Abstract
Background
The interleukin-2 (IL2)-mediated immune response is critical for host defence against infectious pathogens. CISH, a suppressor of cytokine signalling, controls IL2 signalling.
Methods
We tested for association between CISH polymorphisms and susceptibility to major infectious diseases (bacteremia, tuberculosis and severe malaria) in 8402 persons from the Gambia, Hong Kong, Kenya, Malawi, and Vietnam using a case-control design. We have previously tested twenty other immune-related genes in one or more of these sample collections.
Results
We observed associations between variant alleles of multiple CISH polymorphisms and increased susceptibility to each infectious disease in each of the study populations. When all five SNPs (CISH −639, −292, −163, +1320 and +3415) within the CISH-associated locus were considered together in a multi-SNP score, we found substantial support for an effect of CISH genetic variants on susceptibility to bacteremia, malaria, and tuberculosis (overall P=3.8 × 10−11) with CISH −292 being “responsible” for the majority of the association signal (P=4.58×10−7). Peripheral blood mononuclear cells of adult volunteers carrying the CISH −292 variant showed a muted response to IL2 stimulation — in the form of 25-40% less CISH — when compared with “control” cells lacking the −292 variant.
Conclusions
Variants of CISH are associated with susceptibility to diseases caused by diverse infectious pathogens, suggesting that negative regulators of cytokine signalling may play a major role in immunity against various infectious diseases. The overall risk of having one of these infectious diseases was found to be increased by at least 18 percent in individuals carrying the variant CISH alleles.
Introduction
Tuberculosis, malaria, and invasive bacterial disease together account for over five million deaths annually in the developing world. Although a significant proportion of inter-individual variation in disease susceptibility rests on environmental agents such as malnutrition and infection with HIV, a substantial portion is unexplained. Studies of twins and adopted persons suggest a genetic component,1 and the genes responsible for many primary immunodeficiency states have been identified. Such immunodeficiencies are extremely rare, however, and current understanding of common host genetic factors influencing susceptibility to major infectious diseases at broad population levels is limited.
A principal feature of the host immune response to infection by structurally diverse pathogens is the inflammatory cytokine response.2-4 The central pro-inflammatory cytokine interleukin-2 (IL2) determines the magnitude and duration of the T-cell response immediately following antigen encounter,5 and assists in the maturation of macrophages, and the proliferation of B cells and natural killer cells6 in the early stages of the adaptive immune response. IL2 also regulates the evolution of memory T cells after resolution of infection.7 An excessive cytokine-mediated inflammatory response can be harmful to the host, resulting in severe forms of malaria and sepsis.8-11
Control of cytokine signalling in humans is mediated in part via negative feedback by the Suppressor of Cytokine Signalling (SOCS) family of proteins. CISH (cytokine inducible SH-2 domain protein) was the first member of the SOCS family to be described.12,13 CISH has recently been shown to be the gene most consistently up-regulated by IL2 stimulation in humans14 and appears to be critical for T-cell proliferation and survival15 in response to infection. CISH controls the signalling of a variety of cytokines, in particular IL2. Unlike the other members of the SOCS family, CISH binds to the phosphorylated tyrosine residues of cytokine receptors through its SH-2 domain and masks sites at which STAT5 would otherwise dock.12,16-19 Thus, increased CISH activity blocks the cytoplasmic docking and activation of STAT5, and thereby inhibits downstream cytokine signalling.
Given the central role of CISH in controlling IL2 signaling, we hypothesised that variation in CISH influences susceptibility to common infectious disease in humans.
Methods
Patient collections
We analyzed 8,402 persons, making up seven case-control series (Table 1). These comprised Kenyan children with bacteremia,8,20 persons with tuberculosis from Malawi,21 Hong Kong22 and the Gambia, and persons with severe malaria from the Gambia,23 Kenya,24 and Vietnam.8 We obtained written informed consent from the study participants and ethical approval from the relevant national authorities (see Supplementary Appendix) for all study collections. We obtained blood samples from which we extracted DNA (see Supplementary Appendix). Control groups were geographically matched to the cases. Twenty other immune-related genes have been previously studied in one or more of these sample collections (see Supplementary Table 1).
Table 1.
Characteristics of all study cohorts enrolled.
| Cohort | Country | Locality | Disease | Control | Reference |
|---|---|---|---|---|---|
| Kenya Bacteremia (KB) | Kenya | Kilifi Coastal Area | 770 | 560 | 7, 19 |
| Malawi Tuberculosis (MTB) | Malawi | Karonga District | 335 | 450 | 20 |
| Hong Kong TB (HKTB) | China | Hong Kong City | 907 | 784 | 21 |
| Gambian TB (GTB) | The Gambia | Banjul Coastal Area | 1309 | 1427 | 7 |
| Kenyan Malaria (KM) | Kenya | Kilifi Coastal Area | 685 | 560* | 23 |
| Gambian Malaria (GM) | The Gambia | Banjul Coastal Area | 485 | 210 | 22 |
| Vietnam Malaria (VM) | Vietnam | Ho Chi Minh City | 375 | 105 | 7 |
| Total | 4866 | 3536 | 8402 |
Shared controls were used for both the Kenyan bacteremia and Kenyan malaria studies.7
Genotyping
We used standard methods, details of which can be found in Supplementary information.
