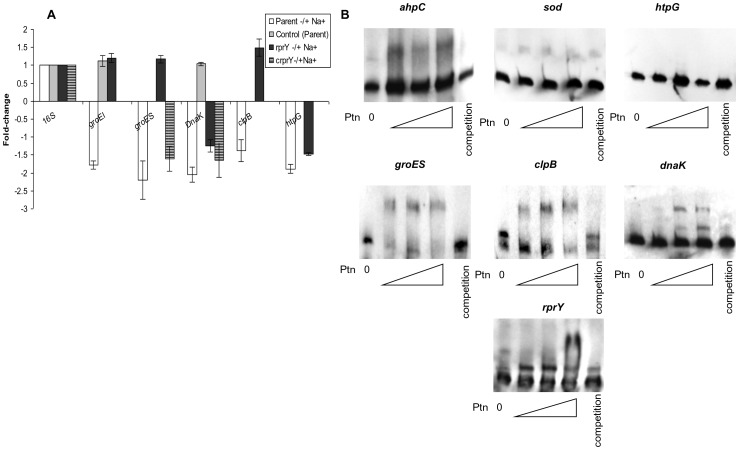
Heat shock chaperones and stress response proteases are expressed during adverse conditions to protect cells by ensuring correct protein folding and removal of denatured proteins. We observed significant downregulation of chaperones (Dnak, GroEL, GroES and HtpG) and ClpB and ClpC proteases due to Na+-depletion in the parent strain (Table 4), however, similar responses were not detected for groESL and clpB in the rprY mutant suggesting an inability to regulate expression of these defence proteins. This differential regulation was confirmed by qPCR with a subset of heat shock genes (Fig. 6A). Complementation of the rprY mutation restored expression of groES and dnaK transcripts to parental levels. Preparation of cells prior to incubation with and without Na+ did not affect expression of these genes (control).
Table 4. Differential regulation of heat shock and chaperone genes under Na+ depleted conditions.
| Parent ATCC 33277−/+Na+ | rprY mutant −/+Na+ | ||||||
| Locus | Common name | Fold exp | P- val | Repeats | Fold exp | P- val | Repeats |
| PG0010 | ATP-dependent Clp protease, ATP-binding subunit ClpC | 1.20 | 0.6093 | 12 | 2.44 | 0.0007 | 12 |
| PG0045 | Heat shock protein HtpG | 0.52 | 0.3206 | 11 | 1.01 | 0.9714 | 11 |
| PG0245 | Universal stress protein family | 0.45 | 0.0002 | 5 | 0.82 | 0.0959 | 10 |
| PG0520 | Chaperonin, 60 kDa | 0.55 | 0.2643 | 12 | 0.90 | 0.2939 | 12 |
| PG0521 | Chaperonin, 10 kDa | 0.34 | 0.0719 | 10 | 0.90 | 0.1981 | 8 |
| PG1118 | ClpB protein | 0.22 | 0.0000 | 5 | 1.44 | 0.2589 | 12 |
| PG1208 | DnaK protein | 0.39 | 0.2538 | 8 | 1.04 | 0.8673 | 12 |
| PG1775 | GrpE protein | 0.18 | 0.0000 | 5 | 0.66 | 0.0031 | 9 |
Figure 6. RprY and expression and regulation of chaperones.
A. Expression of a subset of heat shock proteins was quantified by qRT-RTPCR. Positive- fold values refer to increased and negative values refer to decreased expression under Na+ depleted conditions. B. EMSA showing direct binding of RprY protein (0, 8.8, 17.6, and 35.2 pmoles) to the promoters of clpB, groES, dnaK, and rprY. The promoters of sod and ahpC were used as a negative and positive controls, respectively.
These data suggested that RprY may play a role in regulating the heat shock response in P. gingivalis. We performed electrophoretic mobility shift assays (EMSA) to determine whether the regulation was due to direct binding of RprY to the promoter regions of the genes in question or an indirect effect because of RprY interaction with another regulator. DNA probes were generated by amplification of the 5′ intergenic region upstream of the ORFs. DIG-labeled probes were incubated with increasing amount of phosphorylated rRprY protein (∼0, 8.8, 17.6 and 35.2 pmol). Protein binding specificity was determined by adding 50–100X excess unlabeled probe to the reaction mixes. In these experiments the ahpC and sod promoters were used as positive and negative controls, respectively, as we previously showed that ahpC, but not sod, was a target of RprY [9]. By EMSA we showed that RprY bound to the promoter regions of groES, clpB and dnaK, but did not bind to the promoter region of htpG (Fig. 6B).
