Summary
Mesp1 is regarded as the master regulator of cardiovascular development, initiating the cardiac transcription factor cascade to direct the generation of cardiac mesoderm. To define the early embryonic cell population that responds to Mesp1, we performed pulse inductions of gene expression over tight temporal windows following embryonic stem cell differentiation. Remarkably, instead of promoting cardiac differentiation in the initial wave of mesoderm, Mesp1 binds to the Tal1 (Scl) +40k enhancer and generates Flk-1+ precursors expressing Etv2 (ER71) and Tal1 that undergo hematopoietic differentiation. The second wave of mesoderm responds to Mesp1 by differentiating into PDGFRα+ precursors that undergo cardiac differentiation. Furthermore, in the absence of serum-derived factors, Mesp1 promotes skeletal myogenic differentiation. Lineage tracing revealed that the majority of yolk sac and many adult hematopoietic cells derive from Mesp1+ precursors. Thus, Mesp1 is a context-dependent determination factor, integrating stage of differentiation and signaling environment to specify different lineage outcomes.
Keywords: Mesp1, mesoderm patterning, cardiogenesis, hematopoiesis, skeletal myogenesis
Introduction
Cardiac cell therapy from pluripotent stem cells has received intense interest as a possible treatment of heart diseases (Burridge et al., 2012; Laflamme and Murry, 2011; Ptaszek et al., 2012). Pluripotent stem cells first differentiate through an epiblast state, are subsequently restricted to mesoderm, patterned to become cardiovascular, and eventually develop into cardiovascular progenitors and cardiomyocytes. Much work has been done aiming to facilitate this process by employing various signaling pathway modulators (Burridge et al., 2011; Kattman et al., 2011; Laflamme et al., 2007; Yang et al., 2008). Despite the value of this approach, each pluripotent stem cell line produces different levels of endogenous cytokines during differentiation, hence the reproducibility of methods across cell lines is low (Bock et al., 2011; Osafune et al., 2008). Nonetheless, methods to modulate cardiogenic signaling pathways all converge on the activation of a core cardiogenic transcription network (Olson, 2006). Direct regulation of cardiac transcription factors is therefore a promising strategy to generate cardiomyocytes.
Several transcription factors have been identified to be essential to cardiac development (see Olson, 2006). Among them, Mesp1 is expressed the earliest at E6.5 along the primitive streak, and marks the cardiac mesodermal population that gives rise to both the primary and the secondary heart fields (Saga et al., 1996). Severe cardiac defects are observed in Mesp1-null embryos, resulting in lethality by E10.5 (Saga et al., 1999). Furthermore, Mesp1/Mesp2 (a homolog of Mesp1) double knockout cells fail to contribute to heart development (Kitajima et al., 2000). In the embryonic stem (ES) cell / embryoid body (EB) system, Mesp1 overexpression was shown to induce an array of cardiogenic transcription factors and to promote cardiovascular differentiation at the expense of other lineages, for example blood (Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008). These observations were interpreted to mean that Mesp1 is responsible for specifying a cardiovascular fate within mesoderm by acting as a master regulator at the apex of a hierarchy of cardiac transcription factors. It is thus interesting that in the context of reprogramming fibroblasts into cardiomyocytes, Mesp1 is entirely dispensable (Ieda et al., 2010). As mentioned above, distinct cell populations arise during ES/EB differentiation in a temporally-regulated manner and they react diversely to endogenous and exogenous cues. However, in previous overexpression studies (Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008), Mesp1 was induced either constitutively or over a prolonged period of time. Hence, the cell population that undergoes cardiac differentiation upon Mesp1 induction remains undefined, and the possibility that different cell populations may respond differently to Mesp1 in different contexts has not been addressed.
In order to better define the role of Mesp1 in promoting cardiac differentiation, we undertook a temporal induction study in which differentiating ES progeny were exposed to Mesp1 for short time windows and in different signaling environments. We demonstrate in vitro that responsiveness of cells to Mesp1 is determined in a context-dependent manner, and that the spectrum of outputs includes hematopoietic and skeletal myogenic differentiation. Mesp1-Cre lineage tracing in embryos and adults reveals that indeed the majority of yolk sac hematopoietic cells and large fractions of both the adult hematopoietic stem cell pool and the adult skeletal muscle satellite cell pool derive from Mesp1-expressing precursors. We further show that Mesp1-marked adult bone marrow cells and satellite cells are transplantable and differentiate into multiple hematopoietic lineages and muscle fibers respectively. Our results thus redefine the role of Mesp1 as a context-dependent coregulator of at least three types of mesoderm, namely cardiac, hematopoietic and skeletal myogenic.
Results
Temporal mapping reveals hematopoietic and cardiogenic Mesp1-responsive windows
Mesp1 has been reported to act as a master regulator of cardiovascular specification and its overexpression over several days of EB differentiation increases cardiomyocyte production in vitro (Bondue et al., 2008, 2011; David et al., 2008; Lindsley et al., 2008). However in development, Mesp1 expression is extremely transient, and primarily restricted to cells within and immediately egressing from the primitive streak (Saga et al., 1999). To elucidate the key cell population that responds to Mesp1, we generated a mouse ES cell line in which Mesp1 can be induced by doxycycline (Dox) (Figure 1A and 1B). In contrast to previous reports (Bondue et al., 2008; Lindsley et al., 2008), we made use of a second-generation tetracycline-responsive element (Agha-Mohammadi et al., 2004) which minimizes leakiness and allows tighter regulation of Mesp1 expression (Figure 1C). We applied 24-hr pulses of Dox over the course of EB differentiation (except for the first 2 days where a 48-hour pulse was employed) to determine the optimal time frame for generating cardiomyocytes. The EB is composed of continually evolving cell populations: epiblast cells form by day 2 (i.e., 48 hr – EB formation is considered day 0), the first mesoderm shortly thereafter, and lineage committed cells on day 4 and beyond (Ismailoglu et al., 2008; Keller et al., 1993). Unexpectedly, we observed opposite outcomes of an early (day 2-3 / 48-72 hr) pulse of Mesp1 induction versus a later (day 3-4 / 72-96 hr) pulse (Figure 1D and Supplementary Figure S1A-C).
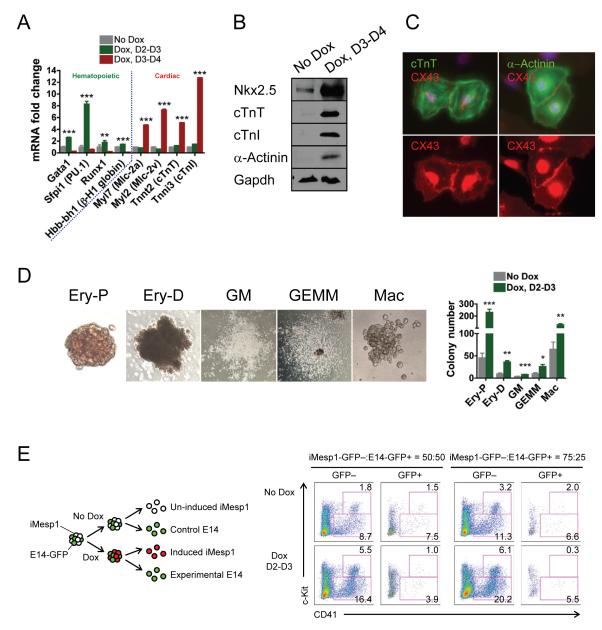
Figure 1. Opposing outcomes of early versus late induction of Mesp1.
