Abstract
We describe strand-specific, base-resolution detection of 5-hydroxymethylcytosine (5-hmC) in genomic DNA with single-molecule sensitivity, combining a bioorthogonal, selective chemical labeling method of 5-hmC with single-molecule, real-time (SMRT) DNA sequencing. The chemical labeling not only allows affinity enrichment of 5-hmC–containing DNA fragments but also enhances the kinetic signal of 5-hmC during SMRT sequencing. We applied the approach to sequence 5-hmC in a genomic DNA sample with high confidence.
The base 5-hydroxymethylcytosine (5-hmC) is a newly discovered DNA modification in mammalian cells and, along with 5-methylcytosine (5-mC), is believed to be an important epigenetic mark involved in many critical cellular functions, including embryonic stem cell differentiation, normal myelopoiesis as well as zygotic development1–3. Understanding the biological functions of 5-hmC requires the development of sensitive sequencing methods to reveal locations of this base modification in the genome, as existing sequencing methods, such as bisulfite sequencing, cannot be used to differentiate 5-hmC from 5-mC (ref. 4).
Recently, we developed a selective chemical labeling technology for 5-hmC, in which 5-hmC is first modified with an azide-substituted glucose using β-glucosyltransferase followed by a click chemistry reaction to install a biotin tag5. Using this method, 5-hmC in genomic DNA has been enriched for deep sequencing to provide the genomic distribution of this base modification. However, this and other methods5–11 do not currently give information about the exact genomic locations of 5-hmC. Progress in the understanding of 5-hmC biology has been hampered by the lack of a method for high-throughput, strand-specific, base-resolution sequencing of 5-hmC.
Single-molecule, real-time (SMRT) DNA sequencing is a third-generation sequencing technology that uses individual DNA polymerase molecules to perform DNA synthesis, monitoring the continuous incorporation of phospholinked nucleotides12. The real-time recording of nucleotide incorporations, detected as fluorescent pulses, generates not only the sequence readout but also valuable information about the polymerase kinetics, which can be used to identify DNA base modifications. Typically, the polymerase rate at and around the modified base position in the DNA template is slowed compared to unmodified DNA. This can be expressed quantitatively by comparing the time between incorporation events, the interpulse duration (IPD), for each template position. Using this method, we have demonstrated that SMRT sequencing can be used to directly detect DNA methylation including N6-methyladenine, 5-mC and 5-hmC (ref. 13).
