SUMMARY
microRNAs regulate developmental cell fate decisions, tissue homeostasis and oncogenesis in distinct ways relative to proteins. Here, we show that the tumor suppressor microRNA miR-34a is a cell fate determinant in early stage dividing colon cancer stem cells (CCSCs). In pair-cell assays, miR34a distributes at high levels in differentiating progeny, while low levels of miR34a demarcate self renewing CCSCs. Moreover, miR34a loss of function and gain of function alters the balance between self-renewal and differentiation both in vitro and in vivo. Mechanistically, miR34a sequesters Notch1 mRNA to generate a sharp threshold response where a bimodal Notch signal specifies the choice between self-renewal versus differentiation. In contrast, the canonical cell fate determinant Numb regulates Notch levels in a continuously graded manner. Taken together, our findings highlight a unique microRNA regulated mechanism that converts noisy input into a toggle switch for robust cell fate decisions in CCSCs.
INTRODUCTION
microRNAs silence gene expression by binding to the 3′ untranslated regions (3′UTRs) of target mRNAs, inhibiting their translation or marking them for degradation (Pauli et al., 2011). microRNAs often target genes that regulate cell fate decisions (Ivey and Srivastava, 2010; Pauli et al., 2011). Recent studies show that microRNAs confer robustness to biological processes in distinct ways relative to proteins, such as suppressing fluctuations in gene regulation (Ebert and Sharp, 2012). microRNAs also frequently form feedback and feedforward loops with other microRNAs and proteins to enhance robustness (Osella et al., 2011; Tsang et al., 2007). microRNAs are globally downregulated in tumors relative to normal tissues, which potentially contributes to the lack of control for differentiation and arrest in cancer cells (Loboda et al., 2011).
Originally identified as a p53 target, the microRNA miR-34a acts as a tumor suppressor in many types of solid tumors (He et al., 2007; LaPointe et al., 2008; Li et al., 2007; Liu et al., 2011; Wurbel et al., 2011; Youn et al., 2001). miR-34a also regulates multiple developmental cell fate mechanisms, including differentiation of mouse and human embryonic stem cells and somatic cell reprogramming, among others (Choi et al., 2011; Guardavaccaro and Clevers, 2012; Sampieri and Fodde, 2012; Sikandar et al., 2010). Among the regulatory mechanisms targeted by miR-34a, the Notch pathway plays a prominent role in cell fate determination during development and oncogenesis (Alison et al., 2012). miR-34a binds to the 3′UTR mRNA sequences of Notch receptors, which causes reduced Notch protein levels and dampens downstream Notch signaling (Li et al., 2009).
The Notch pathway is a critical regulator of asymmetric division in many types of normal stem cells. Asymmetric cell division is a mechanism commonly used by stem cells to generate both a daughter stem cell for self-renewal and a more differentiated daughter cell to create cellular diversity (Neumuller and Knoblich, 2009). Stem cells perform asymmetric division to maintain stem cell number and tissue homeostasis in a robust and precise way (Sanchez-Tillo et al., 2011). Certain types of cancer cells also perform asymmetric division (Dey-Guha et al., 2011; Lathia et al., 2011; O’Brien et al., 2012; Pece et al., 2010; Pine et al., 2010). Similar to the situation in normal stem cells, disruption of asymmetric division can alter the balance between self-renewal and differentiation in cancer stem cells and impact tumor growth (Cicalese et al., 2009; Sugiarto et al., 2011). However, despite the importance of the Notch pathway in the regulation of asymmetric division, it remains unknown whether individual microRNAs such as the Notch-targeting miR-34a play any role in determining cell fate asymmetry in normal and cancer stem cells.
Notch signaling is known to play essential roles in promoting self-renewal of intestinal and colon stem cells (ISCs) and in specifying the choice of absorptive or secretory lineage differentiation (de Sousa et al., 2011; Taketo, 2011; van Es et al., 2005). ISCs undergo both symmetric and asymmetric divisions, probably at different stages, during development and crypt homeostasis (Goulas et al., 2012; Itzkovitz et al., 2012; Potten et al., 2002; Quyn et al., 2010). Colon cancer stem cells (CCSCs) from colorectal cancer (CRC) are thought to arise from, or at least share common properties with, normal colon stem cells (Arrowsmith, 2011a; Clevers, 2011; Dalerba et al., 2007; O’Brien et al., 2007; Ricci-Vitiani et al., 2007). Tumors formed by xenotransplanted CCSCs show heterogeneity in morphology and are populated by cell types reflecting the histopathology of the parental tumor. Like ISCs, CCSC self-renewal also requires Notch signaling (Sikandar, 2010; van Es et al., 2005).