Cell stimulation and gene expression study
We purified peripheral blood mononuclear cells (PBMCs) from whole blood drawn from human volunteers, cultured these cells, and then stimulated them with IL2 or IL3. We harvested the PBMCs at 0, 0.5 hr, 1 hr and 2 hr time-points after addition of interleukin, extracted RNAusing a standard method and synthesized and assayed cDNA using real time PCR (comparative method). Details can be found in Supplementary information.
Statistical analysis
Power calculations for all case-control studies can be found in Supplementary Figure 1. Comparison of allele frequencies between cases and controls was performed with the Pearson’s χ2 test under the different genetic modes of inheritance. The most likely mode of inheritance was determined by best model fit via logistic regression. Detailed descriptions of the statistical procedures, as well as of the various modes of inheritance are presented in Supplementary Information. Analysis of pair-wise linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) was performed using the r2 algorithm within HaploView v3.2.25 Multi-SNP score was analysed as previously described for case-control populations.26 Individuals were coded accordingly with the number of risk alleles they carry (0, 1, 2, 3, or ≥4). A trend test for association was then performed.27 Gene expression data analysis is located in Supplementary information.
Results
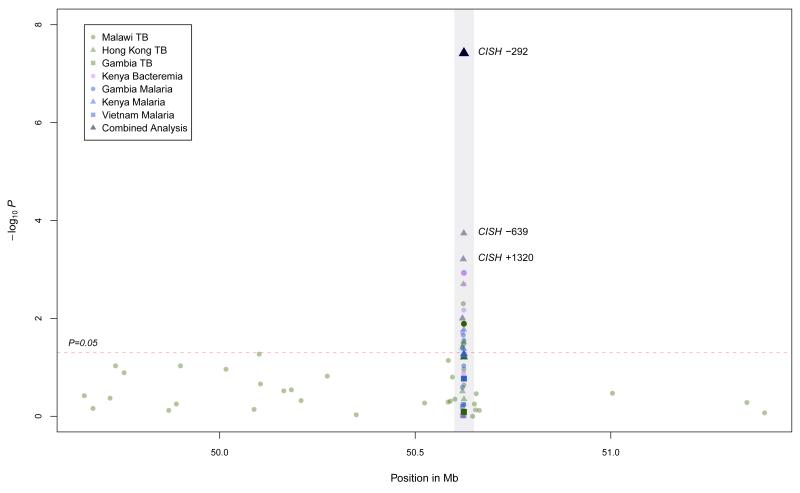
CISH comprises four exons, of which exons 2 to 4 encode the CISH protein. We sequenced CISH (1,000 base-pairs upstream of the transcription start to the end of exon 4, including introns) in 24 cases and 24 controls from the Kenyan Bacteraemia (KB) study. Power to detect SNPs of minor allele frequency (MAF)=0.05 was 99.27% (Supplementary Figure 2). We identified eight SNPs with a minor allele frequency greater than 5% (Supplementary Figure3a) and did not detect any novel coding changes or predicted splice-junction variants. These eight SNPs were then genotyped in the KB case-control study: four of these SNPs (at positions −639, −292, −163, and +3415) showed evidence of association (P=0.019–1.0×10−3; Figure 1, Table 2a); the variant alleles at each SNP were associated with increased susceptibility to bacteremia. Adjusting for HIV status, malnutrition and age did not significantly affect these associations.
Figure 1.
Log P-value plot for SNPs typed within CISH (grey highlight) and its 2 Mb flanking region.
Table 2a.
Results of single SNP analysis of CISH −639, −292, −163 and +3415 (per study and pooled).