We combined the selective chemical labeling of 5-hmC and SMRT sequencing to provide a high-throughput, base-resolution 5-hmC detection method. The selective chemical labeling enables enrichment of 5-hmC–containing DNA to reduce the amount of sequencing required. In addition, the larger size of the tag on 5-hmC yields larger kinetic signals in SMRT sequencing for confident assignments of the modified base at lower sequencing coverage.
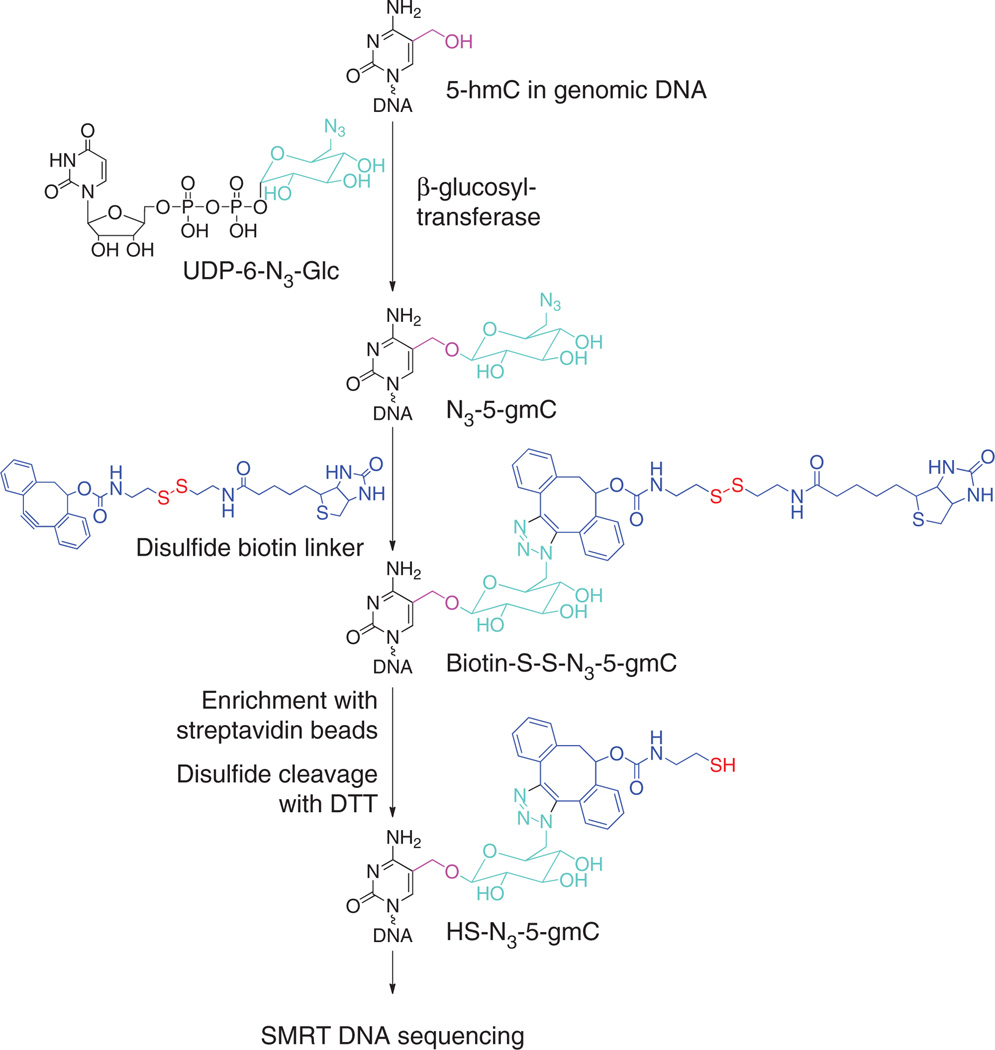
To achieve targeted enrichment and SMRT sequencing of 5-hmC (Fig. 1), we first used β -glucosyltransferase to transfer azide-glucose to 5-hmC, yielding β-6-azide-glucosyl-5-hydroxymethyl-cytosine (N3-5-gmC), as described previously5. For the second step, we developed a new cleavable biotin-containing capture agent with a disulfide linker as the click reaction partner to form biotin-S-S-N3-5-gmC to accommodate the sequencing protocol, as that method also uses biotin (for immobilization of DNA polymerase). After selectively capturing 5-hmC–containing DNA fragments from genomic DNA by streptavidin beads, a simple dithiothreitol (DTT) treatment releases the bound DNA fragments of interest, with 5-hmC modified as HSN3-5-gmC. Using synthetic DNA templates, we found this DTT-mediated cleavage to be quantitative and confirmed efficient conversion of all other reaction steps, as described previously5 (Supplementary Fig. 1). The pulldown yield for DNA fragments containing only a single 5-hmC was ~50%, consistent with the much lower density-dependence of biotin-based pulldown methods, compared to antibody-based immunoprecipitation8. The approach is highly specific to 5-hmC, as no 5-mC–containing DNA was pulled down as measured by UV absorbance (Supplementary Table 1). The disulfide-reduction strategy to release desired DNA fragments was also less time-consuming and more efficient than the previous monomeric avidin column–based purification method5, increasing the pulldown efficiency by two- to threefold (Supplementary Table 1).
Figure 1.
Principle of selective chemical labeling of 5-hmC followed by SMRT DNA sequencing.