The down regulation of chaperones in response to sodium depletion in the parent strain and the lack thereof in the rprY mutant suggested that RprY may be a negative regulator (repressor), and prompted an investigation of whether RprY was self-regulated. By EMSA we showed rRprY bound to its own promoter, lending support to the notion that RprY function was autoregulated, and possibly in response to the same stimulus of Na+ depletion.
Discussion
Porphyromonas gingivalis is an opportunistic pathogen found in relatively low numbers in the healthy gingival crevice. During infection, bacteria are exposed to variable oxygen tension, ions, and proteins from saliva, serum and blood, and have evolved mechanisms to sense and respond to environmental changes and ensure survival. Conditions during periodontal infection such as low redox and the presence of host proteins and peptides favour growth of P. ginigvalis. As an obligate anaerobe, it is not surprising that the organism has developed redundant pathways to sense and respond to oxygen tension [24]. Previously, we identified gene targets of response regulator RprY by DNA-binding assays and comparative transcriptional profiling of parent and mutants strains grown under anaerobic conditions in complete medium (Na+ replete). Among the targets identified were NQR, the primary sodium pump; alkyl hydroperoxidase; ruberythrin; a ferrous iron transporter; and an iron -sulfur assembly system. From these results we hypothesized that RprY might play a role in the oxidative stress response, acting as both an activator and repressor [9].
Although there is high homology between regulator RprY from P. gingivalis and oral and enteric Bacteroidetes, the RprX sensor from these genera shares only weak homology with P. gingivalis histidine kinases, specifically at their C-termini. RprY does not contain transmembrane domains and must rely on a partner sensor kinase for perception of environmental cues and activation signal relay. Candidates for this function are still being sought, but include an orphan sensor kinase (PG0052) and possibly those paired with other response regulators. To circumvent the problem of the unknown sensor kinase, we used a reporter protein fusion approach to identify conditions that activate RprY using a construct in which the rprY promoter region and 41 bp of N-terminal coding nucleotides (13 amino acids) were fused to lacZ. Protein fusion expression was detected in E. coli implying a sensing and phosphorelay system that could activate the P. gingivalis rprY promoter. Interestingly, RprY expression in E. coli was dramatically induced in medium without NaCl and we demonstrated that specifically depletion of Na+ induced expression (Fig. 1). In P. gingivalis, omission of NaCl from culture media lead to a significant lag in the growth of the parent strain compared to growth in complete medium. The lag observed when the rprY mutant was grown in complete medium was also followed by growth recovery, however, the growth was not restored in the absence of NaCl. These differences in growth in Na+ -replete and -depleted media may result from disruption of Na+-dependent metabolic processes such as protein folding, transport of growth substrates, and energy pathways. However, an oxyR mutant of P. gingivalis showed a similar growth phenotype in Na+ -depleted medium (Fig. 5), suggesting that condition induced an oxidative stress response that could not be controlled in the mutant.
In the present study comparative transcription profiling was carried out under more specific conditions, i.e. 2 hr in medium with and without Na+ (Table 2). Under Na+-depleted conditions both parent and mutant strains appeared to undergo oxidative stress as evidenced by up-regulation of genes known to be involved in the response [9], [22], [23], [25]. In most cases the relative levels of expression were greater in the rprY mutant than in the parent indicating that the regulator controls expression of genes important for growth and survival under these conditions. The preparation (centrifugation) of cells for Na+ stress experiments did not itself induce oxidative stress, consistent with the view that Na+ depletion was the cause. That the oxidative stress response was greater in the rprY strain than in the parent, suggests either that the mutant produced more oxidation products or the response could not be controlled adequately in the absence of RprY, which possibly functions as a repressor.
The proposal that Na+ depletion induced an oxidative stress response in P. gingivalis was supported by analyzing the production of small molecule metabolites (Table 3). Levels of DAP increased almost four-fold in the rprY mutant under normal growth conditions and decreased in the Na+-depleted condition. Statistically, this result indicated a gene-specific effect rather than treatment, suggesting that an RprY-regulated function is limiting under Na+ stress. An important component of the bacterial cell wall, DAP can play a role in protection against unfavorable conditions, e.g. higher levels of DAP have been observed in cells grown in biocide -supplemented media [26]. Accumulation of DAP was possibly associated with the long lag growth phase (10 to 20 hr) of the mutant in normal medium. Eventually the mutant appeared to adapt and grow in normal medium, while in the absence of Na+ oxidative conditions preclude recovery.