| SNP ID |
Study | Sample size |
Best model |
Risk allele frequency (affected)d |
Risk allele frequency (controls) |
P-value | Odds Ratio (OR) |
95% CI for OR |
Pooled P-valueb |
Pooled OR |
95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| −639 | KB | 1326 | allelic | 0.064 | 0.04 | 6.8 × 10−3 | 1.64 | 1.14-2.35 | |||
| MTB | 607 | allelic | 0.036 | 0.021 | 0.11 | 1.76 | 0.87-3.53 | ||||
| HKTB | - | - | - | - | - | - | - | ||||
| GTB | 2690 | allelic | 0.025 | 0.024 | 0.88 | 1.03 | 0.68-1.57 | 1.8 × 10−4 | 1.49 | 1.22-1.83 | |
| KM | 1212 | allelic | 0.062 | 0.04 | 0.017 | 1.57 | 1.08-2.28 | ||||
| GM | 660 | allelic | 0.031 | 0.015 | 0.093 | 2.11 | 0.87-5.11 | ||||
| VM | - | - | - | - | - | - | - | ||||
| −292 | KB | 1293 | allelic | 0.412 | 0.349 | 1.1 × 10−3 | 1.31 | 1.11-1.55 | |||
| MTB | 749 | allelic | 0.454 | 0.389 | 0.013 | 1.30 | 1.06-1.61 | ||||
| HKTB | 1677 | allelic | 0.41 | 0.37 | 0.031 | 1.17 | 1.01-1.34 | ||||
| GTB | 2696 | allelic | 0.282 | 0.285 | 0.81 | 0.99 | 0.88-1.11 | 4.58×10−7 | 1.19 | 1.12-1.25 | |
| KM | 1174 | allelic | 0.39 | 0.349 | 0.043 | 1.19 | 1.01-1.41 | ||||
| GM | 675 | allelic | 0.282 | 0.224 | 0.027 | 1.36 | 1.03-1.78 | ||||
| VM | 471 | dominant | 0.41 | 0.34 | 0.033 | 1.60 | 1.01-2.52 | ||||
| −163 | KB | 1006 | allelic | 0.173 | 0.122 | 2.0 × 10−3 | 1.50 | 1.16-1.95 | |||
| MTB | 629 | allelic | 0.16 | 0.135 | 0.23 | 1.21 | 0.88-1.68 | ||||
| HKTB | 1316 | allelic | 0.17 | 0.18 | 0.45 | 0.93 | 0.76-1.13 | ||||
| GTB | 2690 | allelic | 0.20 | 0.19 | 0.64 | 1.04 | 0.88-1.22 | c | c | c | |
| KM | 995 | allelic | 0.235 | 0.122 | 3.6×10−10e | 2.20 | 1.72-2.84 | ||||
| GM | 649 | allelic | 0.127 | 0.116 | 0.58 | 1.11 | 0.76-1.63 | ||||
| VM | - | - | - | - | - | - | - | ||||
| +1320 | KB | 1200 | allelic | 0.16 | 0.16 | 0.99 | 1.00 | 0.74-1.35 | |||
| MTB | 819 | recessive | 0.17 | 0.16 | 5.0 × 10−3 | 2.99 | 1.27-7.18 | ||||
| HKTB | 1691 | allelic | 0.36 | 0.31 | 2.0 × 10−3 | 1.25 | 1.08-1.44 | ||||
| GTB | 2736 | allelic | 0.063 | 0.050 | 0.033 | 1.28 | 1.02-1.62 | 6.1×10−4 | 1.17 | 1.07-1.28 | |
| KM | 1024 | allelic | 0.16 | 0.16 | 0.99 | 1.00 | 0.78-1.28 | ||||
| GM | 689 | allelic | 0.064 | 0.034 | 0.022 | 1.98 | 1.09-3.57 | ||||
| VM | 479 | allelic | 0.33 | 0.31 | 0.58 | 1.10 | 0.79-1.53 | ||||
| +3415 | KB | 1257 | allelic | 0.242 | 0.202 | 0.019 | 1.26 | 1.04-1.53 | |||
| MTB | 780 | allelic | 0.287 | 0.24 | 0.039 | 1.27 | 1.01-1.59 | ||||
| HKTB | 1677 | allelic | 0.055 | 0.063 | 0.31 | 0.86 | 0.65-1.15 | ||||
| GTB | 2702 | allelic | 0.22 | 0.22 | 0.96 | 1.00 | 0.88-1.13 | 0.010 | 1.11 | 1.03-1.20 | |
| KM | 1189 | allelic | 0.237 | 0.202 | 0.040 | 1.23 | 1.01-1.50 | ||||
| GM | 684 | allelic | 0.233 | 0.205 | 0.26 | 1.18 | 0.89-1.56 | ||||
| VM | 478 | allelic | 0.057 | 0.049 | 0.63 | 1.19 | 0.59-2.42 |
We observed low pair-wise LD between SNPs at positions −639 and +3415, between these SNPs and those that lie between them, and between the SNPs that lie between them (r2<0.50; Supplementary Figure 3b). We also observed associations between multiple SNPs and susceptibility to disease and, bearing in mind the low pair-wise LD between these SNPs, hypothesized that risk alleles at these SNPs confer susceptibility independent from one another. To investigate whether risk of disease increased in a dose-dependent manner with respect to number of risk alleles, we performed multi-SNP scoring for the five SNPs.26 The risk of bacteremia was found to be proportionate with the number of risk alleles carried (P=5.1×10−5; Table 2b). Haplotypic analysis was uninformative due to the low inter-marker LD. We observed no significant evidence of interaction between the five SNPs, and went on to genotype them in the remaining six case-control studies.