(A) Quantitative RT-PCR (n=3) and (B) immunoblot showing Mesp1 induction by Dox.
(C) Immunoblot demonstrating that Mesp1 induction is tightly regulated by Dox.
(D) FACS profile and (E-H) quantification of mesoderm (Flk-1/PDGFRα), hematopoietic (c-Kit/CD41, c-Kit/CD45) and cardiac markers (cTnT) during EB differentiation. Dox (500 ng/ml) was applied over 24 hr as indicated by red boxes.
(E) The late 24 hr pulse of Mesp1 induction from day 3-4 increased Flk-1+ PDGFRα+ presumptive cardiac mesoderm at day 4 (left, n=15) and cTnT+ cardiomyocytes at day 8 (right, n=15).
(F) The early 24 hr pulse of Mesp1 induction from day 2-3 promoted Flk-1+ PDGFRα+ presumptive early unpatterned mesoderm at day 3.25 (left, n=5) and Flk-1+ PDGFRα− presumptive lateral plate mesoderm at day 4 (right, n=10).
(G,H) Mesp1 day 2-3 induction increased several hematopoietic progenitor populations at day 6, including c-Kit+ CD41+ (G, left, n=10), c-Kit+ CD45+ (G, right, n=8), c-Kit− CD41+ (H, left, n=10) and c-Kit− CD45+ (H, right, n=8).
Mean ± SEM is shown in panels A, E, F, G and H. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox.
See also Figure S1
The late 24 hr pulse of Mesp1 induction from day 3, corresponding to the window in which cardiovascular mesoderm is normally specified (Kattman et al., 2006; Kouskoff et al., 2005), produced the anticipated increases in both Flk-1+ PDGFRα+ cells, the presumptive cardiac mesoderm population at day 4, and cTnT+ (cardiac troponin T) cardiomyocytes by day 8 (Figure 1D, E).
A 24 hr pulse of Mesp1 induction in the earlier window (from day 2) also had dramatic effects on mesoderm formation. Presumptive early unpatterned mesoderm, the Flk-1+ PDGFRα+ population at day 3.25, and presumptive lateral plate mesoderm, the Flk-1+ PDGFRα− population at day 4, were increased by 3-fold and 2-fold respectively (Figure 1D,F). In contrast to the late pulse, this early pulse of Mesp1 did not increase cardiomyocyte production at day 8 (Figure 1D, bottom row). Rather, this early Mesp1 pulse unexpectedly increased hematopoietic progenitor populations, c-Kit+/CD41+ and /CD45+ cells by 3- and 7-fold, respectively, at day 6 (Figure 1D and 1G). More differentiated hematopoietic cells including c-Kit− CD41+ and c-Kit− CD45+ were also elevated (by 2- and 13-fold respectively, Figure 1D and 1H). The different phenotypes produced by the early and late pulses of Mesp1 are unlikely a direct consequence of variable Mesp1 dosages, as similar levels of Mesp1 were induced in both windows (Supplementary Figure S1D).
We next investigated whether Flk-1 and PDGFRα are direct targets of Mesp1, which may explain the rapid generation of Flk-1+ and PDGFRα+ cells. EBs cultured in the absence of serum do not express Flk-1 or PDGFRα (Supplementary Figure S1E, left column). Upon Mesp1 induction from day 3, PDGFRα+ cells appeared within 6 hours (day 3.25) and were greatly increased in 24 hours (day 4), while all cells remained Flk-1–(Supplementary Figure S1E, left column). These results, and ChIP for Mesp1 on Pdgfra (Supplementary Figure S1F-G) suggest PDGFRα, but not Flk-1, as a potential direct target of Mesp1.
The unanticipated hematopoiesis contrasts with previous work (Bondue et al., 2008; Lindsley et al., 2008), in which hematopoiesis was inhibited by Mesp1 induction. However, those studies used an extended period of Mesp1 induction while the 24 hr pulses used here more closely approximate the transient pattern of Mesp1 expression in the embryo (Saga et al., 1999). Therefore, our results suggest that instead of solely specifying cardiovascular mesoderm, Mesp1 has distinct effects in different cell populations, with cardiogenic and pro-hematopoietic effects occurring in temporally separate windows.
Fully differentiated hematopoietic cells and cardiomyocytes specified by early or late pulses of Mesp1
To further characterize the differentiation outputs of the two Mesp1 induction windows, we measured the expression levels of key genes specific to these two lineages at day 6 (Figure 2A). We observed significant upregulation of hematopoietic-specific genes, such as Gata1, Sfpi1 (PU.1), Runx1 and Hbb-bh1, by the early pulse but not by the late pulse. In contrast, the late pulse upregulated cardiac-specific markers, including Myl7 (Mlc-2a), Myl2 (Mlc-2v), Tnnt2 (cTnT) and Tnni3 (cardiac troponin I, cTnI), while the early pulse did not. At the protein level, the late pulse of Mesp1 elevated the expression of Nkx2.5, a cardiogenic transcription factor, and downstream markers, cTnT, cTnI and sarcomeric α-actinin (Figure 2B). Furthermore, after plating on adherent surfaces, adjacent Mesp1-induced ES cell-derived cardiomyocytes exhibited synchronous contractions, and expressed gap junction protein connexin 43 (CX43) at cell-cell interfaces, indicating electrical coupling (Figure 2C). These data are all consistent with the later window of Mesp1 induction driving cardiac specification.
Figure 2. Characterization of differentiated cell types promoted by Mesp1 in early or late mesoderm.
(A) Quantitative RT-PCR for hematopoietic- and cardiac-specific markers for day 6 EBs (n=3). Note that the early pulse (day 2-3) of Mesp1 induction (green bars) upregulated hematopoietic but not cardiac specific markers (versus No Dox, gray bars), whereas the late pulse (day 3-4, red bars) did the opposite.
(B) Immunoblot demonstrating the upregulation of cardiac-specific proteins in day 8 EBs subjected to the late pulse of Mesp1 induction.
(C) Immunostaining for cTnT (green) or α-actinin (green) and CX43 (red) in Mesp1- induced EB-derived cardiac cells.
(D) Hematopoietic colonies induced by Mesp1 (left) and quantification (right, n=3-6). Note that the early pulse of Mesp1 induction increased the numbers of both primitive (Ery-P) and definitive (Ery-D, GM, GEMM, Mac) hematopoietic cell types.
(E) Scheme for determining cell autonomy of Mesp1-induced hematopoiesis (left) and FACS analysis of hematopoietic markers (c-Kit/CD41) in day 6 EBs (right). Note that both c-Kit+ CD41+ progenitors and c-Kit− CD41+ differentiated cells were increased in the GFP− population, but neither was increased in the GFP+ population, suggesting that Mesp1 induction of hematopoiesis is cell autonomous.
Ery-P, primitive erythroid; Ery-D, definitive erythroid; GM, granulocyte-macrophage; GEMM, granulocyte-erythrocyte-megakaryocyte-macrophage; Mac, macrophage
Mean ± SEM is shown in panels A and D. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox.