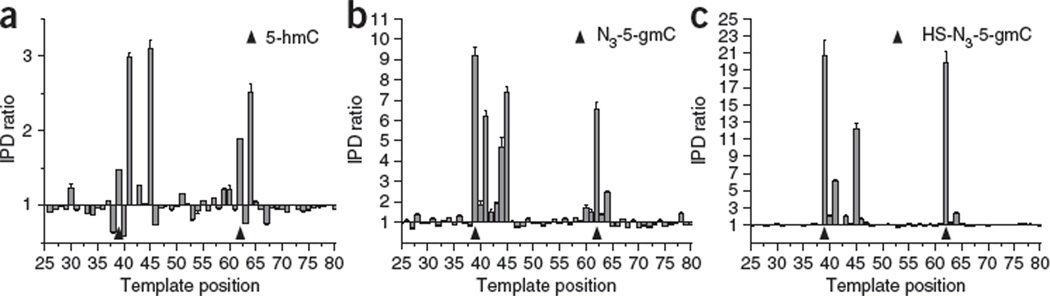
To test the method, we subjected synthetic DNA templates with known 5-hmC positions to this selective chemical labeling protocol and tested the effect of the various modifications on kinetic signatures during SMRT sequencing. The 5-hmC itself gave an increase in IPD values of about two- to threefold compared to an unmodified control template (Fig. 2a), as observed previously13. The addition of azide-glucose to form N3-5-gmC resulted in a substantial increase in the kinetic signature with IPD ratios of ~7–9 (Fig. 2b). The cleaved biotin linker adduct HS-N3-5-gmC resulted in an even stronger kinetic signature with IPD ratios of ~7–25 (Fig. 2c), allowing the clearest detection of the modification. As previously described, the kinetic signatures extended over a region around the 5-hmC position (over a range starting ~1 base before the modified base and ending ~7 bases after it) and were sequence context–dependent13. For the final adduct, HS-N3-5-gmC, we observed characteristic secondary peaks for most sequence contexts 2 and 6 bases downstream of the 5-hmC position. This information can be used algorithmically to increase the confidence of 5-hmC assignments.
Figure 2.
Effects of 5-hmC modifications on polymerase kinetics in SMRT DNA sequencing. (a–c) Using synthetic DNA templates with known positions of 5-hmC (triangles), the graphs show IPD ratios, at each template position, of the modified DNA template over a control DNA template of identical sequence but lacking 5-hmC. Conditions were 5-hmC untreated (a), upon coupling with glucose azide (b) and upon additional coupling with the disulfide-containing biotin linker, followed by cleavage of the disulfide bond (c).
Because the forward and reverse strand of a double-stranded DNA template molecule are interrogated independently in a SMRT sequencing read, our approach can differentiate hemi- and fully hydroxymethylated DNA. To demonstrate this, we sequenced synthetic DNA templates with known 5-hmC modifications on either or both strands and found that kinetic signals were strictly limited to sequencing reads obtained from template strands carrying the modification (Supplementary Fig. 2).
To test the feasibility of this approach for biological samples, we applied this method to detect 5-hmC in genomic DNA from mouse embryonic stem cells (mESC), one of the first cell types in which 5-hmC had been detected by bulk methods2, 5. After selectively labeling and enriching 5-hmC–containing DNA fragments, we prepared them into SMRTbell DNA libraries14 for shotgun SMRT sequencing and analyzed them for kinetic signatures. We detected clear signals for HS-N3-5-gmC (Supplementary Table 2), as shown by the example of a hemi-hydroxylated position on mouse chromosome 1 (Fig. 3). Because of the greatly increased magnitude of the kinetic signal, assignments of 5-hmC positions can be made for single-molecule reads and do not require comparison with an amplified, unmodified control sample.
Figure 3.
Example of 5-hmC detection by SMRT sequencing from mESC genomic DNA. (a) The raw SMRT sequencing read. A.u., arbitrary units. (b) Sequencing subreads from the SMRTbell template, mapped onto mouse chromosome 1 over the sequencing time course. Pauses appear as discontinuities as the polymerase temporarily stops progressing along the DNA template. F, forward strand reads; R, reverse strand reads. (c) Subreads are grouped and annotated by IPD values in a heat-map scale, identifying a hemi-hydroxymethylated CG position in this DNA molecule. Arrows indicate consistent pausing of the polymerase (that is, large IPD value) at the same genomic position across multiple intramolecular subreads, indicating the presence of a 5-hmC adduct.