In Gram-negative bacteria, a response to nutrient limitation is production of polyhydroxyalkanoates, storage compounds that are produced in unbalanced metabolic conditions [27], [28]. In the present study, rprY mutant cells contained high levels of undecanoate, a subunit of a polyhydroxyalkanoate, which appeared to be a gene effect. Interestingly, polyhydroxyalkanoate production decreased the oxidative stress response in a psychrophilic Pseudomonas species by modulating the levels of reducing equivalents [29]. Thiamine levels were approximately five-fold lower in the mutant strain than in the parent, another effect of the rprY mutation. A link between thiamine and oxidative stress was provided by the observation that thiamine supplementation restored the growth of Salmonella enterica on paraquat, a superoxide-generator, the interpretation being that oxidative stress directly caused the thiamine requirement [30]. Our data would suggest that low levels of thiamine in the rprY mutant result from oxidative stress induced by the mutation, even in normal media.
It has been hypothesized that, as in eukaryotes, sphingolipids, may be involved in sensing and signalling in response to stress in Bacteriodes fragilis [31]., Stationary phase cells of B. fragilis either chemically or nutritionally deprived of sphingolipids were less able to survive oxidative stress and DNA crosslinking, leading to the proposal that sphingolipid-rich domains in the periplasmic membrane may transmit environmental information by signalling relays to transcription factors that regulate appropriate responses [31]. Other Bacteroidetes, including Porphyromonas, also contain sphingolipids [32]. 3-Ketosphinganine is a direct presursor of sphinganine, and we observed that upon Na+ depletion, both parent and rprY mutants cells contained significantly lower amounts of this intermediate (approximately six-fold reductions). The statistics suggested that this was a treatment effect, the absence of Na+ having induced stress, however, we do not yet know whether the low levels of 3-ketosphinganine are a direct result of Na+ depletion.
During Na+ stress both parent and mutant cells contained higher levels of intracellular methionine sulfoxide, attributable to the treatment rather than a mutation. Methionine is modified to methionine sulfoxide by reactive oxygen species and to repair oxidative damage the sulfoxide is converted back to methionine by methionine sulfoxide reductases. Interestingly, in the response of B. fragilis to air exposure methionine sulphoxide reductase was induced more than ten-fold [33], however, there were no significant changes in the expression of this gene in P. gingivalis strains under Na+ stress (data not shown). As an asaccharolytic organism, P. gingivalis preferentially uses dipeptides as carbon, nitrogen and energy sources. The metabolite data indicated that in addition to the preferred aspartate and glutamate containing peptides, Na+ stressed cells showed an increased ability to use diverse peptides, notably alanyl peptides with leucine and glutamate. Whether this expansion of growth substrates is an adaptation that offsets growth limiting conditions is as yet unknown.
The oxyR mutant also failed to grow in Na+-depleted medium, implying that this condition mimics oxidative stress (Fig. 5). Approximately half the genes up regulated in both ATCC 33277 parent and rprY mutant strains under Na+ stress were reported to be associated with the OxyR regulon [18]. It was demonstrated that expression of several of these genes was maximal during normal anaerobic growth and were downregulated in an oxyR mutant grown under the same conditions, consistent with OxyR was a positive regulator of these genes [18]. Our data show that under Na+ stress expression of these genes was higher in the mutant consistent with RprY being a repressor. A possible scenario is that under anaerobic conditions OxyR constitutively activates oxidative stress genes and RprY, acting as a repressor, fine- tunes the expression of a subset of these to maintain a balance that prevents a superoxidative stress response. Sodium depletion disrupts the balance triggering induction of RprY in order to restore homeostasis and eventual growth as in the parent strain. However, in the absence of RprY a superoxidative stress response ensues from which the mutant cannot recover and grow.
Transcriptome comparisons between parent and rprY mutant strains in Na+-depleted conditions also revealed differential expression of certain chaperones and heat shock proteins that were down- regulated in the parent but not in the rprY mutant (Table 4); we confirmed these differences in a subset of genes by QRT-PCR (Fig. 6). Molecular chaperones are important for maintaining the quality of the proteins expressed in the cells through stabilization and correct folding primarily supported by trigger factor followed by the DnaKJ and GroESL chaperone systems [34], [35]. Certain chaperones also play a role in disaggregation and refolding of misfolded proteins, e.g. protein aggregation induces sets of chaperones (DnaKJ, GroESL, ClpB, HtpG etc.) and proteases (ClpXP, Lon etc), collectively termed heat shock proteins (HSP).