Here, we show that, similar to ISCs, CCSCs from early stage, well-differentiated CRC tumors can perform both self-renewing symmetric division (producing two CCSC daughter cells) and asymmetric division (producing a CCSC daughter cell and a differentiated, non-CCSC daughter cell). The decision of a CCSC to perform either symmetric or asymmetric division is tightly controlled by the miR-34a level. High miR-34a levels dampen Notch signaling and promote daughter cells to become non-CCSCs, whereas low miR-34a levels upregulate Notch signaling and promote daughter cells to remain CCSC. Investigation of regulation kinetics demonstrated a critical role for miR-34a to convert “noisy” signaling inputs into clean bimodal Notch levels that enables robust binary daughter cell fate decisions. This role of miR-34a is distinct from that of the canonical cell fate determinant protein Numb, which regulates Notch levels in a continuously graded manner. These studies provide new insights into asymmetric cell division mechanisms, highlighting unique regulatory roles performed by microRNAs.
RESULTS
Characterization of Early Stage, Well-Differentiated CCSCs
Using established CCSC markers CD133, CD44 and Aldehyde Dehydrogenase 1 (ALDH1) (Emmink et al., 2011; Huang et al., 2009; Ricci-Vitiani et al., 2007; Todaro et al., 2007), we isolated two CCSC lines, CCSC1 and CCSC2, from well-differentiated, early stage CRC patient specimens (Table 1) (see the Experimental Procedures). Consistent with previous reports (Huang et al., 2009; O’Brien et al., 2007; Sikandar, 2010), both CCSC1 and CCSC2 efficiently formed xenograft tumors that maintained the histopathology of their primary human CRCs upon xenografting in immunodeficient mice (Figure S1A).
Table 1.
| ID | Age/Sex | Stage | miR-34a* | Numb* | p53 mutation |
Kras mutation |
% of C/C** |
% of C/D** |
% of D/D** |
% of am** |
|---|---|---|---|---|---|---|---|---|---|---|
| CCSC1 | 57/M | I | 16.7 | 7.2 | NO | NO | 57.6 | 14.1 | 26.1 | 2.2 |
| CCSC2 | 51/M | II | 14.3 | 5.8 | NO | NO | 59.3 | 12.7 | 24.5 | 3.5 |
| CCSC3 | 74/F | I | 13.2 | 4.7 | NO | YES | 61.4 | 13.4 | 25.2 | 0 |
| CCSC4 | 54/M | III | 3.7 | 3.5 | NO | NO | 89.3 | 0 | 9.5 | 1.2 |
| CCSC5 | 61/M | IV | 1.0 | 1.0 | YES | NO | 96.3 | 0 | 3.2 | 0 |
| CCSC6*** | 47/M | II | 11.3 | 4.2 | NO | NO | 41.2 | 19 | 31.8 | 8 |
| CCSC7*** | 87/M | III | 2.5 | 4.1 | NO | NO | 82.6 | 0 | 11.3 | 6.1 |
| CCSC8*** | 50/M | III | 5.2 | 2.2 | YES | YES | 69.7 | 6.1 | 18.2 | 6 |
| CCSC9*** | 86/M | IV | 1.7 | 3.1 | NO | NO | 80.8 | 0 | 19.2 | 0 |
Expression level of miR-34a and Numb were measured by RT-qPCR, relative to CCSC5 sphere.
The pair-cell assay was performed in CCSC spheres by co-immunofluorescence of ALDH1 and CK20. C/C, symmetric self-renewal (CCSC/CCSC); C/D, symmetric division (CCSC/Differentiated cell); D/D, symmetric differentiation (Differentiation cell/Differentiation cell).
The cell lines are freshly isolated CCSCs from primary CRC.