See also Figure S2
Considering that PDGFRα is a potential target of Mesp1, and that the early pulse of Mesp1 appears to promote the Flk-1+ PDGFRα+ population at day 4 without inducing cardiac differentiation (Figure 1D, second and bottom rows), we wished to investigate the cardiogenic potential of day 4 Flk-1+ PDGFRα+ cells. Supporting our previous observation, day 4 sorted Flk-1+ PDGFRα+ cells previously subjected to the late pulse of Mesp1 exhibited an upregulation of cardiac genes (Myl7, Myl2, Tnnt2 and Tnni3) after 4 days of additional culture in cardiogenic medium (Supplementary Figure S2A). Immunostaining further showed that the Flk-1+ PDGFRα+ fraction, in contrast to the Flk-1− PDGFRα− fraction, is enriched in generating cTnT+ cardiomyocytes (Supplementary Figure S2B).
To confirm that the early pulse of Mesp1 indeed promotes hematopoiesis, colony-forming assays were performed. The early Mesp1 pulse resulted in a 2- to 5-fold increase in all types of hematopoietic colonies assayed (Figure 2D), demonstrating that Mesp1 induction during early mesoderm specification promotes both the first wave of primitive erythroid as well as later multi-lineage hematopoiesis.
Mesp1 acts cell autonomously in inducing hematopoietic differentiation
Mesp1 has been shown to act cell autonomously in inducing cardiomyocyte differentiation (Bondue et al., 2008). To determine whether Mesp1 acted cell autonomously in promoting hematopoiesis, we generated chimeric EBs by mixing the inducible Mesp1 ES (iMesp1) cells with wild type, GFP-labeled, ES cells (E14-GFP) at defined ratios and provided a hematopoietic-inducing pulse of Mesp1 at day 2 (Figure 2E, left panel). We observed increases in CD41+ cells only in the GFP− (iMesp1) fraction (Figure 2E, right panel). These data strongly suggest that the early pulse of Mesp1 promotes hematopoiesis specifically in those cells that express Mesp1, that is, in a cell autonomous fashion.
Mesp1 induces hematopoiesis via regulation of Etv2 (ER71) and Tal1 (Scl)
As discussed above, Mesp1 produced distinct mesoderm populations at different stages of EB development (Figure 1D), and generated different lineage-specific gene expression signatures (Figure 2A). Consequently, we profiled temporal expression patterns of major pro-hematopoietic and cardiogenic transcription factors and discovered that the two Mesp1 induction windows activated discrete mesoderm patterning programs. In unstimulated cells, Etv2 (ER71) and Tal1 (Scl), two critical modulators of hematopoietic/endothelial specification (Ferdous et al., 2009; Kallianpur et al., 1994; Lee et al., 2008; Robb et al., 1995; Shivdasani et al., 1995), show strongest expression from day 3.25 to day 4 and gradually decrease to day 6 (Figure 3A, gray lines). The early Mesp1 pulse accelerated this process by markedly inducing Etv2 (13-fold) and Tal1 (22-fold) as early as at day 3.25 (Figure 3A, green lines). Other pro-hematopoietic transcription factors such as Gata2, Lmo2 and Runx1 were also increased by early Mesp1 induction (Supplementary Figure S3A). While Etv2 has been suggested to be expressed by some Mesp1+ cells (Bondue et al., 2008, 2011), Tal1 is not reported to be a direct target of Mesp1. The elevated Tal1 expression at day 3.25 and 4 is particularly interesting, because 24-hr pulses of Tal1 induction were shown to produce phenotypes very similar to what we have observed from the early pulse of Mesp1: dramatic increases in Flk1+ PDGFRα− presumptive lateral plate mesodermal cells at day 4 and c-Kit+ CD41+/CD45+ hematopoietic progenitors at day 6 (Ismailoglu et al., 2008). Overexpression of Etv2 produces similar phenotypes (Koyano-Nakagawa et al., 2012). Therefore, coordinate upregulation of Etv2 and Tal1 likely explains the pro-hematopoietic effects of the early pulse of Mesp1 expression.
Figure 3. Mesp1 promotes hematopoiesis by regulating Etv2 and Tal1.
(A) Quantitative RT-PCR for regulatory hematopoietic and cardiogenic transcription factors during EB differentiation (n=3). Note that EBs treated with the early Mesp1 pulse (green line) rapidly upregulated hematopoietic transcription factors Etv2 and Tal1 at day 3.25 (versus No Dox, gray line), with Tal1 continuing to be elevated by day 4, but expression levels fell back to or below the control (No Dox) level by day 6. On the other hand, the late pulse of Mesp1 (red line) led to a gradual and sustained upregulation of cardiogenic transcription factors Gata4, Tbx5 and Nkx2-5 from day 4 to day 6.
(B) Schematic showing the +40k enhancer of Tal1 and the E-box motifs therein (left). The relative locations of a nearby gene Pdzk1ip1 (Map17) and the primer set for PCR detection are also shown. ChIP analysis on day 3 EBs (right, n=3) illustrates that Mesp1 binds to this Tal1 cis-regulatory element. Gapdh was used as a control.
(C) Electrophoretic mobility shift assay analysis showing that neither Mesp1 (left) nor its co-factor E12 (an isoform of E2A) (middle) alone bound to the E-boxes of the Tal1 +40k enhancer (left), but their dimerization complex could (right). Specific binding and supershift signal are indicated by the arrowhead and the star respectively.
ctrl, control (cell lysate from empty vector for lane 2 and IgG antibody for lane 6); mt, mutant (E-box motif mutant competitor); wt, wildtype (wildtype competitor)
Mean ± SEM is shown in panels A and B. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox.
See also Figure S3
Mesp1 dimerizes with E12 to bind to the Tal1 +40k enhancer
Several Tal1 enhancers have been identified, but only the +40k enhancer region contains E-box motifs (Ogilvy et al., 2007), potential binding sites for Mesp1 (Figure 3B, left panel). We performed ChIP in day 3 EBs after a prior 24 hr Mesp1 pulse and found that Mesp1 was enriched at this cis-regulatory element (Figure 3B, right panel) with a concurrent upregulation of Tal1 expression (Supplementary Figure S3B). We further verified this interaction by testing a Gal4-Mesp1 fusion for transcriptional activity and found that it is a transactivator in multiple cell types (Supplementary Figure S3C). We next performed electrophoretic mobility shift assays to confirm the direct interaction between Mesp1 and the Tal1 +40k enhancer. Typically for a class I bHLH factor, Mesp1 by itself did not bind to the E-box motif of the Tal1 +40k enhancer (Figure 3C, left). However, upon heterodimerization with E12 (an isoform of E2A), Mesp1 could bind to the Tal1 E-box motif (Figure 3C, right). E12 alone did not bind (Figure 3C, middle). Mesp1-E12 binding was further verified by its reduction by wild-type competitor but not an E-box motif mutant, and the presence of a supershift signal (Figure 3C, right).