Table 2b.
Meta-SNP analysis of CISH −639, −292, −163, +1320 and +3415 in all study cohorts. Trend tests were performed for the increasing number of CISH risk alleles (0, 1, 2, 3, or ≥4) carried and increasing odds of disease susceptibility for each population.
| Population specific disease odds ratios | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| No. of risk alleles carried |
KB | MTB | HKTB | GTB | All TB | GM | KM | VM | All Malaria | All cohorts |
| N~1200 | N~750 | N~1600 | N~2700 | N~5050 | N~650 | N~1150 | N~480 | N~2300 | N~8000 | |
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1.79 | 1.00 | 0.9 | 1.28 | 1.12 | 1.24 | 0.64 | 1.58 | 1.01 | 1.18 |
| 2 | 1.37 | 0.88 | 1.22 | 1.28 | 1.20 | 2.13 | 0.74 | 1.65 | 1.32 | 1.26 |
| 3 | 2.08 | 1.54 | 1.2 | 1.28 | 1.29 | 1.92 | 1.34 | 1.38 | 1.50 | 1.46 |
| ≥4 | 2.36 | 1.77 | 1.43 | 1.28 | 1.40 | 4.19 | 1.70 | - | 2.60 | 1.81 |
|
| ||||||||||
| Overall Trend P |
5.1×10−5 | 0.030 | 0.011 | 0.034 | 8.7×10−4 | 4.7×10−3 | 1.7×10−3 | 0.13 | 1.96×10−6 | 3.8×10−11 |
KB: Kenyan Bacteraemia study
MTB: Malawian tuberculosis study
HKTB: Hong Kong tuberculosis study
GTB: Gambian tuberculosis study
GM: Gambian malaria study
KM: Kenyan malaria study
VM: Vietnam malaria study
- denotes that the marker was non-polymorphic in that specific population.
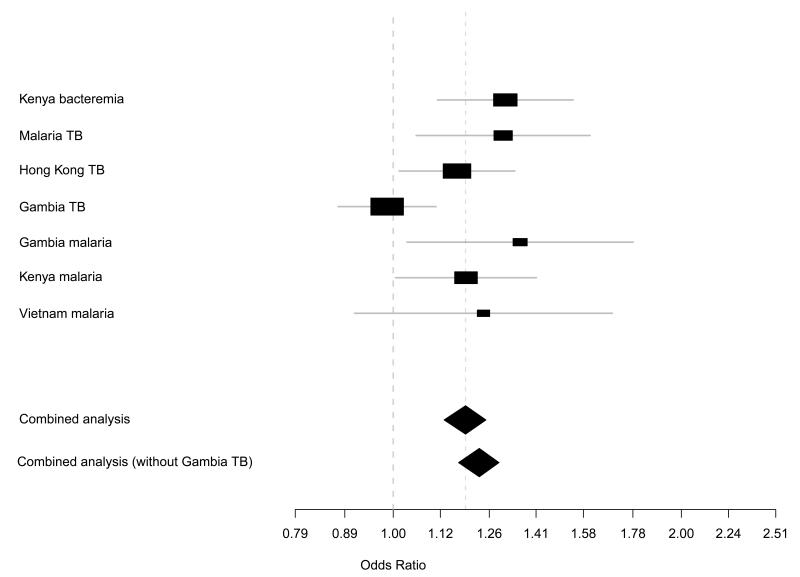
Both the Tarone and Breslow-Day’s test for homogeneity was not significant across all populations for CISH −639, −292 and +3415 (P>0.4), indicating that the pooled odds ratio and accompanying pooled P was applicable across all populations. However, for CISH −292, inclusion of the GTB set causes the tests of homogeneity to be borderline significant (P=0.059). As such, we performed a secondary analysis for −292 which did not include the GTB set (P = 3.9 × 10−8, OR = 1.23, 95%CI: 1.19-1.34).
The Tarone and Breslow-Day’s test for homogeneity was markedly significant (P<0.001) for CISH −163, and thus pooling should not be performed for this SNP. However, if pooling was performed anyway for the sake of information, P = 3.0×10−7.
Risk allele frequency referred to the minor allele frequency for each SNP genotyped. They were the actual allelic frequencies and were not the result of prediction programs. It was calculated as number of mutant alleles divided by total number of alleles.
As this was an apparently highly significant single-point observation, we confirmed the direct sequencing calls by an independent and blinded person. The concordance rate was absolute, arguing against genotyping error as the cause of this observation.
