Abstract
Extensive contact with DNA via multiple zinc fingers allows highly specific DNA-binding of zinc-finger-class transcription factors, but can also slow the target search process. Here we introduce recent insights into how zinc-finger proteins can rapidly scan DNA. Potential application of the new knowledge to the zinc-finger-based technology is also discussed.
Keywords: zinc-finger transcription factors, DNA-scanning, target search, gene control, kinetics, dynamics, zinc-finger nucleases
Introduction
Cys2-His2 type zinc-finger proteins are the most common family of transcription factors with > 500 genes encoded in the human genome. These proteins recognize target DNA sequences via three or more zinc-finger domains. Their extensive contact with DNA via multiple zinc fingers enables high stability of the sequence-specific complexes (with typical dissociation constants, Kd, in the pM to nM range) that regulate gene transcription. The zinc-finger protein family includes both constitutive transcription factors (e.g., Sp1) and inducible transcription factors (e.g., Egr-1). Expression levels of constitutive transcription factors in the nuclei are constantly high. For example, Sp1’s concentrations in the nuclei were estimated to be as high as ~100 nM. Inducible transcription factors are short-lived proteins whose expressions are induced only by certain stimuli to the cells. Within their short lifetimes (~½ – 1 h for Egr-1),1 inducible transcription factors must rapidly scan DNA, find targets, and activate or repress particular sets of genes so that the cells can rapidly respond to the stimuli. Although both stability in target binding and rapidity in target search are important, these two are mutually opposing issues. This is a problem known as the speed-stability paradox.2 In the present article, we introduce recent insights into how zinc-finger proteins efficiently scan DNA and resolve the speed-stability paradox. The new knowledge seems useful for understanding natural zinc-finger transcription factors as well as for improving artificial zinc-finger proteins.
Egr-1: An inducible zinc-finger transcription factor that must act fast
Egr-1 (also known as Zif268) is one of the best-characterized zinc-finger proteins. The crystal structure of Egr-1 bound to its target DNA was solved in 1991,3 which greatly helped researchers not only to understand DNA-recognition by zinc-finger proteins but also to develop the zinc-finger technology. Egr-1 plays crucial roles particularly in the brain and cardiovascular systems in mammals. In the brain, Egr-1 is induced by synaptic signals in an activity-dependent manner and activates genes for long-term memory formation and consolidation.4,5 In the cardiovascular system, Egr-1 is a stress-inducible transcription factor that activates the genes for initiating defense responses against vascular stress and injury.1,6 Egr-1 recognizes a 9-bp target DNA sequence, GCGTGGGCG, via three zinc fingers. Egr-1 induced by the cellular stimuli activates more than 50 genes. For example, Egr-1 induced by vascular stress activates genes of pivotal signaling proteins TNFα, ICAM-1, CD44, PDGF A/B chains, TGFβ and M-CSF.1,6,7 Gene activation by Egr-1 is rapid. Expression of these proteins is found ~30 min after Egr-1 expression.7 Considering this time gap, together with the times typically necessary for transcription (~5 min), RNA processing and export (~20 min),8 and translation (~5 min),9 gene activation by Egr-1 seems to occur in less than ~2–3 min. In this short time, Egr-1 must find its target DNA sites and recruit RNA polymerase II for transcription.
Speed-stability paradox in DNA scanning
Transcription factors have to initiate their role by searching for its target DNA sites among billions of DNA base pairs in the nucleus. While scanning DNA, they should discriminate the target sites from an enormous amount of nonspecific sites based on relatively small difference. In the target search process, the transcription factors perpetually change their locations on DNA in a stochastic manner. Three major mechanisms for proteins’ translocation on DNA are known: 1) sliding, 2) dissociation and re-association, and 3) intersegment transfer (also known as direct transfer).10-12 When considering the DNA-scanning process, one should remember that DNA concentration in the nuclei is extremely high: ~100 mg/ml.13 This corresponds to ~5 mM for 30-bp segments. With typical affinities for nonspecific DNA (Kd around 0.1 – 10 μM), transcription factors in the search process in the nuclei should be mostly bound to nonspecific DNA sites, rather than in the free state, before reaching their target sites.12 Although this can in principle shorten the search time via reduced dimensionality,14 trapping by the nonspecific DNA sites can substantially slow the search process.10,15
In their 2004 paper on theoretical considerations on kinetics of protein-DNA interactions,2 Slutsky and Mirny demonstrated that rapidity in target search and stability in target binding are difficult to simultaneously be achieved without a special mechanism. This speed-stability paradox is highly relevant to zinc-finger proteins that involve several zinc fingers as DNA-binding domains. Extensive contact with DNA via multiple zinc fingers is obviously advantageous for high stability of the specific complex with a target site. However, the extensive contact with DNA can slow the search because the protein molecule has to break a larger number of interactions with DNA whenever it moves from one site to another on nonspecific DNA for scanning (Fig. 1A). Despite this problem, zinc-finger transcription factors such as Egr-1 achieve both rapid search and high affinity for target sites. How do the natural proteins resolve the speed-stability paradox?