Mesp1-expressing progenitors contribute to both embryonic and adult hematopoiesis in vivo
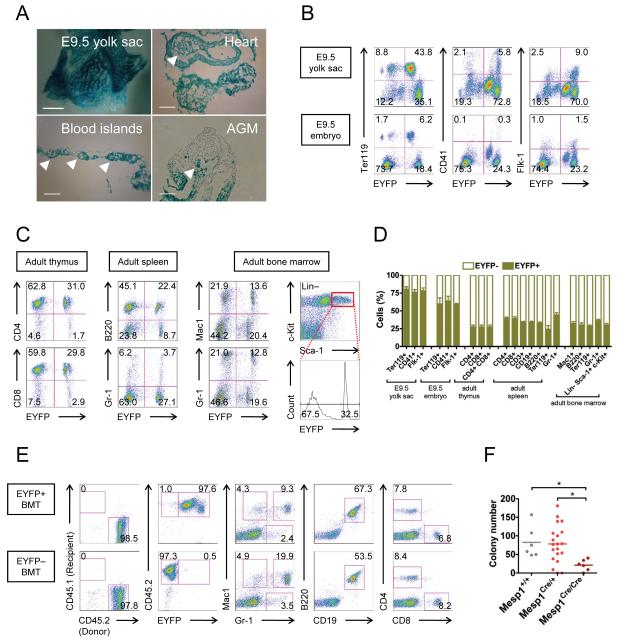
The results above are entirely in vitro, and previous Mesp1-Cre lineage tracing experiments indicated that Mesp1+ precursors do not contribute to adult hematopoiesis (Lindsley et al., 2008). Because ES/EB system recapitulates yolk sac hematopoiesis, we reasoned that the pro-hematopoietic effects of Mesp1 may have been restricted to transient early yolk sac hematopoiesis, which had not previously been evaluated. We crossed the Mesp1-Cre mice (Mesp1Cre/+) with a floxed-stop lacZ reporter line (R26fl-stop-lacZ/fl-stop-lacZ) to trace the progeny of Mesp1-expressing cells during embryogenesis. At E9.5, Mesp1Cre/+;R26fl-stop-lacZ/+ embryos showed extensive lacZ staining in the heart (Figure 4A) as expected (Saga et al., 1999). Remarkably, most hematopoietic cells inside the heart chamber were also positive for lacZ (Figure 4A, white arrowheads). In the yolk sac, the majority of hematopoietic cells in the blood islands were lacZ+, as were the underlying endothelial cells, but not the yolk sac endoderm (Figure 4A, white arrowheads). Moreover, some lacZ+ cells were also visible in the aortic endothelium of the aorta-gonad-mesonephros (AGM) region (Figure 4A, white arrowheads). To quantify this phenomenon, we further crossed the Mesp1Cre/+ mice with a fluorescent floxed-stop reporter line (R26fl-stop-EYFP/fl-stop-EYFP) and performed FACS analysis. Evaluation of E9.5 yolk sacs and embryos revealed that a majority of Ter119+ primitive erythroid cells, CD41+ hematopoietic progenitor cells and Flk-1+ endothelial cells in both yolk sacs and embryos were EYFP+ (Figure 4B and 4D). These observations thus imply that Mesp1-expressing precursors contribute extensively to embryonic hematopoiesis.
Figure 4. Mesp1 marks progenitors that contribute to both embryonic and adult hematopoiesis.
(A) Whole-mount lacZ staining of Mesp1Cre/+;R26fl-stop-lacZ/+ E9.5 embryos and yolk sacs and sections thereof. Note that lacZ staining was observed in cells of mesodermal origin but not cells in the neural crest and the yolk sac endoderm layer. In particular, lacZ+ blood cells (white arrowheads) were found within the heart chambers, the AGM region and the yolk sac blood islands. Bar = 500 μm (upper left) or 100 μm (others).
(B) FACS profiles of hematopoietic (Ter119, CD41) and endothelial (Flk-1) markers in Mesp1Cre/+;R26fl-stop-EYFP/+ E9.5 yolk sacs (top row) and embryos (bottom row).
(C) FACS profiles of hematopoietic markers in the thymus (far left), the spleen (middle left) and the bone marrow (middle and far right) of adult Mesp1Cre/+;R26REYFP/+ animals.
(D) Quantification of EYFP expression by FACS in hematopoietic fractions of various origins in E9.5 and adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals (n=5-7). Note that all hematopoietic lineages examined contain a significant EYFP+ fraction.
(E) Mesp1-marked bone marrow cells repopulated and contributed to multiple hematopoietic lineages. EYFP+ (top row) and EYFP− (bottom row) total bone marrow cells from adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals were transplanted to NSG-CD45.1/CD45.1 recipient mice. FACS analysis of the peripheral blood 4 months later showed that almost all blood cells derived from the donor (CD45.2+) but not from the host (CD45.1+) (far left). Note that Mesp1-unlabelled transplanted cells remained EYFP− (bottom row, near left), indicating that Mesp1 is not re-expressed in the adult hematopoietic system.
(F) Hematopoietic colony-forming assay showing that yolk sacs of Mesp1Cre/Cre knockout embryos (E8.25-E8.5, equivalent to 6-8 somite pairs) produced fewer hematopoietic colonies than those of Mesp1+/+ and Mesp1Cre/+ embryos. *, p<0.05
AGM, aorta-gonads-mesonephros; BMT, bone marrow transplantation; Lin, lineage cocktail comprising CD3, CD4, CD8, Mac1, B220, Gr-1 and Ter119; HSC, hematopoietic stem cells; NSG-CD45.1/CD45.1, CD45.1/CD45.1 homozygous on the NOD scid gamma background.
Mean ± SEM is shown in panel D.
See also Figure S4
These data strongly suggest a role for Mesp1 in yolk sac primitive hematopoiesis. The contribution of the yolk sac to the adult hematopoietic system remains controversial (Cumano et al., 1996, 2001; Huang and Auerbach, 1993; Lux et al., 2008; Medvinsky and Dzierzak, 1996; Palis et al., 1999; Samokhvalov et al., 2007). To evaluate whether Mesp1+ progenitors are restricted to yolk sac hematopoiesis, we analyzed the labeling of different hematopoietic lineages in 6-week old mice from the same crosses. FACS analysis of the thymus, spleen and bone marrow of these animals revealed considerable EYFP expression in all hematopoietic populations examined in the thymus, spleen and bone marrow (Figure 4C, 4D and Supplementary Figure S4A-C). Most importantly, the bone marrow hematopoietic stem cell compartment (Lin− Sca-1+ c-Kit+) was 30% EYFP+ (Figure 4C far right, and 4D).
Mesp1+ bone marrow cells repopulate the hematopoietic compartment of irradiated hosts
To conclusively demonstrate that Mesp1-marked bone marrow contained HSCs, we sorted the EYFP+ fraction from total bone marrow cells of Mesp1Cre/+;R26fl-stop-EYFP/+ animals and transplanted it into irradiated NSG-CD45.1/CD45.1 recipient mice. The NSG-CD45.1/CD45.1 mice were homozygous for the CD45.1 allele, and thus facilitated simultaneous analysis of host (CD45.1+) or donor (CD45.2+) derived hematopoietic cells. Remarkably, the peripheral blood of recipients was mostly donor-derived at 4 weeks (5/5, 89±7% chimerism), 8 weeks (4/4, 96±2% chimerism) and 4 months (4/4, 96±4% chimerism) in every lineage examined (Figure 4E, top row). We further demonstrated, by transplanting Mesp1-unlabeled (EYFP− CD45.2+) bone marrow cells, that Mesp1 was not re-expressed in the adult hematopoietic system, as there were no EYFP+ cells in the peripheral blood (Figure 4E, bottom row). Therefore, our data provide clear evidence that Mesp1+ precursors contribute to both embryonic and adult hematopoiesis, and by correlation, that Mesp1 expression per se does not inhibit hematopoiesis in vivo as previously suggested.
