Abstract
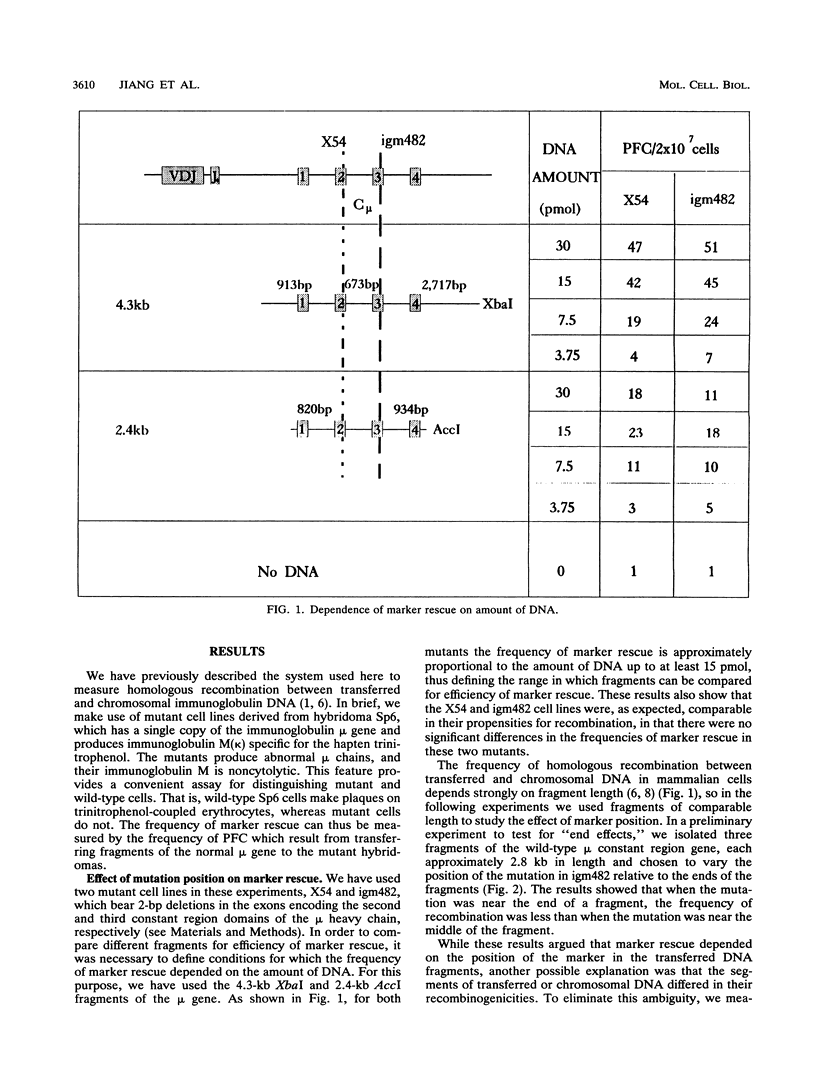
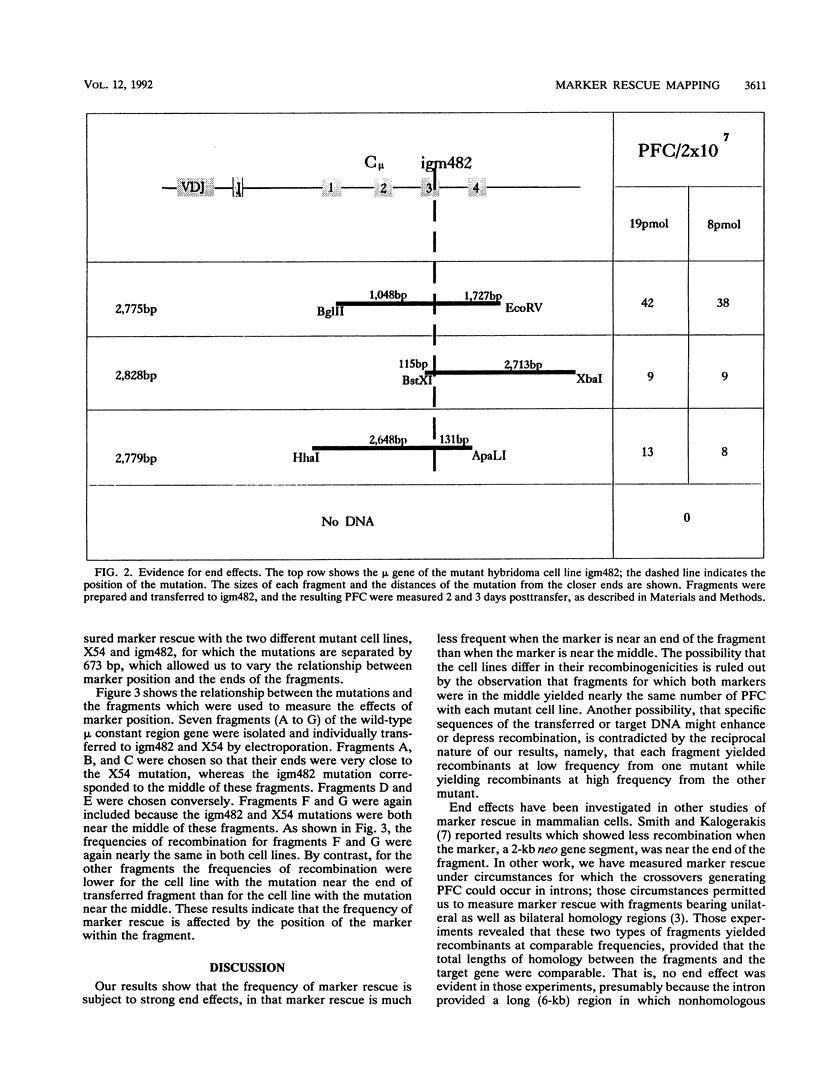
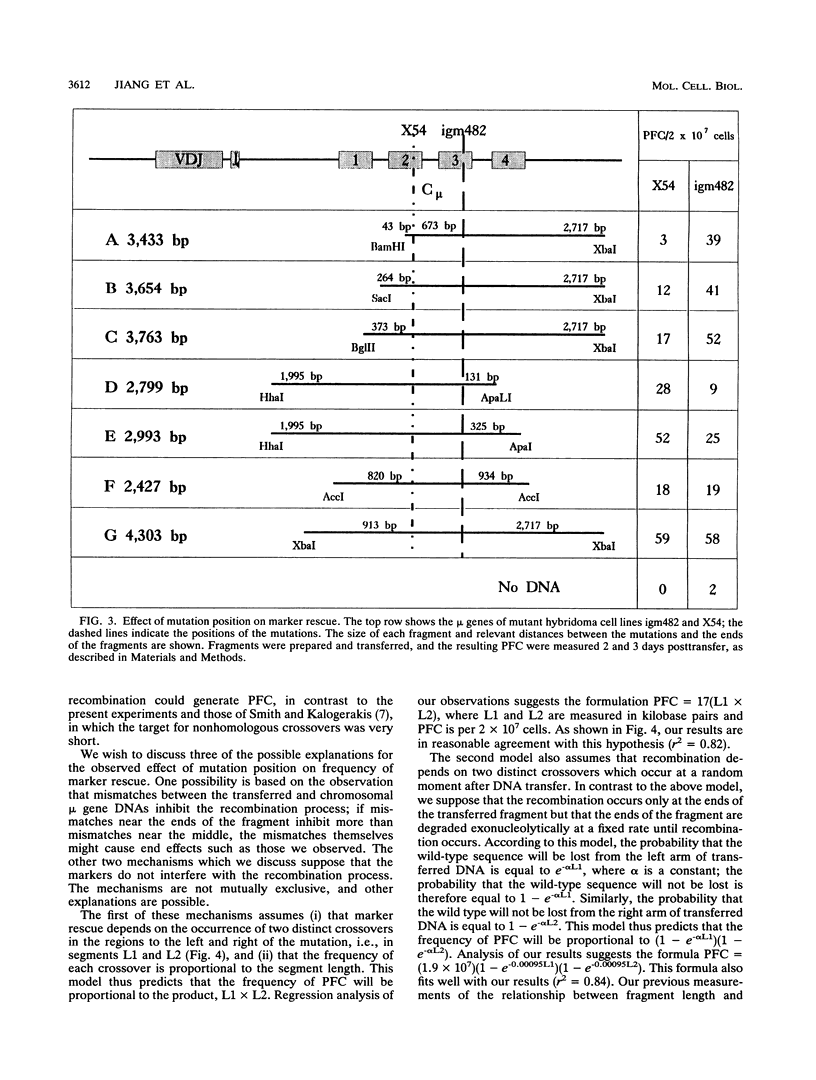
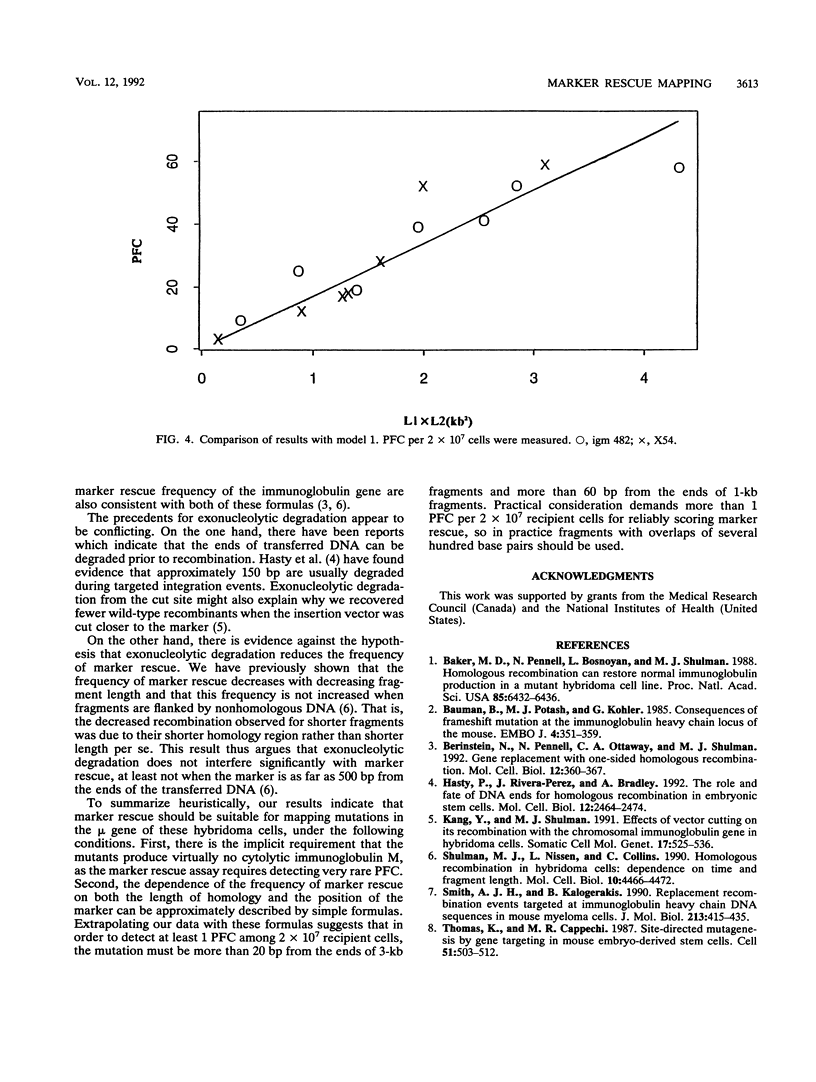
Homologous recombination between transferred and chromosomal DNA can be used for mapping mutations by marker rescue, i.e., by identifying which segment of wild-type DNA can recombine with the mutant chromosomal gene and restore normal function. In order to define how much the fragments should overlap each other for reliable mapping, we have measured how the frequency of marker rescue is affected by the position of the chromosomal mutation relative to the ends of the transferred DNA fragments. For this purpose, we used several DNA fragments to effect marker rescue in two mutant hybridomas which bear mutations 673 bp apart in the exons encoding the second and third constant region domains of the immunoglobulin mu heavy chain. The frequency of marker rescue decreased