Both CCSC1 and CCSC2 propagate as spheres in ultralow-attachment flasks and are capable of generating cellular diversity in vitro. From dissociated spheres, fluorescence-activated cell sorting (FACS) identified a CD133+CD44+ CCSC subpopulation and a CD133−CD44− non-CCSC subpopulation (Figures S1B). Isolated CCSCs became heterogeneous again in spheres and reached a similar equilibrium between CCSCs (CD133+CD44+) and non-CCSCs (CD133−CD44−) as they proliferated (Figures S1B). Consistent with previous studies of ALDH1 and CCSCs (Huang et al., 2009), FACS analysis confirmed that CCSCs were ALDH1+ whereas non-CCSCs were ALDH1− (Figure S1C).
We then compared the tumorigenic capacity of CCSCs and non-CCSCs using the limiting dilution assay. Also consistent with previous studies of CCSC, for both CCSC1 or CCSC2, as few as 1000 CCSCs were sufficient to form subcutaneous xenograft tumors, while non-CCSCs failed to form tumors during the observed period (2 months) even when up to 1×106 cells were injected (Figure S1D and data not shown). Sphere propagation assays confirmed that, unlike CCSCs, non-CCSCs were incapable of forming and serially propagating spheres in vitro (Figures S1E–S1G). To evaluate the differentiation potential of CCSCs, dissociated sphere cells were cultured in differentiation medium (DMEM with 10% FBS). After 10 days in culture, CD133 and CD44 expression significantly decreased while expression of cytokeratin 20 (CK20), a marker of mature normal colonocytes and non-CCSCs, increased (Figures S1H–S1K). Consistent with these findings, the tumorigenic ability of CCSCs cultured in differentiation medium was greatly reduced (Figure S1L).
miR-34a Inhibits CCSC Self-Renewal In Vitro
microRNA profiling previously identified miR-34a, but not miR-34b or -34c, as expressed in cultured CRC spheres (Jahid et al., 2012). Since miR-34a can cause cell differentiation by inhibiting Notch signaling, we examined how miR-34a expression levels differ between CCSCs and non-CCSCs. RT-qPCR studies showed that miR-34a expression was downregulated in CCSCs and upregulated in non-CCSCs (Figure 1A). Infection of CCSC1 and CCSC2 sphere cells with lentivirus driving miR-34a constitutive over-expression (miR-34a OE) increased the proportion of non-CCSCs relative to CCSCs (Figures 1B and 1C). Overall, these data are consistent with miR-34a promoting CCSC differentiation into non-CCSCs.
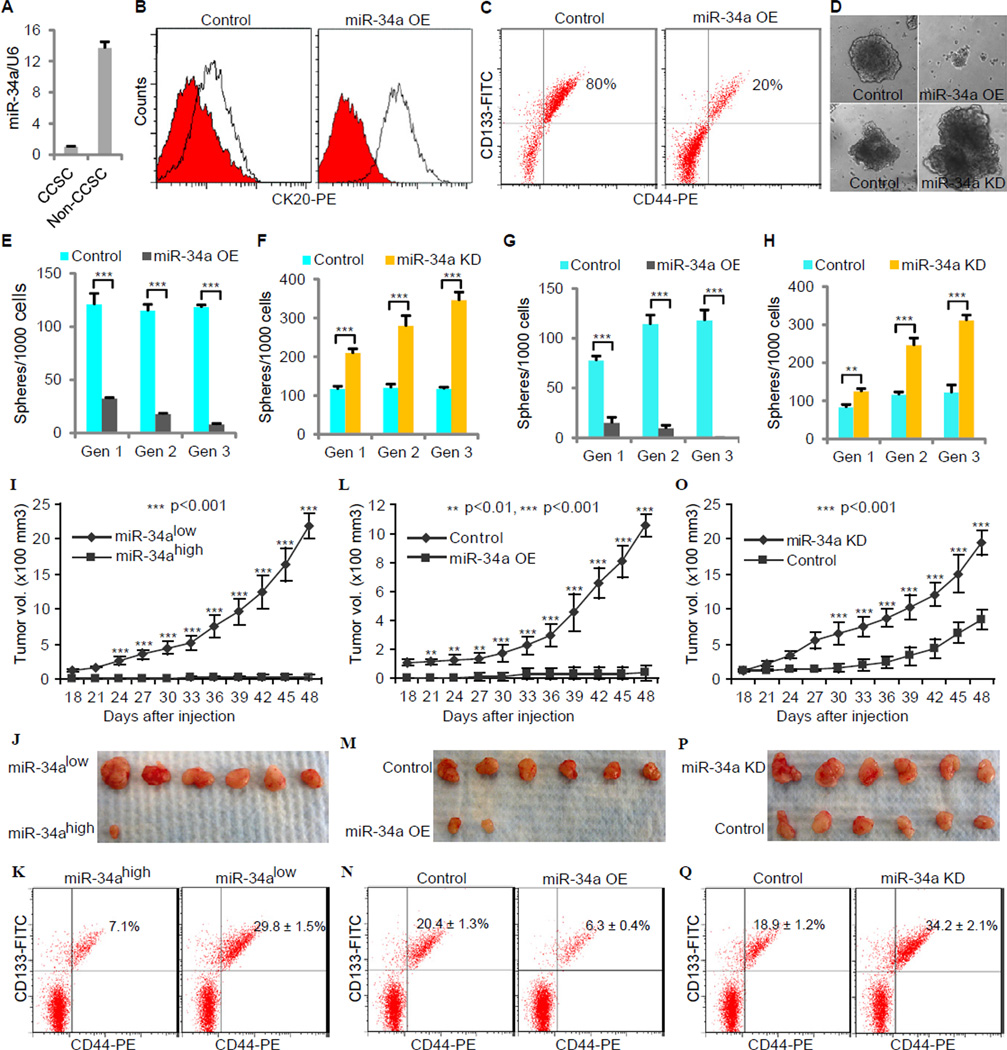
Figure 1. miR-34a Regulates CCSC self-renewal and Tumor Formation. Also see Figure S1 and Figure S2.