Mesp1 is necessary for normal yolk sac hematopoiesis
To address whether Mesp1 is required for yolk sac hematopoietic development, we took advantage of the fact that Mesp1-Cre is a null allele (Saga et al., 1999). We intercrossed Mesp1Cre/+ mice to generate Mesp1Cre/Cre, which have no functional Mesp1. Mesp1Cre/Cre animals had severe cardiac defects and did not survive beyond E10.5 (our observation and Saga et al., 1999). Because fluid shear stress is known to regulate embryonic hematopoiesis (Adamo et al., 2009; North et al., 2009), we analyzed the Mesp1Cre/Cre embryos at E8.25-E8.5 (6-8 somite pairs) prior to circulation to avoid secondary effects from blood flow. Using the colony-forming assay, we observed that the yolk sacs of Mesp1Cre/Cre embryos produced significantly fewer hematopoietic colonies than those of Mesp1+/+ and Mesp1Cre/+ embryos (Figure 4F). Therefore, Mesp1 is necessary for normal yolk sac hematopoiesis.
Mesp1 promotes paraxial mesoderm and myogenic derivatives in the absence of serum-derived factors
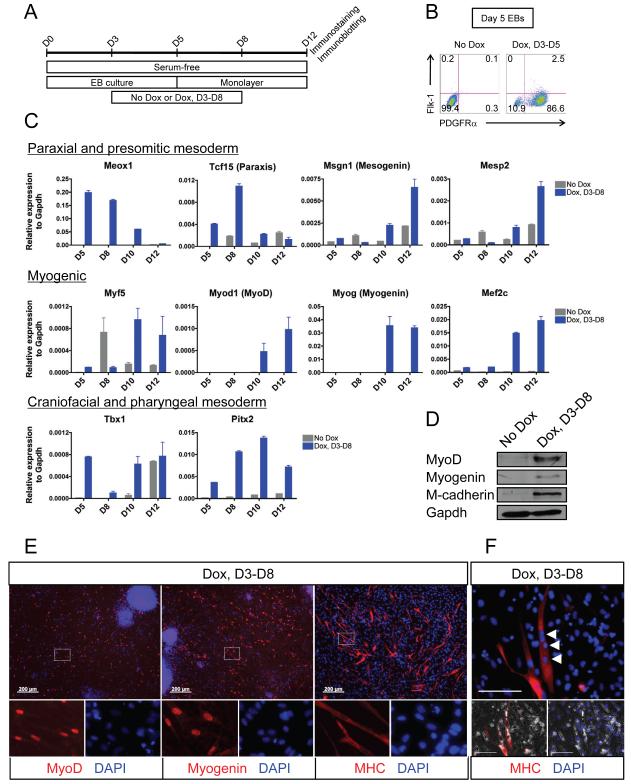
Mesp1 is known to label the craniofacial mesodermal population that develops into the cranial musculature (Harel et al., 2009; Saga et al., 1999, 2000), but Mesp1 has not been demonstrated to induce cranial myogenesis in ES cells, and indeed skeletal myogenesis is notoriously absent in conventional ES/EB differentiation (Darabi et al., 2008). Because serum-derived factors are required for both hematopoietic and cardiac differentiation, we investigated what effect Mesp1 induction would have in the absence of serum (Figure 5A). Mesoderm formation is abolished in EBs grown in the absence of serum, as indicated by a lack of Flk-1+ or PDGFRα+ cells in day 5 EBs (Figure 5B). However, continuous Mesp1 induction from day 3 promoted the emergence of Flk-1− PDGFRα+ cells, suggesting a presumptive paraxial mesoderm population (Darabi et al., 2008; Sakurai et al., 2006) (Figure 5B). Further gene expression characterization of Mesp1-induced cells showed that Mesp1 induction from day 3-8 led to the upregulation of paraxial and presomitic mesodermal genes including Meox1, Tcf15 (paraxis), Msgn1 (mesogenin) and Mesp2 (Figure 5C, top row). Pan-myogenic genes such as Myf5, Myod1 (MyoD), Myog (myogenin) and Mef2c, and myogenic genes pertaining to craniofacial and pharyngeal muscles such as Tbx1 and Pitx2 were also upregulated (Figure 5C, middle and bottom row). Interestingly, these myogenic genes remained elevated for at least 4 days after Dox removal (assayed at day 12, Figure 5C, middle and bottom row), indicating that Mesp1 promoted the initiation of, but was dispensable for the maintenance of, the skeletal myogenic program. Immunoblotting further verified that Mesp1 induction promoted the generation of skeletal myogenic progenitors, as demonstrated by the presence of MyoD, myogenin and M-cadherin (Figure 5D). Immunohistochemistry indicated the presence of myoblasts, characterized as MyoD+, myogenin+, sarcomeric myosin heavy chain (MHC)+ (Figure 5E), as well as multi-nucleated MHC+ myotubes under conditions of differentiation (Figure 5F). Thus, we have demonstrated that an extended window of late Mesp1 induction in the absence of serum-derived factors promotes paraxial mesoderm and myogenic derivatives from ES cells.
Figure 5. Mesp1 promotes paraxial mesoderm and myogenic derivatives in the absence of serum-derived factors.
(A) Scheme depicting the protocol used to examine the effect of Mesp1 on ES differentiation in serum-free conditions.
(B) FACS analysis of mesoderm markers Flk-1 and PDGFRα in day 5 EBs grown under serum-free conditions.
(C) Quantitative RT-PCR for various lineage-specific markers in serum-free culture conditions in which Mesp1 was induced from day 3 to day 8 (blue bars) or not induced (gray bars) (n=3).
(D) Immunoblot showing upregulation of myogenic proteins upon Mesp1 induction from day 3 to day 8 under serum-free conditions.
(E) Immunostaining for myogenic markers, MyoD (left), Myogenin (middle) and MHC (right), in EB-derived cells induced by Mesp1 from day 3-8 cultured in the absence of serum. Areas depicted by the white dotted rectangle are magnified to demonstrate the localization of the nucleus (DAPI) and the nuclear (MyoD and Myogenin) or cytoplasmic (MHC) markers of interest (bottom panels). No MyoD+, Myogenin+ or MHC+ cells were observed in control (No Dox) wells (not shown). Bar = 200 μm
(F) Immunostaining showing the presence of multiple nuclei (white arrowheads) in Mesp1-induced ES cell-derived MHC+ myotube. The red channel (MHC) and the blue channel (DAPI) are individually merged with the phase contrast channel to indicate that multiple DAPI+ nuclei are present in the same MHC+ myogenic cell (bottom panels). Bar = 100 μm
Mean ± SEM is shown in panel C.