In the Malawian tuberculosis (MTB) study, the minor alleles of three SNPs (-292, +1320, and +3415) were associated with increased susceptibility to TB (P=0.013-5.0×10−3; Figure 1, Table 2a). For CISH +1320 (overall P=6.0×10−3), the effect of the risk allele appeared strongest with a recessive mode of inheritance. Individuals homozygous for the risk allele were significantly more susceptible to tuberculosis compared to matched controls (5.6% of cases were homozygous for the risk allele, in contrast with 1.9% of controls; P=5.0×10−3). Adjusting for the potential confounding effects of age, gender, and ethnicity did not affect the degree of statistical significance and ORs for each SNP. The trend test for increasing disease risk with increasing number of risk alleles was also positive in this study (P=0.030, Table 2b). In the Hong Kong TB study (HKTB), those with TB were more likely to carry a couple of variant CISH alleles (at positions −292 and +1320) than unaffected persons (P=0.031 and P=2.0×10−3 respectively). In the case-control TB study from the Gambia (GTB), we observed association between the CISH +1320 variant allele (but not the −292 SNP variant allele) and susceptibility to clinical tuberculosis (P=0.033). The positive trend test for TB observed in the MTB study was replicated in both HKTB (P=0.011) and GTB (P=0.034).
We genotyped the five CISH SNPs in three severe malaria collections from Gambia (GM), Kenya (KM), and Vietnam (VM). The variant alleles at positions −639, −292, +1320 and +3415 showed significant association with increased susceptibility to disease. In particular, the minor allele of −292 showed significant association with susceptibility to severe malaria in GM, KM, as well as the smaller VM study (Table 2a). As was observed in the bacteremia and tuberculosis studies, the trend test for increasing disease risk with increasing carriage of CISH risk alleles replicated across all three malaria studies (Table 2b).
To investigate the possibility that the associated SNPs might be in LD with a true causative variant elsewhere we performed exclusion mapping by genotyping 28 additional evenly-spaced SNPs spanning 2,000,000 base-pairs in the Chr3p21 region surrounding CISH in the Malawi TB study population. None of the additional SNPs typed displayed evidence of association with TB (Figure 1), suggesting that the observed associations are specific to CISH.
On pooled analysis, carriage of CISH risk alleles at −639, −292, −163, +1320, and +3415 associated with increased susceptibility to the infectious diseases studied (Table 2a), with −292 accounting for the majority of the association signal (P=4.58×10−7, Table 2a and Figure 2). Multi-SNP scoring of the five SNPs in each case-control study revealed that carriage of ≥1 CISH risk allele is accompanied by increasing risk of disease in each study (trend P=3.8×10−11, Table 2b).
Figure 2.
Forest plots analyses for CISH −292 (rs414171) in all study cohorts. Results of the overall pooled analysis and case-control analyses were generated using the allelic test. Plots include the disease odd ratio for the variant allele in each study.
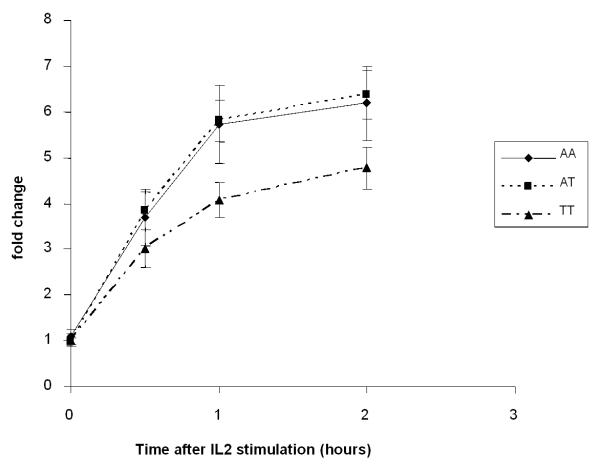
We studied the functional effects of the CISH promoter variants because these SNPs showed the strongest associations with disease status and they are more likely to affect gene expression than intra-genic SNPs. We genotyped the promoter SNPs, SNPs −639, −292 and −163 in 400 healthy adult volunteers of Chinese descent. The observed risk allele frequency of CISH −292 and −163 was 41.5% and 6.1% respectively, with only a single volunteer found to be homozygous for the variant allele at position −163; CISH −639 was non-polymorphic in these individuals, consistent with observations from the Hong Kong Chinese case-control studies. We first examined the individual effects of SNPs −292 and −163 on CISH gene expression in human PBMCs following stimulation with IL2 at a final concentration of 100U/mL. As expression levels of the wild-type CISH −292A/A and carrier CISH −292A/T genotypes were similar (P>0.9), we questioned whether this SNP might exert a recessive effect. As shown in Figure 3a, individuals homozygous for the variant CISH −292T/T risk genotype (n=10) showed significantly lower levels of CISH when compared with either those homozygous for alternative allele (A/A)(n=5) or heterozygous carriers (A/T)(n=10) at 0.5, 1 and 2 hours post-stimulation with IL2. We did not observe any difference in CISH expression (with respect to CISH −292 genotype) following stimulation with IL3. Nor did we observe a genotype-specific difference (according to allelic variation at position −163) after stimulating cells with either IL2 or IL3 (P>0.8, data not shown).