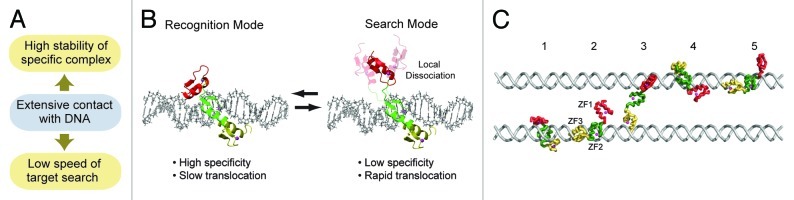
Figure 1. (A) Speed-stability paradox. (B) The search and recognition modes found in the DNA-scanning process of the zinc-finger protein Egr1. In the search mode, a zinc finger (red) is mainly dissociated while the other zinc fingers (green and yellow) are nonspecifically bound to DNA. In the recognition mode, all zinc fingers are in contact with DNA. (C) Intersegment transfer observed in the coarse-grained molecular dynamics simulations.
Some theoretical research groups considered a model known as the “conformational switch model” as a mechanism that can resolve the speed-specificity paradox.2,16-18 In this model, the speed-stability paradox is resolved via rapid conformational transitions of the proteins between the search and recognition modes. The search mode is suitable for the proteins’ rapid translocation but unsuitable for high stability and specificity, whereas the recognition mode is unsuitable for rapid translocation but suitable for high stability and specificity. By taking these two modes dynamically while bound to nonspecific DNA sites, the proteins can efficiently scan DNA in this model. Owing to recent advances in experimental and computational biophysics that have enabled investigations of sub-nanoscale dynamics in the DNA-scanning processes,19-22 examining the theoretical model has become easier.
The search and recognition modes of Egr-1
By using NMR spectroscopy and coarse-grained molecular dynamics simulations, Zandarashvili et al. found that the conformational switch model explains well how Egr-1 can achieve both high specificity in binding and rapidity in search.23 Egr-1 binds to its target DNA via three homologous zinc fingers ZF1, ZF2 and ZF3 (connected with two 5-residue linkers).3 The NMR investigations on Egr-1 bound to nonspecific DNA indicated that Egr-1 undergoes highly dynamic domain motions when scanning DNA.23 Egr-1’s ZF1 in the nonspecific DNA complex is mainly dissociated from DNA and undergoes independent collective motions with ZF2 and ZF3 being nonspecifically bound to DNA. The dynamic behavior of ZF1 in the DNA-scanning process was unexpected, because crystallographic studies of the specific complex indicated that all of Egr-1’s three zinc fingers are equally involved in binding to the 9-bp target sequence.3 In light of the conformational switch model, the dynamic state of Egr-1 bound to nonspecific DNA via only two zinc fingers corresponds to the search mode (Fig. 1B); the state observed in the crystal structure of the specific complex with all three zinc finger domains bound to DNA corresponds to the recognition mode. Domain motions seem to permit the transitions between the search and recognition modes of Egr-1. Because ZF1’s domain motions occur on a nanosecond timescale, transitions between the two modes can occur more rapidly than translocation. Owing to ZF1’s local dissociation, Egr-1 can rapidly transfer from one nonspecific site to another in the search mode. By kinetic experiments and coarse-grained molecular dynamics (CGMD) simulations, Zandarashvili et al. found that ZF1’s local dissociation in the search mode can facilitate intersegment transfer between two DNA duplexes via an intermediate where an Egr-1 molecule transiently bridges two DNA molecules (Fig. 1C).23 This process follows the “monkey-bar” mechanism.22 In fact, Egr-1’s intersegment transfer between two nonspecific DNA duplexes was found to occur very efficiently with a second-order rate constant of 3.6 × 106 M−1 s−1. Based on the CGMD data and the distance between two DNA ends of a nucleosome, they speculated that Egr-1 might bypass nucleosomes via intersegment transfer effectively and carry out continuous scanning for an efficient target search.