Positions of HS-N3-5-gmC are visible as extended pauses in a raw sequencing read (Fig. 3a). Pauses corresponding to 5-hmC positions can be distinguished from occasional stochastic pauses15 by taking advantage of circular consensus sequencing14, in which the same base of a DNA molecule is interrogated multiple times in consecutive subreads on the topologically circular SMRTbell template. This allows for multiple occurrences of 5-hmC detection for high-confidence assignments; in the example shown, the polymerase pauses consistently at a particular template position in all reverse-strand subreads (Fig. 3b). The absence of pausing at this CG position in the corresponding forward strand subreads implies that this genomic location was hemi-hydroxymethylated. Grouping of forward- and reverse-strand subreads facilitates strand-specific 5-hmC identification by kinetic consensus and eliminates the effects of raw subread errors by virtue of generating a circular consensus sequence14 (Fig. 3c). From initial sequencing of this sample, we found high-confidence genomic positions displaying 5-hmC signatures across many different sequence contexts containing CG (Supplementary Table 2). The sequenced DNA fragment sizes ranged from ~100 bases to ~400 bases, allowing the observation of multiple 5-hmC occurrences on the same DNA molecule (Supplementary Table 2). We are now refining the 5-hmC detection algorithm by characterizing sequence-context influences and effects of closely spaced 5-hmC positions on the kinetic signatures.
We also validated four high-confidence hits in the CCGG context as well as an instance of the CCGG context that we observed to have no 5-hmC using a low-throughput commercial 5-hmC detection assay. The assay is based on PCR amplification of a specific sequence after glucosylation-sensitive restriction digestion (Supplementary Fig. 3).
In conclusion, we presented to our knowledge the first high-throughput sequencing method for 5-hmC by taking advantage of selective chemical labeling of 5-hmC and SMRT sequencing. We anticipate that the method can be combined with the detection of 5-mC, which also causes a kinetic signature in SMRT sequencing, albeit currently at a smaller amplitude so that more fold coverage is required for high-confidence assignments. We are now applying this method to sequence 5-hmC across the entire mESC genome. With an estimated abundance of ~1,500,000 5-hmC positions (~0.05% of all nucleotides) and considering ~100-bp DNA fragments sequenced, the total genomic fraction amounts to ~150 Mb. This compares well with a current SMRT sequencing throughput of ~50–100 Mb per run. The above estimate does not take into account likely clustering of 5-hmC, which would reduce the overall sequencing required. Additional improvements in the throughput of sequencing and increases in read length and accuracy are also expected to reduce the number of sequencing runs. We anticipate that this technology will be a powerful method to explore the biology of 5-hmC.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
ONLINE METHODS
Preparation of genomic DNA, synthetic DNA templates and β-glucosyltransferase
Mouse feeder-free E14Tg2A embryonic stem cell (mESC) genomic DNA was prepared as previously described5. Oligonucleotides were synthesized and synthetic DNA templates were prepared as previously described13, 16. β-glucosyltransferase (β-GT) was prepared as previously described5.
The 5-hmC labeling reaction and chemistry
For synthetic DNA, the 5-hmC labeling reactions were performed in a 20-µl solution containing 50 mM HEPES buffer (pH 7.9), 25 mM MgCl2, 2 µg DNA template, 100 µM UDP-6-N3-Glc and 1 µM β-GT. The reactions were incubated for 1 h at 37 °C. After the reaction, the DNA substrates were purified by Qiagen Nucleotide removal kit and eluted in H2O. For mESC DNA, the 5-hmC labeling reactions were performed in a 400-µl solution containing 50 mM HEPES buffer (pH 7.9), 25 mM MgCl2, 200 µg sonicated genomic DNA (100–500 bp), 100 µM UDP-6-N3-Glc and 2 µM β-GT. The reactions were incubated for 1 h at 37 °C. After the reaction, the DNA substrates were purified by Bio-Rad Micro Bio-Spin 6 spin column and eluted in H2O. The click chemistry was performed with addition of 150 µM dibenzocyclooctyne with disulfide biotin linker into the DNA solution, and the reaction mixture was incubated for 2 h at 37 °C. The DNA samples were then purified by Qiagen nucleotide removal kit and eluted in H2O.