(A) RT-qPCR showing miR-34a expression in CCSCs and non-CCSCs. Error bars denote the s.d. between triplicates. (B and C) FACS plots showing CK20, CD44 and CD133 levels in spheres after ectopic miR-34a expression (miR-34a OE). In (B), the red histograms represent isotype controls, and the blank histograms represent CK20+ cells. (D) Representative images of CCSC spheres after ectopic miR-34a expression (miR-34a OE, top) and miR-34a knockdown (miR-34a KD, bottom). (E and F) Sphere formation during serial passages after ectopic miR-34a expression (E) and miR-34a knockdown (F). Error bars denote the s.d. between triplicates. (G and H) Serial sphere formation of CCSCs from xenografts of miR-34a OE (G) and miR-34a KD (H) cells. Equal number of cells was passaged for 3 generation to form spheres. Error bars denote the s.d. between triplicates. (I and J) miR-34alow sphere cells were more tumorigenic than miR-34ahigh sphere cells in vivo, shown by tumor growth curves (I) and images of xenograft tumors (J). Error bars denote the s.d. derived from 6 mice per group. (K) FACS showing the percentages of tumor cells that are CCSCs. (L and M) Ectopic expression of miR-34a (miR-34a OE) reduces tumorigenicity, shown by tumor growth curves (L) and images of xenograft tumors (M). (N) FACS showing the percentages of tumor cells that are CCSCs. Error bars denote the s.d. derived from 6 mice per group. (O and P) Knockdown of miR-34a (miR-34a KD) enhances tumorigenicity, shown by tumor growth curves (O) and images of xenograft tumors (P). Error bars denote the s.d. derived from 6 mice per group. (Q) FACS showing the percentages of tumor cells that are CCSCs. Gen, generation. **, p<0.01; ***, p<0.001.
We then performed serial sphere propagation assays to examine the impact of miR-34a on self-renewal. For both CCSC1 and CCSC2 lines, we analyzed cells with the stably integrated lentiviral miR-34a OE expression cassette described in the preceding paragraph. In addition, we also created CCSC1 and CCSC2 lines with a stably integrated miR-34a “sponge” construct (miR-34a KD). This construct drives transcription of a decoy mRNA containing multiple tandem binding sites for miR-34a, which reduces levels of free miR-34a available to bind its endogenous mRNA targets (Ebert et al., 2007). The efficiency of the miR-34a KD construct was validated by a luciferase miR-34a reporter assay (Figure S2A). After selection for cells containing the miR-34a KD cassette, single cells were allowed to form spheres in vitro. Subsequently, spheres containing the miR-34a KD or control cassettes were dissociated and passaged for several generations (Figure 1D). While spheres from CCSC1 and CCSC2 cells with the control sequence maintained a stable level of sphere-forming ability, miR-34a KD cells had significantly increased sphere-forming ability; in contrast, spheres from CCSC1 and CCSC2 cells with the stably integrated miR-34a OE cassette had diminished sphere-forming activity (Figure 1E, 1F, Figure S2B and S2C). Furthermore, cells from miR-34a OE expressing spheres lost the ability to form new spheres serially after being passaged for several generations, while cells from untransduced or miR-34a KD expressing spheres could be passaged significantly longer (for at least 12 months).