Mesp1-expressing precursors give rise to satellite cells of craniofacial skeletal muscles
Based on these in vitro results, we next examined the relative contribution of Mesp1-expressing progenitors in cranial and trunk myogenesis, identifying satellite cells as Lin−(CD31− and CD45−) α7-integrin+ CD34+ or Lin− α7-integrin+ VCAM-1+ (Biressi and Rando, 2010). We further verified that these gating strategies (Figure 6A) using Pax7-ZsGreen mice in which all satellite cells are ZsGreen+ (Bosnakovski et al., 2008). Satellite cells from six different muscle groups (facial, masseter, diaphragm, triceps, back and hindlimb muscles) of adult Mesp1Cre/+;R26fl-stop-YFP/+ mice were analyzed by FACS for EYFP expression (Figure 6B and Supplementary Figure S5). Satellite cell populations in facial and masseter muscles that are of craniofacial mesodermal origin were mainly EYFP+ (73% in the facial muscles and 76% in the masseter, Figure 6B far and middle left, and 6C). The diaphragm, potentially of mixed origin of both trunk paraxial and cranial splanchnic mesoderm (Ackerman and Greer, 2007), contained a small but obvious and reproducible fraction of EYFP+ satellite cells (12%, Figure 6B middle right, and 6C). In contrast, hindlimb- and trunk-derived satellite cells rarely co-expressed EYFP (3% in both muscles, Figure 6B far right, and 6C). To confirm the skeletal myogenic potential of the Mesp1-marked prospectively isolated satellite cells, we cultured them under myogenic differentiation conditions (Bosnakovski et al., 2008). Sarcomeric MHC+ myotubes were readily observed in all samples (Figure 6D). These results therefore demonstrate that Mesp1 induction of skeletal myogenesis is physiologically significant, particularly to anterior muscle groups.
Figure 6. Mesp1-expressing progenitors contribute to the adult satellite cell pool.
(A) FACS analysis of TA muscle from Pax7-ZsGreen mice confirming that more than 90% of Lin− α7-integrin+ VCAM-1+ cells are ZsGreen+ (Pax7+).
(B) FACS profiles showing the satellite cell marker-gated population (Lin− α7-integrin+ VCAM-1+) and the EYFP content thereof in the facial muscles (far left), the masseter (middle left), the diaphragm (middle right) and the hindlimb muscles (far right) of adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals.
(C) Quantification of EYFP expression by FACS in satellite cell marker-gated populations (left: Lin− α7-integrin+ CD34+; right: Lin− α7-integrin+ VCAM-1+) from various muscle groups in adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals (n=4-5). Note that EYFP+ satellite cells are predominant in the facial muscles and masseter, rare in the diaphragm and triceps, and almost undetectable in the back and hindlimb muscles.
(D) Immunostaining for MHC in EYFP+ sorted populations gated on satellite cell markers (Lin− α7-integrin+ VCAM-1+) from various muscle groups. Multinucleated MHC+ myotubes co-expressing EYFP+ are observed in all sorted populations, indicating that the gated populations of Mesp1 origin are indeed skeletal myogenic progenitors, and not a non-myogenic subpopulation. Bar = 100 μm.
(E) Mesp1-marked satellite cells engrafted and differentiated into muscle fibers. EYFP+ satellite cells (Lin− α7-integrin+ VCAM-1+) isolated from the masseter and TA of Mesp1Cre/+;R26fl-stop-EYFP/+ animals (dystrophin+) were transplanted into the TA of NSG-mdx4Cv recipients (dystrophin−). Immunostaining revealed the presence of dystrophin+ (red) muscle fibers, indicating engraftment, in transplanted (left and middle) but not in control non-transplanted (right) tissues. Bar = 100 μm
Lin, lineage cocktail comprising CD31 (endothelial) and CD45 (hematopoietic).
Mean ± SEM is shown in panel C.
See also Figure S5
Mesp1+ satellite cells engraft and form muscle fibers in mdx4Cv mice
To conclusively demonstrate that Mesp1-marked, Lin− α7-integrin+ VCAM-1+ cells were truly muscle stem cells, we purified and transplanted these cells into immune-deficient, dystrophin-deficient NSG-mdx4Cv animals and used dystrophin staining to identify donor-derived myofibers in vivo. Twenty-five hundred and 200 EYFP+ satellite cells from the masseter and the tibialis anterior (TA), respectively, were transplanted into cardiotoxin-injured, irradiated recipient TA muscles. The lower number of marked cells from the donor TAs reflects the very low percentage of TA satellite cells that are marked by Mesp1. Six weeks post-transplant, immunostaining revealed large clusters of dystrophin+ myofibers in transplanted but not control recipient TAs (Figure 6E), indicating that both the abundant Mesp1-marked satellite cells from the masseter, and the rare Mesp1-marked cells from the TA are bona fide muscle stem cells.
Discussion
Mesp1 patterns mesoderm into different fates depending on the stage of differentiation and signaling environment
Mesp1 has been considered the cardiac master regulator, driving cardiovascular specification and inhibiting other mesodermal lineages (Bondue et al., 2008, 2011; David et al., 2008; Lindsley et al., 2008). In this report, we challenge this paradigm and provide evidence that Mesp1 actually patterns mesoderm into multiple mesodermal lineages, including cardiac, hematopoietic, and skeletal muscle, in a context-dependent manner.
Mesp1 is expressed in the nascent mesoderm but is abruptly downregulated as the newly formed mesoderm migrates out of the primitive streak (Saga et al., 1999). Mesp1-expressing cells were found to give rise to all cardiac lineages (Kitajima et al., 2000; Saga et al., 1999, 2000). Interestingly, some marking was also observed in extraembryonic mesoderm at early-streak stage (E6.5-6.75), and Mesp1 was postulated to be expressed earlier in this mesodermal subpopulation than in cardiac mesoderm (Saga et al., 1999). By looking at the yolk sac in E9.5 embryos, we show that Mesp1 labels extraembryonic hematopoietic and endothelial cells extensively. In line with this observation, we demonstrate in vitro that Mesp1 can induce hematopoietic differentiation, and this effect is most pronounced during a window that slightly proceeds that in which Mesp1 drives cardiac differentiation.
In conventional ES/EB differentiation, mesoderm is postulated to arise in distinct waves, with the first wave contributing primarily to the hematopoietic and endothelial lineages and the second contributing to cardiac and endothelial lineages (Irion et al., 2010; Kattman et al., 2006; Kouskoff et al., 2005). Further work suggested that the first wave generates a Flk-1+ PDGFRα− presumptive lateral plate mesoderm population while the second wave gives rise to a Flk-1+ PDGFRα+ presumptive cardiac mesoderm population (Hirata et al., 2007; Sakurai et al., 2006). It is interesting that we observed similar temporal specificities of Mesp1 induction in which an early 24 hr pulse (starting at 48 hrs) promoted a Flk-1+ population and hematopoiesis, whereas a later pulse (starting at 72 hrs) promoted a PDGFRα+ population and cardiogenesis. The stage of differentiation thus governs the responsiveness of cells to Mesp1.
The patterning activity of Mesp1 is modulated by the signaling environment. In the absence of serum, Mesp1 induces paraxial mesoderm that goes on to differentiate into skeletal myogenic derivatives, remarkable because this lineage is virtually absent in conventional EB differentiation (Darabi et al., 2008). The impact of signaling on mesodermal patterning has been extensively studied (Arnold and Robertson, 2009; Tam and Loebel, 2007). As Mesp1-expressing cells leave the primitive streak, they are subjected to various signaling gradients including BMP4, known to induce cardiogenesis but inhibit cranial myogenesis (Tirosh-Finkel et al., 2006). Our results suggest that serum-derived factors recapitulate this effect and direct Mesp1-responsive cells towards the cardiac lineage while the lack thereof in serum-free conditions leads to paraxial mesoderm/skeletal myogenic differentiation.