greatly when the mutation was located near one of the ends of the fragments, the results indicating that fragments should be designed to overlap by at least several hundred base pairs. Possible explanations for this "end effect" are considered.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker M. D., Pennell N., Bosnoyan L., Shulman M. J. Homologous recombination can restore normal immunoglobulin production in a mutant hybridoma cell line. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6432–6436. doi: 10.1073/pnas.85.17.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash M. J., Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berinstein N., Pennell N., Ottaway C. A., Shulman M. J. Gene replacement with one-sided homologous recombination. Mol Cell Biol. 1992 Jan;12(1):360–367. doi: 10.1128/mcb.12.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P., Rivera-Pérez J., Bradley A. The role and fate of DNA ends for homologous recombination in embryonic stem cells. Mol Cell Biol. 1992 Jun;12(6):2464–2474. doi: 10.1128/mcb.12.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Shulman M. J. Effects of vector cutting on its recombination with the chromosomal immunoglobulin gene in hybridoma cells. Somat Cell Mol Genet. 1991 Nov;17(6):525–536. doi: 10.1007/BF01233617. [DOI] [PubMed] [Google Scholar]
- Shulman M. J., Nissen L., Collins C. Homologous recombination in hybridoma cells: dependence on time and fragment length. Mol Cell Biol. 1990 Sep;10(9):4466–4472. doi: 10.1128/mcb.10.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J., Kalogerakis B. Replacement recombinant events targeted at immunoglobulin heavy chain DNA sequences in mouse myeloma cells. J Mol Biol. 1990 Jun 5;213(3):415–435. doi: 10.1016/s0022-2836(05)80205-0. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987 Nov 6;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]