Balance between the search and recognition modes
Zandarashvili et al. demonstrated that the balance between the search and recognition modes could be modulated via mutagenesis.23 ZF1’s weaker interactions with DNA and with ZF2 appear to be the determinants of ZF1’s unique behavior in DNA-scanning (Figs. 2A and B). In fact, T23K/Q32E mutations that strengthen these interactions suppressed ZF1’s domain motions in DNA-scanning to a level comparable to those for ZF2 and ZF3. In other words, these mutations increase the population of the recognition mode, and decrease the population of the search mode. This caused significantly lower efficiency in intersegment transfer. It is probably possible to lower the population of the recognition mode by weakening ZF-DNA and ZF-ZF interactions. Such artificial modulation of the search and recognition modes could be useful in the zinc-finger-based technology (see below).
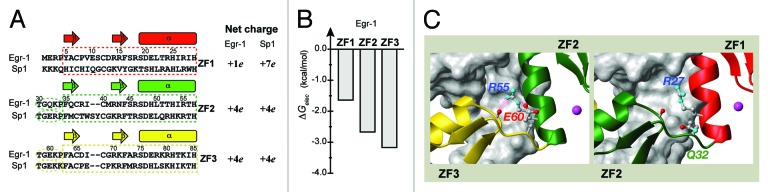
Figure 2. (A) The amino-acid sequences and net charges of the three zinc-finger domains of Egr-1 and Sp1. (B) Electrostatic binding free energies for Egr-1’s three zinc finger domains (C) Inter-domain interfaces in the crystal structure of the Egr-1 / target DNA complex. Two hydrogen bonds involving a salt bridge stabilize inter-domain interactions between ZF2 and ZF3, whereas there is no corresponding stabilization between ZF1 and ZF2.
The balance between the search and recognition modes for natural zinc-finger proteins may depend on whether they are inducible or constitutive transcription factors. Due to their high expression levels, constitutive transcription factors do not have to exhibit rapid search for rapid gene activation. So the balance between speed and stability for constitutive transcription factors may be shifted toward stability in target binding. Interestingly, comparison of amino-acid sequences of Sp1 and Egr-1 zinc fingers supports this idea (Fig. 2A). Together with its electrostatic free energy profile (Fig. 2B), Egr-1’s charge distribution (ZF1, +1e; ZF2, +4e; ZF3, +4e) suggests weak ZF1-DNA interaction, whereas that of the constitutive transcription factor Sp1 (ZF1, +7e; ZF2, +4e; ZF3, +4e) suggests strong ZF-DNA interactions for all zinc fingers. Furthermore, all amino-acid residues involved in inter-domain hydrogen bonds and salt bridge, which are found for ZF2-ZF3 interface of Egr-1 (Fig. 2C), are conserved for both ZF1-ZF2 and ZF2-ZF3 interfaces of Sp1. These facts implicate a low population of the search mode and a high population of the recognition mode for Sp1. This should be investigated in further studies.
Relevance to the zinc-finger-based technology
Artificial modulation of the balance between the search and recognition modes may allow improving the zinc-finger-based technology. Engineering of Cys2-His2-type zinc-finger proteins has been one of the most popular strategies for artificial gene control and in vivo gene manipulation.24-27 The methodology to change DNA-binding specificity of the zinc-finger proteins has been well established. It makes use of affinity-based selection from a pool of zinc-finger domains with random mutations at residues responsible for DNA recognition.28 Combining the engineered zinc-finger domains with other functional domains enable creating the artificial zinc-finger proteins that can control or manipulate particular genes. Artificial zinc-finger transcription factors with transactivation or repression domains have been successfully created for controlling disease-relevant genes.24,25 Zinc-finger nucleases (ZFNs) with a nuclease domain permit gene modifications via sequence-specific double-strand break followed by endogenous repair processes.26,27 Because the current zinc-finger-based technology relies on affinity-based selection that can give a strong bias toward stability of the complexes with target sites, it is possible that the current artificial zinc-finger proteins do not necessarily locate their target DNA rapidly.
In fact, there are some recent reports on defects of artificial zinc-finger proteins despite their high affinities for the target DNA sites.29-31 Shimizu et al. found that adding zinc fingers to ZFN substantially reduced the sequence-specific DNA-cleavage activity of ZFN.31 This is understandable in light of the speed-stability paradox, because additional zinc fingers would certainly render slower search for the target sites. Pattayanak et al. showed that off-target cleavage, which will cause toxicity in therapeutic applications of ZFN, becomes more significant with excessive DNA-binding energy.30 If the balance between stability and speed is shifted too much toward stability, the protein’s residence times at each nonspecific site should become longer, which may let cleavage happen. Engineering to optimize the balance between binding stability and search speed could improve artificial zinc-finger proteins for therapeutic applications.