Figure 3.
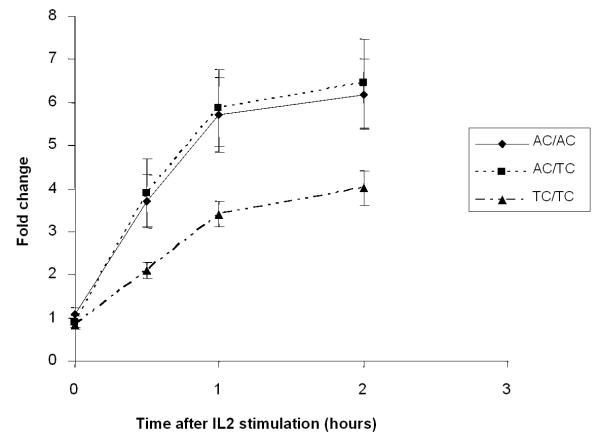
a) Differential expression of wild-type (AA; n=5), heterozygote (AT; n=10) and homozygous mutant (TT; n=10) forms of CISH −292 in response to IL-2. b) Differential expression of wild-type (AC/AC; n=5), heterozygote (AC/TC; n=5) and homozygous mutant (TC/TC; n=5) forms of CISH −292 in response to IL-2 in individuals who are wild-type homozygous (CC) at the −163 locus.
To test for an interactive effect between CISH −292 or CISH −163, we determined two-SNP (−292 and −163) diplotypes (with chromosomal phase determined via subcloning and sequencing for individuals whose phase was uncertain) for the subjects analysed for CISH gene expression, and CISH expression was then compared between the −292 genotypes in response to IL2 stimulation using −163 as the conditioning locus. For individuals who were wild-type at −163 (C/C), carrying the variant −292T/T genotype resulted in markedly lower overall gene expression in response to IL2 stimulation at all time points (Figure 3b). We observed no significant differences in CISH expression following stimulation by IL2 or IL3 when comparing the other CISH diplotypes with one another (data not shown).
Discussion
We identified a panel of five CISH SNPs associated with increased susceptibility to bacteremia, tuberculosis and malaria in human populations, and estimated that the overall risk of having one of these infectious diseases was increased by 18 percent in persons carrying a single “risk” CISH allele, increasing to 81 percent in those with four or more risk alleles (Table 2b).
Two important considerations in genetic association studies are population stratification and multiple testing. To assess the presence of population stratification, we examined 28 independent markers in the 2-Mb region flanking CISH and did not detect significant inflation of test statistics (λgc = 1.03). Furthermore, the consistency of the association across multiple ethnicities argues against the results being a product of population stratification. Regarding multiple testing, we evaluated CISH in the context of 20 other immune-related genes (analyzing a total of 187 SNPs) previously tested in one or more of these sample collections (Supplementary Table 1); the single-point observation with CISH −292 (P=4.58×10−7) remains significant following correction for all the genes and SNPs tested (Threshold after correction for multiple-testing: 0.05/500 = 10−4; 187 SNPs tested a total of 500 times cumulatively). The false-positive-report-probability28 for −292 is ≤10−4 even at prior probability levels of ≤10−5. Further confidence is lent by the very low P-value (<5×10−11) observed with the multi-SNP score and the level of replication between study groups.
The pattern of association with CISH −292 was consistently reproducible across six out of the seven study groups, with the exception being the Gambian TB study. One possible explanation for this heterogeneity is that disease susceptibility was accounted for by more than one SNP within the five-SNP panel, thus rendering single SNP analysis incomplete. A second reason might relate to underlying population structure, where different CISH SNPs may tag the informative variants in each distinct population. A third possibility is that there remain unidentified functional SNPs within the region of association delineated by the five-SNP panel that also account for association with disease. This last possibility is unlikely however, as direct sequencing did not detect additional putatively functional polymorphisms. To explore the first and second possibilities, we utilized multi-SNP scoring for all five associated SNPs. Risk of disease increased markedly with an increase in the number of risk alleles carried in each population. As this multi-SNP analysis was more informative than single-point analysis, the above two possibilities remain plausible explanations. The mechanisms underlying an association between the CISH multi-SNP score and accompanying dose-dependant effect on disease susceptibility likely reflect the potential regulatory effect of these polymorphisms within a ‘multi-hit model’, whereby each ‘hit’ affects gene expression cumulatively in aggregate. Such a process is in contrast to structural variants, where the presence of one deleterious mutation may be sufficient to account for disease.