Mesp1 does not invariably suppress hematopoiesis
Mesp1 has been reported to directly suppress hematopoiesis in vitro (Bondue et al., 2008, 2011; Lindsley et al., 2008) and to not be expressed in the precursors of the hematopoietic lineage in vivo (Lindsley et al., 2008). However, the broad distribution of Mesp1-expressing cells in the yolk sac at its hemogenic prime merits reevaluation of this assumption. Lineage tracing revealed that a large fraction of hematopoietic progenitors in embryos and hematopoietic stem cells in adults is of Mesp1 origin. A larger number of Mesp1-labeled hematopoietic cells in embryos than in adults may indicate the emergence of a Mesp1-unlabeled hematopoietic progenitor population later (>E9.5) during development. Although our lineage tracing data in adult mice contradicts that of a previous report, the two studies used different reporters: EGFP in the former (Lindsley et al., 2008), versus EYFP, which generates a more intense fluorescent signal (Shaner et al., 2005), in the current study. Furthermore, Mesp1 expression is not merely passively marking hematopoietic progenitors: embryos deficient for Mesp1 suffer a severe reduction in yolk sac hematopoietic progenitor frequency. It was recently shown that Mesp1 is required for expression of Meis1 in endothelial cells derived from ES cells, interesting because of the pro-hematopoietic effects of Meis1 (Cai et al, 2012). Thus, in the appropriate context, Mesp1 promotes and is required for hematopoiesis.
We found that an early pulse of Mesp1 induction transiently induced the transcription factors Etv2 and Tal1, and resulted in significantly increased numbers of both primitive erythroid and multi-lineage hematopoietic progenitors by day 6. Importantly, within this early window, we discovered that Mesp1 directly regulates the transcription of Tal1, and we identified E12 as a co-factor that allows binding of Mesp1 to the +40k cis-regulatory element of Tal1. This interaction is particularly interesting in light of a recent report demonstrating that in the absence of Tal1, yolk sac cells can differentiate into cardiomyocytes (Van Handel et al., 2012). This and previous work (Ismailoglu et al., 2008) shows that the simple presence of Tal1 drives the hematopoietic program in cells otherwise fated to become cardiac. The lineage tracing of Mesp1 to the yolk sac suggests that in the absence of Tal1, Mesp1 promotes a cardiac program in these earliest mesodermal cells. It will be interesting to determine what prevents Mesp1 from activating Tal1 outside of this unique early window.
Regulation of Mesp1 is a developmentally-relevant approach to generating craniofacial myogenic precursors
Certain muscle disorders manifest differently between the head and the trunk muscles (Emery, 2002). For instance, Duchenne muscular dystrophy affects the limb muscles more severely than those in the head (Duchenne, 1868), while in facioscapulohumeral muscular dystrophy the facial muscles are almost always profoundly affected but distal limb muscles are relatively spared (Landouzy and Déjérine, 1885). Although this discrepancy is not fully understood, the cranial muscles are known to have different muscle fiber type compositions (Schiaffino and Reggiani, 2011) and genetic signatures (Bothe et al., 2011; Bryson-Richardson and Currie, 2008; Shih et al., 2008) compared to their trunk counterparts. Examination of the regenerative potentials of satellite cells from different head and trunk muscle groups revealed that the satellite cells of the head muscles show different proliferative properties but engraft equally well in limb muscles (Ono et al., 2010; Sambasivan et al., 2009). Therefore, deriving craniofacial myogenic precursors may be useful for studying disease processes specific to these muscles. Considering the substantial contribution of Mesp1+ precursors to the adult head muscle fibers (Harel et al., 2009) and the adult muscle stem cell pool (our data), it is conceivable that Mesp1 plays a key role in activating the skeletal myogenic program. In fact, our work constitutes a method to generate skeletal myogenic precursors from ES cells by Mesp1 induction, a feat previously only accomplished by Pax3 or Pax7 expression (Darabi et al. 2008, 2011, 2012). These myogenic precursors can fuse into myotubes, and contain elevated expression of Tbx1 and Pitx2 which are the hallmark genes of pharyngeal and craniofacial muscles (Sambasivan et al., 2009). Therefore, Mesp1 induction is a developmentally-relevant approach to derive craniofacial myogenic precursors from pluripotent cells.
In this report, we provide the evidence that Mesp1 contributes to the specification of multiple mesoderm lineages in a context-dependent manner. In particular, besides cardiac, Mesp1 also drives the specification of the hematopoietic and skeletal myogenic lineages, and marks precursors committed to these lineages. This observation not only redefines the role of Mesp1, but also postulates a context-dependent and dynamic nature to the Mesp1 transcriptional network.
Experimental procedures
Detailed experimental procedures can be found in the Supplementary Material section.
Generation of doxycycline-inducible Mesp1 mouse ES cell line
The Dox-inducible Mesp1 ES cell line was generated using the inducible cassette exchange strategy as previously reported (Iacovino et al., 2011).
ES cell culture and differentiation
ES cells were cultured and differentiated via EBs formation into hematopoietic and cardiac lineages using standard methods. Protocol for skeletal myogenic differentiation is illustrated in Figure 5A.
Gene expression analysis
Quantitative RT-PCR were conducted using Taqman probes (Applied Biosystems, Carlsbad, CA) (Supplementary Table S1).
Flow cytometry analysis
FACS analysis was performed using BD FACSAriaII (BD Biosciences, San Diego, CA). Antibodies used were listed in Supplementary Table S2.
ChIP – PCR analysis
ChIP was performed as described previously (Peng et al., 2009). The primer pair for the Tal1 +40k enhancer was: forward: 5’-CGCCAAGACCCTCTTCCTTAT-3’ and reverse: 5’-CCAGCTGGTGCGTTATCAGTT-3’.
Electrophoretic mobility shift assay
Details about the electrophoretic mobility shift assay were described previously (Shi and Garry, 2010). The oligonucleotide harboring the E-box motif in Tal1 +40k enhancer is 5’-CTGATAACGCACCAGCTGGGCCCCCCACCA-3’.
Mice
All animal procedures were performed according to the University of Minnesota Institutional Animal Care and Use Committee guidelines and approved protocols. Mesp1Cre/+ mice were obtained from Dr. Yumiko Saga (through Dr. Kenneth Murphy). R26fl-stop-lacZ/fl-stop-lacZ (B6.129S4-Gt(ROSA)26Sortm1Sor/J), R26fl-stop-EYFP/fl-stop-EYFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J), NOD scid gamma (NSG) (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ), mdx4Cv (B6Ros.Cg-Dmdmdx-4Cv/J) and CD45.1/CD45.1 (CBy.SJL(B6)-Ptprca/J) mice were obtained from Jackson Laboratories (Bar Harbor, ME). Pax7-ZsGreen mice were described previously (Bosnakovski et al., 2008).