Future Perspectives
Although target search by DNA-binding proteins has been studied over three decades, the field has been largely just descriptive, lacking the maturity required to enable engineering for improving the search kinetics of natural or artificial proteins. This was primarily because there were no suitable methods for investigating the DNA-scanning process at an atomic level. This critical barrier has been cleared now by recent methodological advances that have enabled obtaining sub-nanoscale information on DNA-scanning. We expect that further studies will enable optimizing the balance between stability and speed for artificial zinc-finger proteins. In particular, more information on the determinants of target search speed for zinc-finger proteins would help establish the strategy to create artificial zinc-finger proteins that can rapidly locate their target DNA sites. Further studies on DNA-scanning of zinc-finger proteins can thus lead to substantial improvement of artificial gene control and gene manipulation, and may boost the field of zinc-finger-based gene therapy, which is currently at Phases 1 and 2 of clinical trials.
Acknowledgments
This work was supported by Grant 12BGIA8960032 from the American Heart Association (to J.I.), and Grant 2010424 from the US-Israel binational science foundation (to J.I and Y.L.). We thank Levani Zandarashvili, Dana Vuzman, and Alexandre Esadze for useful discussion.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/23584
References
- 1.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–31. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 2.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–35. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 4.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40:695–701. doi: 10.1016/S0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 5.Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–43. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- 6.Yan SF, Fujita T, Lu J, Okada K, Shan Zou Y, Mackman N, et al. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat Med. 2000;6:1355–61. doi: 10.1038/82168. [DOI] [PubMed] [Google Scholar]
- 7.Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–91. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 8.Hocine S, Singer RH, Grünwald D. RNA processing and export. Cold Spring Harb Perspect Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nierhaus KH, Wilson D. Protein Synthesis and Ribosome Structure: Translating the Genome p Wiley-VCH: Weinheim, 2004. [Google Scholar]
- 10.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–52. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–8. [PubMed] [Google Scholar]
- 12.Mirny L, Slutsky M, Wunderlich Z, Tafvizi A, Leith J, Kosmrlj A. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J Phys A: Math Theor. 2009;42:401335. doi: 10.1088/1751-8113/42/43/434013. [DOI] [Google Scholar]
- 13.Lewin B. Genes VII p Oxford Univ Press: Oxford, 2000. [Google Scholar]
- 14.Adam G, Delbrück M. Reduction of dimensionality in biological diffusion processes. In Structural Chemistry and Molecular Biology, Rich, A.; Davidson, N., Eds. Freeman: New York, 1968; pp 198-215. [Google Scholar]
- 15.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–8. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 16.Marcovitz A, Levy Y. Frustration in protein-DNA binding influences conformational switching and target search kinetics. Proc Natl Acad Sci U S A. 2011;108:17957–62. doi: 10.1073/pnas.1109594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murugan R. Theory of site-specific DNA-protein interactions in the presence of conformational fluctuations of DNA binding domains. Biophys J. 2010;99:353–9. doi: 10.1016/j.bpj.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc Natl Acad Sci U S A. 2011;108:8651–6. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Givaty O, Levy Y. Protein sliding along DNA: dynamics and structural characterization. J Mol Biol. 2009;385:1087–97. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–30. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 21.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci U S A. 2006;103:15062–7. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vuzman D, Azia A, Levy Y. Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails. J Mol Biol. 2010;396:674–84. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 23.Zandarashvili L, Vuzman D, Esadze A, Takayama Y, Sahu D, Levy Y, et al. Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc Natl Acad Sci U S A. 2012;109:E1724–32. doi: 10.1073/pnas.1121500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, et al. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc Natl Acad Sci U S A. 2005;102:11716–21. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tachikawa K, Schröder O, Frey G, Briggs SP, Sera T. Regulation of the endogenous VEGF-A gene by exogenous designed regulatory proteins. Proc Natl Acad Sci U S A. 2004;101:15225–30. doi: 10.1073/pnas.0406473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–82. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 28.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–40. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 29.Isalan M. Zinc-finger nucleases: how to play two good hands. Nat Methods. 2012;9:32–4. doi: 10.1038/nmeth.1805. [DOI] [PubMed] [Google Scholar]
- 30.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–70. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu Y, Şöllü C, Meckler JF, Adriaenssens A, Zykovich A, Cathomen T, et al. Adding fingers to an engineered zinc finger nuclease can reduce activity. Biochemistry. 2011;50:5033–41. doi: 10.1021/bi200393g. [DOI] [PMC free article] [PubMed] [Google Scholar]