By EMSA we established that RprY binds to the promoter regions of groES, dnaK and clpB. We reasoned that Na+ -depletion caused oxidation of metabolites and proteins, and induction of RprY. Parent cells mount a stress response that included activation of OxyR regulated genes, e.g. superoxide dismutase [25] and repression of RprY–regulated genes, e.g. ahpCF [9] as well as heat shock proteins and chaperones to protect cells from protein oxidation and misfolding. The transcription data suggests that RprY acts as a repressor of dnaK, groESL and clpB which may help the parent adapt to Na+ stress. In the mutant lacking RprY, groESL, dnaK and clpB expression cannot be repressed which may contribute to the growth defect upon Na+-depletion. The expression of heat shock proteins is tightly controlled by both positive and negative regulation [36], [37]. The expression of several of HSP chaperones is maintained at basal levels by mechanisms that include negative feedback loops [38] and blocking of promoters by CIRCE sequences and negative regulators [39]. Positive regulation is achieved by expression of alternate sigma factors [36]. Thus HSP expression is tightly coordinated with cell requirements during stress and non-stress conditions to ensure survival. Analysis of Porphyromonas gingivalis HSP promoter regions did not identify the classic CIRCE sequences required for negative regulation, leading to the assumption that Porphyromonas may utilize different control mechanisms.
RprY expression is induced under several environmental conditions including oxidative and nitrate stresses [40] suggesting an important role as a response regulator. The rprY mutant was unable to survive aerobic stress compared to the parent strain (data not shown). The interpretation of our data is that Na+ depletion leads to activation of RprY enabling it to bind to target promoters, including it’s own, and function as a repressor. The role of RprY in the response to Na+- depletion may be explained by two interlinked scenarios. First, absence of RprY leads to increased oxidative stress during Na+ depletion because of lower basal levels of the Na+-dependent ubiquinone oxidoreductase system (NQR) which RprY appears to regulate [9]. It has been proposed that NQR utilizes the sodium gradient it generates to produce energy for the CydAB system to convert oxidative radicals to water [41]; thus, lower levels of NQR may result in inefficient removal of these radicals. Second, mutation in rprY leads to loss of negative control of the optimal response by both oxidative stress and heat shock genes. As reported by Hase et al., many pathogens possess enzyme systems to generate sodium motive force in addition to or as alternatives to H+ motive force [21]. In P. gingivalis it was suggested that NQR may play a buffering role against Na+ released during periodontal infection and bleeding. However, our data imply that RprY is inactive in the presence of Na+, and the health conditions that lead to Na+ depletion in the oral cavity are, as yet, unknown.
Ackowledgments
Acknowledgments
We thank Yumiko Hosogi for the oxyR deletion mutant, and Dan Fraenkel for reading the manuscript and helpful discussions.
Funding Statement
This work was supported by National Institutes of Health grant R01-DE015931 (MJD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Holt SC, Ebersole JL (2005) Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”. a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38: 72–122. [DOI] [PubMed] [Google Scholar]
- 2. Bostanci N, Belibasakis GN (2012) Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett 333: 1–9. [DOI] [PubMed] [Google Scholar]
- 3. Ximenez-Fyvie LA, Haffajee AD, Socransky SS (2000) Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol 27: 648–657. [DOI] [PubMed] [Google Scholar]
- 4. Colombo AV, da Silva CM, Haffajee A, Colombo AP (2007) Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res 42: 236–243. [DOI] [PubMed] [Google Scholar]
- 5. Rudney JD, Chen R (2006) The vital status of human buccal epithelial cells and the bacteria associated with them. Arch Oral Biol 51: 291–298. [DOI] [PubMed] [Google Scholar]
- 6. Lamont RJ, Jenkinson HF (1998) Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev 62: 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS (2010) Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8: 471–480. [DOI] [PubMed] [Google Scholar]
- 8. Rasmussen BA, Kovacs E (1993) Cloning and identification of a two-component signal-transducing regulatory system from Bacteroides fragilis . Mol Microbiol 7: 765–776. [DOI] [PubMed] [Google Scholar]
- 9. Duran-Pinedo AE, Nishikawa K, Duncan MJ (2007) The RprY response regulator of Porphyromonas gingivalis . Mol Microbiol 64: 1061–1074. [DOI] [PubMed] [Google Scholar]
- 10. Bacic MK, Jain JC, Parker AC, Smith CJ (2007) Analysis of the zinc finger domain of TnpA, a DNA targeting protein encoded by mobilizable transposon Tn4555 . Plasmid 58: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JH (1992) A short course in bacterial genetics. Cold Spring Harbor Laboratory Press.