Similarly, miR-34a OE sphere cells had lower proliferation rates (Figure S2D), consistent with high miR-34a levels promoting differentiation (Figures 1B and 1C). These observations are also consistent with non-CCSCs having lower proliferative potential than CCSCs (Figure 2D). Finally, high miR-34a levels also increased cell cycle arrest and senescence (Figures S2E and S2F).
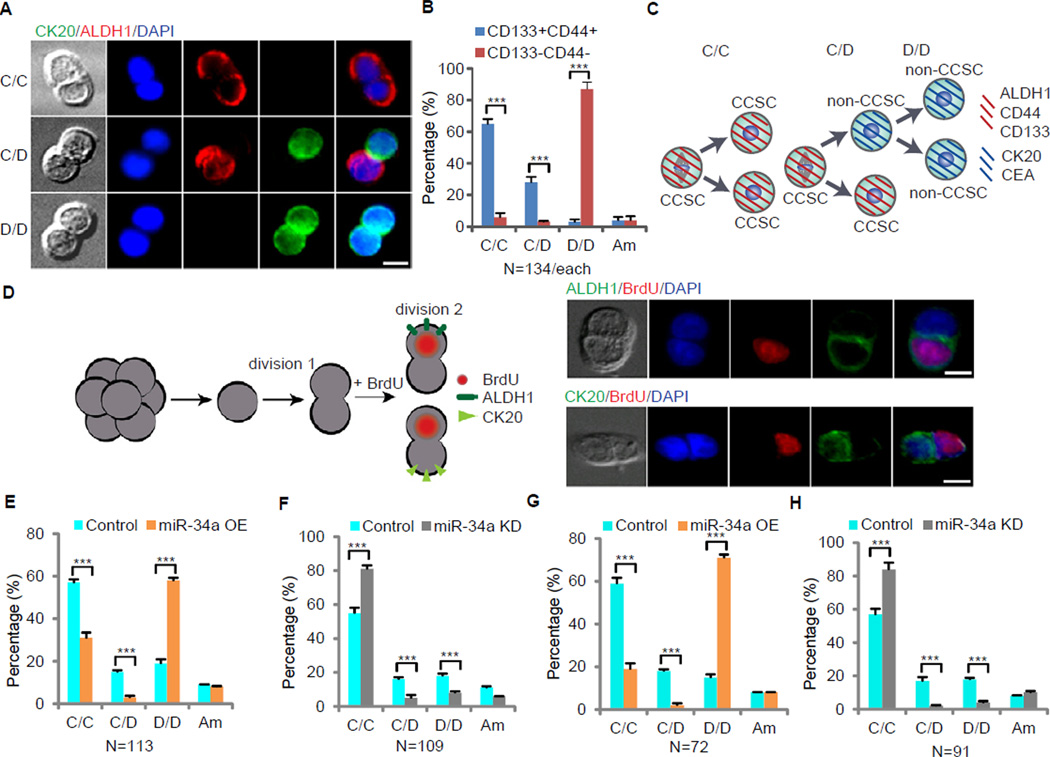
Figure 2. miR-34a Regulates CCSC Division. Also see Figure S3.