In the ex-vivo volunteer study, carriage of the −292 allele reduced CISH expression post-stimulation with IL2. However, this study lacks power to detect significant differences in gene expression in individuals with 0, 1, 2, 3, and ≥4 risk alleles. Although −292 showed associations with multiple infectious diseases and could be a true functional variant, CISH +1320 showed a stronger association with susceptibility to TB, and showed association in each of the “TB” studies. The possibility that variation at +1320 affect transcript expression is possible; this position is located in the untranslated portion of exon 2.29
Further work is required to document the precise mechanisms whereby reduced CISH expression (and presumably consequential disturbed IL2 signaling) predisposes to increased risk of infectious disease. Stimulation by IL2 may enhance microbial and viral replication,30 and its effect may be further dependent on the presence of other immune cells: clinical trials of IL2 infusion in HIV-positive patients have shown different effects on individuals depending on their CD4+ counts.31,32 Although it is perhaps unexpected that common variation within a single gene influences susceptibility to a diverse range of infectious diseases, there is increasing evidence that disparate infections are recognized by a common host inflammatory pathway. 8,33-35
The observation that the risk alleles occur at appreciable allele frequencies in each of the study populations is surprising, given evidence suggestive of evolutionary selective pressure exerted by some infectious diseases.36,37 One explanation may be that the variant alleles could have been concurrent modifiers of human susceptibility against other major causes of mortality in these populations. For example immune modulation at the IL-2 receptor axis may protect against Type 1 diabetes mellitus;38 a possible role of CISH polymorphisms in the development of inflammatory as well as infectious diseases merits further study.
Current clinical management of bacteremia, malaria and TB relies primarily on anti-microbials specifically targeted to the likely pathogen. Our findings implicate CISH in multi-pathogen susceptibility and raise the possibility that pharmacological manipulation of the SOCS pathway may impact the treatment of multiple, diverse infectious diseases. CISH variants may also influence responses to existing immunotherapies, such as IL2 therapy in renal cell cancer, which is associated with wide and largely unexplained variations in inter-individual response rates.39,40 Study of longitudinal immune responses together with responses to antimicrobials and clinical outcome in patients with infectious disease may further define the risk model for CISH SNPs. The integration of such a model with environmental and other host genetic factors should improve the prediction of treatment responses and disease outcomes.
Supplementary Material
Acknowledgements
We thank all the patients and volunteers as well as the many investigators involved in the original case-control studies in The Gambia, Hong Kong, Kenya, Malawi, and Vietnam for their contributions. This work was funded by the Wellcome Trust and the Agency for Science, Technology and Research (A-STAR), Singapore. CCK is a scholar of A-STAR. FOV is supported by the EU FP6 GENOSEPT Grant and by the UK ORS Scheme. SJC is a Wellcome Trust Clinical Research Fellow; TNW, JAS & JAB are Wellcome Trust Clinical Research Fellows, KM is Director of the Wellcome Trust Kenya Major Overseas Programme. DLMG is supported by an A-STAR intramural programme grant, AVSH is a Wellcome Trust Principal Fellow.
Footnotes
Disclosure The authors have no competing financial interest in this manuscript.
References
- 1.Sørensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–32. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 2.Flynn JL, Chan J. Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr Opin Immunol. 2003;15:450–5. doi: 10.1016/s0952-7915(03)00075-x. [DOI] [PubMed] [Google Scholar]
- 3.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 4.Lyke KE, Burges R, Cissoko Y, et al. Serum levels of the proinflammatory cytokines interleukin-1 beta (IL-1beta), IL-6, IL-8, IL-10, tumor necrosis factor alpha, and IL-12(p70) in Malian children with severe Plasmodium falciparum malaria and matched uncomplicated malaria or healthy controls. Infect Immun. 2004;72:5630–7. doi: 10.1128/IAI.72.10.5630-5637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aman MJ, Migone TS, Sasaki A, et al. CIS associates with the interleukin-2 receptor beta chain and inhibits interleukin-2-dependent signalling. J Biol Chem. 1999;274:30266–72. doi: 10.1074/jbc.274.42.30266. [DOI] [PubMed] [Google Scholar]
- 6.Lin JX, Leonard WJ. Signalling from the IL-2 receptor to the nucleus. Cytokine Growth Factor Rev. 1997;8:313–32. doi: 10.1016/s1359-6101(97)00021-x. [DOI] [PubMed] [Google Scholar]