- 12. Horton RM, Cai ZL, Ho SN, Pease LR (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535. [PubMed] [Google Scholar]
- 13. Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, et al. (1995) Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun 63: 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB (1996) Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl Environ Microbiol 62: 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith CJ, Parker A, Rogers MB (1990) Plasmid transformation of Bacteroides spp. by electroporation. Plasmid 24: 100–109. [DOI] [PubMed] [Google Scholar]
- 16. Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- 17. Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075. [DOI] [PubMed] [Google Scholar]
- 18. Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, et al. (2006) Role of OxyR in the oral anaerobe Porphyromonas gingivalis . J Bacteriol 188: 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams SB Jr, Setton LA, Kensicki E, Bolognesi MP, Toth AP, et al. (2012) Global metabolic profiling of human osteoarthritic synovium. Osteoarthritis Cartilage 20: 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, et al. (2008) Analysis of the adult human plasma metabolome. Pharmacogenomics 9: 383–397. [DOI] [PubMed] [Google Scholar]
- 21.Hase CC, Fedorova ND, Galperin MY, Dibrov PA (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65: 353–370, table of contents. [DOI] [PMC free article] [PubMed]
- 22. Lewis JP, Iyer D, Anaya-Bergman C (2009) Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced utilization of lactate. Microbiology 155: 3758–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yanamandra SS, Sarrafee SS, Anaya-Bergman C, Jones K, Lewis JP (2012) Role of the Porphyromonas gingivalis extracytoplasmic function sigma factor, SigH. Mol Oral Microbiol 27: 202–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henry LG, McKenzie RM, Robles A, Fletcher HM (2012) Oxidative stress resistance in Porphyromonas gingivalis . Future Microbiol 7: 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohara N, Kikuchi Y, Shoji M, Naito M, Nakayama K (2006) Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology 152: 955–966. [DOI] [PubMed] [Google Scholar]
- 26. Liaqat I, Sabri AN (2008) Analysis of cell wall constituents of biocide-resistant isolates from dental-unit water line biofilms. Curr Microbiol 57: 340–347. [DOI] [PubMed] [Google Scholar]
- 27. Kim do Y, Kim HW, Chung MG, Rhee YH (2007) Biosynthesis, modification, and biodegradation of bacterial medium-chain-length polyhydroxyalkanoates. J Microbiol 45: 87–97. [PubMed] [Google Scholar]
- 28. Hoefer P (2010) Activation of polyhydroxyalkanoates: functionalization and modification. Front Biosci 15: 93–121. [DOI] [PubMed] [Google Scholar]
- 29. Ayub ND, Tribelli PM, Lopez NI (2009) Polyhydroxyalkanoates are essential for maintenance of redox state in the Antarctic bacterium Pseudomonas sp. 14-3 during low temperature adaptation. Extremophiles 13: 59–66. [DOI] [PubMed] [Google Scholar]
- 30. Thorgersen MP, Downs DM (2009) Oxidative stress and disruption of labile iron generate specific auxotrophic requirements in Salmonella enterica . Microbiology 155: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. An D, Na C, Bielawski J, Hannun YA, Kasper DL (2010) Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc Natl Acad Sci U S A 108 Suppl 14666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nichols FC, Riep B, Mun J, Morton MD, Bojarski MT, et al. (2004) Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis . J Lipid Res 45: 2317–2330. [DOI] [PubMed] [Google Scholar]
- 33. Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, et al. (2008) The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol 67: 129–142. [DOI] [PubMed] [Google Scholar]
- 34. Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696. [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann A, Bukau B, Kramer G (2010) Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta 1803: 650–661. [DOI] [PubMed] [Google Scholar]
- 36. Guisbert E, Yura T, Rhodius VA, Gross CA (2008) Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Narberhaus F (1999) Negative regulation of bacterial heat shock genes. Mol Microbiol 31: 1–8. [DOI] [PubMed] [Google Scholar]
- 38. Tomoyasu T, Ogura T, Tatsuta T, Bukau B (1998) Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli . Mol Microbiol 30: 567–581. [DOI] [PubMed] [Google Scholar]
- 39. Zuber U, Schumann W (1994) CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis . J Bacteriol 176: 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis JP, Yanamandra SS, Anaya-Bergman C (2012) HcpR of Porphyromonas gingivalis is required for growth under nitrosative stress and survival within host cells. Infect Immun 80: 3319–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meuric V, Rouillon A, Chandad F, Bonnaure-Mallet M (2010) Putative respiratory chain of Porphyromonas gingivalis . Future Microbiol 5: 717–734. [DOI] [PubMed] [Google Scholar]
- 42. Hosogi Y, Duncan MJ (2005) Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun 73: 2327–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bernhardt TG, de Boer PA (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52: 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simons RW, Houman F, Kleckner N (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53: 85–96. [DOI] [PubMed] [Google Scholar]
- 45. Spence C, Wells WG, Smith CJ (2006) Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen. J Bacteriol 188: 4663–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]