Bone marrow transplantation
Recipient NSG-CD45.1/CD45.1 mice (6 week-old) were irradiated with 250 cGy 24 hours prior to transplantation. Total bone marrow cells were obtained from femurs and tibia of Mesp1Cre/+;R26fl-stop-EYFP/+ mice. Both EYFP+ and EYFP− fractions were sorted by FACS, and 150,000 cells were injected into recipient mice intravenously. Four weeks, 8 weeks and 4 months after transplantation, peripheral blood samples were analyzed by flow cytometry.
Satellite cell transplantation
NSG-mdx4Cv mice (4 month-old) were subjected to 1200 cGy dose of irradiation two days prior to satellite cells transplantation. Injuries to tibialis anterior (TA) muscle was induced by cardiotoxin one day later. EYFP+ satellite cells (Lin− α7-integrin+ VCAM-1+) isolated from the masseter or TA of Mesp1Cre/+;R26fl-stop-EYFP/+ mice were sorted and injected (2500 cells from the masseter and 200 cells from the TA) into the TA. Tissues were harvested six weeks later.
Statistical analysis
Data are expressed as mean ± SEM. Statistical significance is determined by Student’s t-test and analysis of variance as appropriate. Statistical significance is set as p<0.05.
Supplementary Material
Highlights.
Mesp1 induction can differentiate ES cells into non-cardiac lineages.
Activity of Mesp1 is governed by stage of differentiation and environment.
Mesp1+ progeny contribute to both embryonic and adult hematopoiesis in vivo.
Although necessary for cardiogenesis, Mesp1 is not the cardiac master regulator.
Acknowledgements
We thank Nardina Nash and Cara-lin Lonetree for animal husbandry and genotyping, and Si Ho Choi for technical assistance in ChIP analysis. This work was supported by NIH grants U01 HL100407 and R01 AR055685 and by the American Heart Association: John Holden DeHaan Foundation. RA and AT were supported by NIH T32 AR007612 and T32 HL069764.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am. J. Med. Genet. C Semin. Med. Genet. 2007;145C:109–116. doi: 10.1002/ajmg.c.30128. [DOI] [PubMed] [Google Scholar]
- Adamo L, Naveiras O, Wenzel PL, Kinney-Freeman S, Mack PJ, Gracia-Sancho J, Suchy-Dicey A, Yoshimoto M, Lensch MW, Yoder MC, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459:1131–1135. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agha-Mohammadi S, O’Malley M, Etemad A, Wang Z, Xiao X, Lotze MT. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene Med. 2004;6:817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin. Cell Dev. Biol. 2010;21:845–854. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RCR, Kyba M. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe I, Tenin G, Oseni A, Dietrich S. Dynamic control of head mesoderm patterning. Development. 2011;138:2807–2821. doi: 10.1242/dev.062737. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Langer EM, Gill JG, Satpathy AT, Albring JC, Wumesh KC, Murphy TL, Murphy KM. Dual actions of Meis1 inhibit erythroid progenitor development and sustain general hematopoietic cell proliferation. Blood. 2012;120:335–346. doi: 10.1182/blood-2012-01-403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumano A, DieterlenLievre F, Godin I. Lymphoid potential, probed before circulation in mouse, is restricted to caudal intraembryonic splanchnopleura. Cell. 1996;86:907–916. doi: 10.1016/s0092-8674(00)80166-x. [DOI] [PubMed] [Google Scholar]
- Cumano A, Ferraz JC, Klaine M, Di Santo JP, Godin I. Intraembryonic, but not yolk sac hematopoietic precursors, isolated before circulation, provide long-term multilineage reconstitution. Immunity. 2001;15:477–485. doi: 10.1016/s1074-7613(01)00190-x. [DOI] [PubMed] [Google Scholar]
- Darabi R, Arpke RW, Irion S, Dimos JT, Grskovic M, Kyba M, Perlingeiro RCR. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M, Perlingeiro RCR. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat. Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- Darabi R, Santos FN, Filareto A, Pan W, Koene R, Rudnicki MA, Kyba M, Perlingeiro RC. Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells. 2011;29:777–790. doi: 10.1002/stem.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 2008;10:338–U375. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- Duchenne GB. Recherches sur la paralysie musculaire pseudo-hypertrophique, ou paralysie myo-sclerosique. Archives Generales de Medecine. 1868;11:5–25. 179–209, 305–321, 421–443, 552–588. [Google Scholar]
- Emery AEH. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies and endothelial/endocardial fate in the developing embryo. Proc. Natl. Acad. Sci. USA. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimaraes-Camboa N, Evans SM, Tzahor E. Distinct origins and genetic programs of head muscle satellite cells. Dev. Cell. 2009;16:822–832. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Kawamata S, Murakami Y, Inoue K, Nagahashi A, Tosaka M, Yoshimura N, Miyamoto Y, Iwasaki H, Asahara T, et al. Coexpression of platelet-derived growth factor receptor alpha and fetal liver kinase 1 enhances cardiogenic potential in embryonic stem cell differentiation in vitro. J. Biosci. Bioeng. 2007;103:412–419. doi: 10.1263/jbb.103.412. [DOI] [PubMed] [Google Scholar]
- Huang H, Auerbach R. Identification and characterization of hematopoietic stem cells from the yolk sac of the early mouse embryo. Proc. Natl. Acad. Sci. USA. 1993;90:10110–10114. doi: 10.1073/pnas.90.21.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells. 2011;29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Clarke RL, Luche H, Kim I, Morrison SJ, Fehling HJ, Keller GM. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development. 2010;137:2829–2839. doi: 10.1242/dev.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailoglu I, Yeamans G, Daley GQ, Perlingeiro RCR, Kyba M. Mesodermal patterning activity of SCL. Exp. Hematol. 2008;36:1593–1603. doi: 10.1016/j.exphem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Kallianpur AR, Jordan JE, Brandt SJ. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–1208. [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent Flk-1(+) cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kweon J, Iacovino M, Shi X, Rasmussen TL, Borges L, Zirbes KM, Li T, Perlingeiro RCR, Kyba M, et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells. 2012 doi: 10.1002/stem.1131. doi:10.1002/stem.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landouzy L, Déjérine J. De la myopathie atrophique progressive. Revue de Medecine. 1885;5:81–117. 253–366. [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood. 2008;111:3435–3438. doi: 10.1182/blood-2007-08-107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy S, Ferreira R, Piltz SG, Bowen JM, Gottgens B, Green AR. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol. Cell. Biol. 2007;27:7206–7219. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev. Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptaszek LM, Mansour M, Ruskin JN, Chien KR. Towards regenerative therapy for cardiac disease. Lancet. 2012;379:933–942. doi: 10.1016/S0140-6736(12)60075-0. [DOI] [PubMed] [Google Scholar]
- Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: A novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Era T, Jakt LM, Okada M, Naki S, Nishikawa S, Nishikawa SI. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly R, Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Myogenic regulatory factors transactivate the Tceal7 gene and modulate muscle differentiation. Biochem. J. 2010;428:213–221. doi: 10.1042/BJ20091906. [DOI] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Muscle development: forming the head and trunk muscles. Acta Histochem. 2008;110:97–108. doi: 10.1016/j.acthis.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Loebel DAF. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, et al. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 2012;150:590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR plus embryonic-stem-cell-derived population. Nature. 2008;453:524–U526. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.