Affinity enrichment of the modified 5-hmC DNA fragments (HS-5-N3-gmC)
Invitrogen Dynabeads MyOne Streptavidin C1 was used to pull down the biotinylated DNA following the manufacturer’s instructions with minor modifications. Binding and washing buffer for coupling of nucleic acids was used with the addition of 0.01% Tween-20. We used 100 µl of beads, following the general washing procedure and immobilization of nucleic acids from the manufacturer’s manual. To cleave the disulfide bonds and release the 5-hmC–containing fragments, beads were incubated in 100 µl of 50 mM DTT in H2O for 2 h at room temperature (25 °C) using gentle rotation. Beads were segregated with a magnet, leaving the 5-hmC–containing DNA in solution. The released DNA solution was then applied to Bio-Rad Micro Bio-Spin 6 spin columns to remove DTT. Finally, a Qiagen MinElute PCR Purification kit was used to purify the DNA solution and elute into EB buffer. From 160 µg labeled mESC genomic DNA, 1.3 µg pulldown DNA was obtained for SMRTbell library construction.
Pull-down specificity test
Synthetic 32-mer double-stranded DNA bearing 5-mC or 5-hmC was treated with the labeling process. We purified 2.2 µg of each labeled DNA by 50 µl Invitrogen Dynabeads MyOne Streptavidin C1 as described above. The pulldown DNA was purified using the Qiagen Nucleotide Removal Kit and eluted in 30 µl H2O. We pulled down 1,080 ng of 5-hmC–containing DNA as determined by NanoDrop (pulldown yield was ~50%). No 5-mC–containing DNA was pulled down as measured by NanoDrop. Sequences of the oligonucleotides are listed in Supplementary Table 3.
SMRT DNA sequencing
Synthetic 5-hmC–containing SMRTbells were made by annealing complementary oligonucleotides with specific four-nucleotide overhangs. Annealed oligonucleotides were ligated at 25 °C for 60 min with T4 DNA Ligase (New England Biolabs). Incompletely ligated SMRTbells were degraded with 16 U µg−1 exonuclease III (New England Biolabs) and 1 U µg−1 exonuclease VII (USB) at 37 °C for 30 min. Synthetic SMRTbells were purified using the QIAquick PCR Purification kit (Qiagen). Sequences of the oligonucleotides are listed in Supplementary Table 3. All oligonucleotides contained a 5′ phosphate. Control oligonucleotides contained a standard cytosine in place of the 5-hmC modification. Genomic SMRTbell libraries were generated using the A-tailing method as previously described14. Average insert size was ~200 bp. SMRT DNA sequencing was carried out on the PacBio RS using standard protocols for small insert libraries (Pacific Biosciences). Reads were processed and mapped to the mouse genome (mm9, retrieved from the University of Santa Cruz (UCSC) Genome Browser (http://www.ncbi.nlm.nih.gov/pubmed/20959295) using the basic local alignment with successive refinement (BLASR) mapper (http://www.pacbiodevnet.com/SMRT-Analysis/Algorithms/BLASR) and the Pacific Biosciences SMRT-Analysis pipeline (http://www.pacbiodevnet.com/SMRTAnalysis/Software/SMRT-Pipe) using the standard no-filter mapping protocol. IPDs were measured as previously described13 for all pulses aligned to each position in the genome. For each position with per-strand coverage exceeding 5×, the distribution of IPDs was compared against the expected IPD distribution using a likelihood ratio goodness-of-fit test, applied separately for forward and reverse strand reads. Positions in the genome were ranked according to the outcome of the statistical test on the forward strand and reverse strand (hemi-hydroxymethylation) as well as both strands together (full hydroxymethylation). Results were analyzed visually in the SMRTView genome browser, and a scoring cutoff was selected to generate a set of high-confidence single-nucleotide 5-hmC predictions. The sequencing data with associated kinetic parameters, the analysis tools used to extract 5-hmC positions and detailed instructions on how to use the kinetic analysis tools are available at http://pacificbiosciences.com/devnet/files/datasets/publications/dna-modification-hmc/1.0/index.html.
To validate the consistency of our high-confidence 5-hmC predictions with previously published methods, we surveyed the recent publications on 5-hmC sequencing and compared the identified regions of interest with our reported 5-hmC positions. Hydroxymethylated DNA immunoprecipitation (hMeDIP) sequencing of mouse E14 embryonic stem cells has been published in reference 9, reporting the targets of the TET1 gene and its partner SIN3A complex followed by Illumina sequencing, with a total of ~500 Mb of the mouse genome loci tags, and in reference 6, using ChIP-chip followed by Affymetrix arrays, covering approximately 80 Mb of mouse genome sequence. We cross-checked our predictions with the set of all genetic loci identified in reference 9 and found that 52% of our 5-hmC position predictions overlapped with this set. By comparison, only 35% of the sequence regions identified in reference 6 were covered in reference 9.