(A) Representative images of sphere cell division. Immunofluorescence for ALDH1 (red) and CK20 (green) illustrates three types of division: CCSC/CCSC (C/C), CCSC/non-CCSC (C/D) and non-CCSC/non-CCSC (D/D). (B) Percentages of division types between the CCSC (CD133+CD44+) and non-CCSCs (CD133−CD44−) subpopulations. (C) Schematic illustration of CCSC divisions. (D) A functional assay showing cell fate asymmetry leads to distinct proliferation capacity. Left, schematic representation of the experimental approach. Single sphere cells were allowed to divide once in 24 hours (1st division). Cells were then treated with BrdU for 3 hours to label cells that were entering the 2nd division before co-staining for BrdU/ALDH1 and BrdU/CK20. Right, representative images showing that the CCSC daughter (ALDH1+ or CK20−) was more proliferative and incorporated BrdU. (E) Pair-cell assays with ALDH1 and CK20 co-immunofluorescence showing ectopic miR-34a expression promotes differentiation (D/D) at the expense of asymmetric division (C/D) and symmetric self-renewal (C/C). (F) Pair-cell assay with ALDH1 and CK20 co-immunofluorescence showing miR-34a knockdown increases symmetric self-renewal (C/C) at the expense of asymmetric division (C/D) and differentiation (D/D). (G and H) Immunofluorescence for ALDH1 and CK20 in pair-cell assays showing the percentages of symmetric CCSC/CCSC (C/C), asymmetric (C/D), and non-CCSC (D/D) divisions in sphere cells, which were cultured from CCSCs isolated from the xenografts of miR-34a OE (G) and miR-34a KD (H) spheres. Am, ambiguous. Blue is DAPI staining of the nucleus. Error bars denote the s.d. between triplicates. **, p<0.01; ***, p<0.001.
miR-34a Suppresses CCSC Xenograft Tumor Formation
Using mouse xenograft models, we examined whether miR-34a affects tumor formation in vivo. We constructed a CCSC1 line that stably expresses a lentiviral miR-34a reporter cassette with three miR-34a binding sites cloned into the 3′UTR of a D2EGFP reporter gene. In this reporter line, FACS identified two distinct sphere subpopulations: miR-34ahigh and miR-34alow cells (Figure S2K). When mice were subcutaneously injected with miR-34alow cells, they formed 6/6 tumors. In contrast, mice injected in parallel with miR-34ahigh cells formed only 1/6 tumors and the volume of this one tumor was much smaller than those generated by miR-34ahigh cells (Figures 1I and 1J). Analysis of the disaggregated tumors by FACS showed that tumors developing from miR-34alow cells had a higher percentage of CCSCs than those from miR-34ahigh cells (Figure 1K).
Next, to confirm the inhibitory role of miR-34a in CCSC self-renewal and tumor growth, we subcutaneously injected immunodeficient mice with cells isolated from CCSC1 spheres expressing basal (control), constitutively high (miR-34a OE), or constitutively low (miR-34a KD) miR-34a levels. Only 2/6 mice injected with high miR-34a expressing sphere cells (integrated with the miR-34a OE cassette) developed tumors, whereas 6/6 mice in the control group (injected with cells integrated with the control cassette) developed tumors (Figures 1L and 1M). Additionally, tumors that grew from injected miR-34a high expressing cells were smaller than those arising from control cells. Consistent with these data, the 6/6 tumors that grew from subcutaneously injected miR-34a low-expressing cells (integrated with the miR-34a KD cassette) were consistently larger in size than those in the control group (Figures 1O and 1P). Similar results were observed from xenograft tumors arising from CCSC2 spheres with basal, high, or low miR-34a levels (Figures S2G and S2H). FACS analysis of disaggregated xenograft tumors further showed that high miR-34a levels (miR-34a OE) reduced the ratio of CCSC to non-CCSCs, while low miR-34a levels (miR-34a KD) increased this ratio, suggesting that miR-34a suppressed CCSC self-renewal (Figures 1N and 1Q).
Next, to confirm these findings we isolated CCSCs from xenograft tumors and assayed the impact of miR-34a levels on self-renewal. For both CCSC1 and CCSC2 tumors, serial sphere propagation assays confirmed that low miR-34a expressing CCSCs from miR-34a KD tumors had increased self-renewal ability vs. control CCSCs with basal miR-34a expression levels; in contrast, CCSCs with high miR-34a levels from miR-34a OE tumors had significantly lower self-renewal capacity than control CCSCs (Figure 1G, 1H, Figure S2I and S2J).