- 7.Dooms H, Wolslegel K, Lin P, Abbas AK. Interleukin-2 enhances CD4+ T cell memory by promoting the generation of IL-7R alpha-expressing cells. J Exp Med. 2007;204:547–57. doi: 10.1084/jem.20062381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khor CC, Chapman SJ, Vannberg FO, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 10.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365:63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 11.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 1995;377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimura A, Ohkubo T, Kiguchi T, et al. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–26. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchida K, Yoshimura A, Inazawa J, et al. Molecular cloning of CISH, chromosome assignment to 3p21.3, and analysis of expression in fetal and adult tissues. Cytogenet Cell Genet. 1997;78:209–12. doi: 10.1159/000134658. [DOI] [PubMed] [Google Scholar]
- 14.Jin P, Wang E, Provenzano M, et al. Molecular signatures induced by interleukin-2 on peripheral blood mononuclear cells and T cell subsets. J. Transl Med. 2006;4:26. doi: 10.1186/1479-5876-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol Rev. 2004;202:67–83. doi: 10.1111/j.0105-2896.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto A, Seki Y, Kubo M, et al. Suppression of STAT5 functions in liver, mammary glands, and T cells in cytokine-inducible SH2-containing protein 1 transgenic mice. Mol Cell Biol. 1999;19:6396–407. doi: 10.1128/mcb.19.9.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto A, Masuhara M, Mitsui K, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–54. [PubMed] [Google Scholar]
- 18.Nakajima H, Liu X-W, Wynshaw-Boris A, et al. An indirect effect of Stat5a in IL-2-induced proliferation: A critical role for Stat5a in IL-2-mediated IL-2 receptor a chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 19.Moriggl R, Topham DJ, Teglund S, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–59. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 20.Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 21.Crampin AC, Mwinuka V, Malema SS, Glynn JR, Fine PE. Field-based random sampling without a sampling frame: control selection for a case-control study in rural Africa. Trans R Soc Trop Med Hyg. 2001;95:481–3. doi: 10.1016/s0035-9203(01)90009-4. [DOI] [PubMed] [Google Scholar]
- 22.Tang NL, Chan PK, Hui DS, et al. Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection. Nat Genet. 2007;39:691–2. doi: 10.1038/ng0607-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill AV, Allsopp CE, Kwiatkowski D, et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 24.Marsh K, Forster D, Waruiru C, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332:1399–404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Mira MT, Alcais A, Thuc NV, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 27.Clayton D. In: Handbook of Statistical Genetics. Balding DJ, Bishop M, Cannings C, editors. Wiley; New York: 2003. pp. 939–960. [Google Scholar]
- 28.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Fauci AS. Host Factors in the Pathogenesis of HIV Disease. Program and abstracts of the 39th ICAAC; San Francisco, Calif. September 26-29, 1999; Abstract 614. [Google Scholar]
- 31.Kovacs JA, Baseler M, Dewar RJ, et al. Increases in CD4 T lymphocytes with intermittent courses of interleukin-2 in patients with human immunodeficiency virus infection. A preliminary study. N Engl J Med. 1995;332:567–75. doi: 10.1056/NEJM199503023320904. [DOI] [PubMed] [Google Scholar]
- 32.Kovacs JA, Vogel S, Albert JM, et al. Controlled trial of interleukin-2 infusions in patients infected with the human immunodeficiency virus. N Engl J Med. 1996;335:1350–6. doi: 10.1056/NEJM199610313351803. [DOI] [PubMed] [Google Scholar]
- 33.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–5. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 34.Hawn TR, Dunstan SJ, Thwaites GE, et al. A polymorphism in Toll-interleukin 1 receptor domain containing adaptor protein is associated with susceptibility to meningeal tuberculosis. J Infect Dis. 2006;194:1127–34. doi: 10.1086/507907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summerfield JA, Sumiya M, Levin M, Turner MW. Association of mutations in mannose binding protein gene with childhood infection in consecutive hospital series. BMJ. 1997;314:1229–32. doi: 10.1136/bmj.314.7089.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tishkoff SA, Varkonyi R, Cahinhinan N, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–62. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 37.Barreiro LB, Ben-Ali M, Quach H, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;e1000562 doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chistiakov DA, Voronova NV, Chistiakov PA. The crucial role of IL-2/IL-2RA-mediated immune regulation in the pathogenesis of type 1 diabetes, an evidence coming from genetic and animal model studies. Immunol Lett. 2008;118:1–5. doi: 10.1016/j.imlet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 39.McDermott DF. Update on the application of interleukin-2 in the treatment of renal cell carcinoma. Clin Cancer Res. 2007;13(2):716s–720s. doi: 10.1158/1078-0432.CCR-06-1872. [DOI] [PubMed] [Google Scholar]
- 40.Hoyer KK, Dooms H, Barron L, Abbas AK. Interleukin-2 in the development and control of inflammatory disease. Immunological Reviews. 2008;226:19–28. doi: 10.1111/j.1600-065X.2008.00697.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.