Combined glucosylation and restriction analysis validation of 5-hmC hits
One microgram unsonicated mESC genomic DNA was glucosylated with regular glucose using β-GT. MspI digestion was performed following the manufacturer’s manual (New England Biolabs). PCRs were performed in a 50-µl solution containing 50 ng genomic DNA, 2.5 units REDTaq DNA polymerase (Simga-Aldrich), 1× reaction buffer, 200 µM each dNTP and 0.2 µM of each primer (primer sequences in Supplementary Table 3). PCR cycling conditions were as follows: 94 °C for 3 min and then 25 cycles of PCR at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, followed by a final extension step at 72 °C for 3 min. The PCR products were separated by electrophoresis on 3% NuSieve 3:1 agarose gels (Lonza).
Chemical synthesis
UDP-6-N3-Glc was prepared as described previously5. The disulfide biotin linker was chemically synthesized from biotinyl cystamine (Santa Cruz Biotech) as described previously5. Chemical shifts (δ) were reported in p.p.m. All coupling constants (J values) were reported in hertz. Split pattern abbreviations are as follows: singlet (s), doublet (d), triplet (t), multiplet (m) and doublet of doublets (dd). 1H NMR (500 MHz, CD3OD): δ 7.50 (1 H, aromatics), 7.34–7.23 (7 H, aromatics), 5.35, (1 H, s, CHO), 4.36, 4.17 (m, 2 H, CHNH), 3.39 (m, 1 H, CHS), 3.36 (t, 2 H, J = 6.75 Hz, CH2S2), 3.23 (m, 4 H, CH2NH), 3.15, (1 H, dd, J1 = 15 Hz, J2 = 2 Hz, CH2), 3.08 (m, 1 H, CHHexoS), 2.81, (1 H, dd, J = 4.5, 12.5 Hz, CH2), 2.74 (t, 2 H, S2CH2), 2.60 (d, 1 H, J = 12.5 Hz, CHHendoS), 2.10 (t, 2H, J = 7.5 Hz, CH2CO), 1.5 (m, 6 H, biotin-CH2). 13C NMR (125 MHz, CD3OD): δ 174.8, 164.7, 156.6, 152.2, 151.0, 129.7, 128.0, 127.9, 126.9, 126.6, 125.8, 125.5, 123.6, 121.0, 112.4, 109.6, 76.7, 61.9, 60.2, 55.6, 45.8, 39.8, 39.6, 38.0, 37.6, 37.4, 35.3, 28.3, 28.0, 25.4; high-resolution mass spectroscopy (HRMS): m/z 625.1990 [M + H+]. Calculated for C31H36N4O4S3 m/z 624.1899.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported partly by US National Institutes of Health grants GM071440 (to C.H.) and 1RC2HG005618-01 (National Human Genome Research Institute to S.T.) and The University of Chicago. We thank K. Spittle, M. Boitano, J. Eid, J. Wegener and K. Luong for assistance in data acquisition and figure generation.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
C.H., C.-X.S., J.K. and S.W.T. designed experiments. C.-X.S. performed the labeling and pulldown of synthetic template and mESC sample. T.A.C. prepared library constructs and conducted the sequencing experiments. A.K. analyzed data. Q.D., C.-X.S. and X.-Y.L. carried out the chemical synthesis. X.-Y.L. validated mESC hits. C.H., C.-X.S. and J.K. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the fulltext HTML version of the paper at http://www.nature.com/naturemethods/.
References
- 1.Kriaucionis S, Heintz N. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, et al. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhutani N, Burns DM, Blau HM. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, et al. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song CX, et al. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, et al. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ficz G, et al. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 8.Pastor WA, et al. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams K, et al. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, et al. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munzel M, Globisch D, Carell T. Angew. Chem. Int. Ed. 2011;50:6460–6468. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- 12.Eid J, et al. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 13.Flusberg BA, et al. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korlach J, et al. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 16.Dai Q, Song CX, Pan T, He C. J. Org. Chem. 2011;76:4182–4188. doi: 10.1021/jo200566d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.