Early Stage CCSCs Perform Both Symmetric and Asymmetric Division
To understand the mechanism of miR-34a suppression of CCSC self-renewal, we used the in vitro pair-cell assay to assess how CCSC and non-CCSC cells divide (Bultje et al., 2009) (Figure S3A). When CCSCs were plated as single cells and allowed to progress through one cell division, co-immunofluorescence staining for ALDH1 and CK20 revealed that 65% of cell divisions were symmetrical, producing two CCSC (ALDH1+) daughter cells, whereas 28% were asymmetrical, producing one CCSC daughter and one non-CCSC (CK20+) daughter cell. In contrast, 87% of non-CCSCs plated in parallel divided to give rise to two non-CCSC daughter cells (Figures 2A and 2B). The few ‘non-CCSCs’ that produced CCSC daughter cells were presumably CCSCs with borderline CD44 and CD133 expression that were sorted into the non-CCSC population by FACS. These findings demonstrate that early stage CCSCs can perform both symmetric and asymmetric division whereas non-CCSCs largely divide into non-CCSCs (Figure 2C). This result was confirmed by additional pair-cell assays with immunofluorescence staining for other CCSC and differentiation markers, including the ISC marker Lgr5 (Arrowsmith, 2011b) (Figures S3B–3G). Furthermore, co-immunofluorescence staining for ALDH1 and CD44 or CD133 confirmed that expression of CCSC markers in daughter cells was consistent with each other during symmetric and asymmetric division, as the CCSC daughter cells always express CD44, CD133 and ALDH1 (Figures S3H and S3I).
To understand whether the balance between symmetric and asymmetric division changes during CRC tumor progression, we performed pair-cell assays on three other CCSC lines (CCSC3–5) and CCSCs sorted from primary cells freshly isolated from CRC tumors (CCSC6–9). Asymmetric divisions of CCSCs happen more frequently in early stage CRC tumors than in late stage CRC tumors (Table 1 and Figure S3J). Hence asymmetric division is negatively correlated with tumorigenicity and invasiveness.
We then examined whether CCSC and non-CCSC daughters have different proliferation rates (Sugiarto et al., 2011). After culturing CCSC1 and CCSC2 spheres in proliferative medium (DMEM with 10% FBS) for 24 hours, we plated single cells and allowed them to divide once in proliferative medium for another 24 hours (1st division). We then treated cells with BrdU for 3 hours to label the cells entering the 2nd division before co-staining for BrdU/ALDH1 and BrdU/CK20. The CCSC (ALDH1+) daughter cells entered the 2nd division immediately and incorporated BrdU; in contrast, the non-CCSC (CK20+) daughter cells did not immediately enter the 2nd division and did not incorporate BrdU (Figure 2D and Figure S3K). This experiment indicates a higher proliferative rate of CCSC vs. non-CCSC daughters, similar to the rapidly dividing Lgr5+ ISCs in the intestine (Arrowsmith, 2011b).
Both High and Low miR-34a Levels Inhibit Asymmetric Division
Pair-cell assays with CCSC1 and CCSC2 cells showed that high miR-34a levels (miR-34a OE) decreased both symmetric CCSC-CCSC division and asymmetric division (Figure 2E and Figure S3L), while low miR-34a levels (miR-34a KD) increased symmetric CCSC-CCSC division but still decreased asymmetric division (Figure 2F and Figure S3M). To verify that miR-34a regulates primary tumor cells in the same way, we isolated CCSCs from xenografts arising from high- and low-miR-34a expressing CCSC1 and CCSC2 spheres. These tumor-derived CCSCs were cultured as spheres and plated as single cells in the pair-cell assay. These experiments confirmed that CCSCs isolated from low miR-34a expressing (miR-34a KD) tumors more frequently performed symmetric CCSC-CCSC division, while CCSCs from high miR-34a expressing (miR-34a OE) tumors less frequently performed symmetric CCSC-CCSC division. Interestingly, both performed lower rates of asymmetric division than CCSCs isolated from basal miR-34a expressing (i.e. control) xenografts (Figure 2G, 2H, Figure S3N and S3O). These data support a model in which miR-34a balances self-renewal and differentiation in CCSC as they populate growing tumors: Higher miR-34a levels promote differentiation to produce non-CCSCs while lower miR-34a levels promote self-renewal through symmetric CCSC-CCSC divisions.
CCSCs from late stage CRC tumors have lower miR-34a expression levels than CCSCs from early stage tumors, according to RT-qPCR (Table 1). Consistent with our data from CCSC1 and CCSC2 that miR-34a knockdown promotes symmetric, CCSC-CCSC division while suppressing asymmetric division, CCSCs from late stage CRC tumors have higher rates of symmetric CCSC-CCSC division and lower rates of asymmetric division than CCSCs from early stage CRC tumors (Table 1).
miR-34a Levels Correlate with Cell Fate Asymmetry
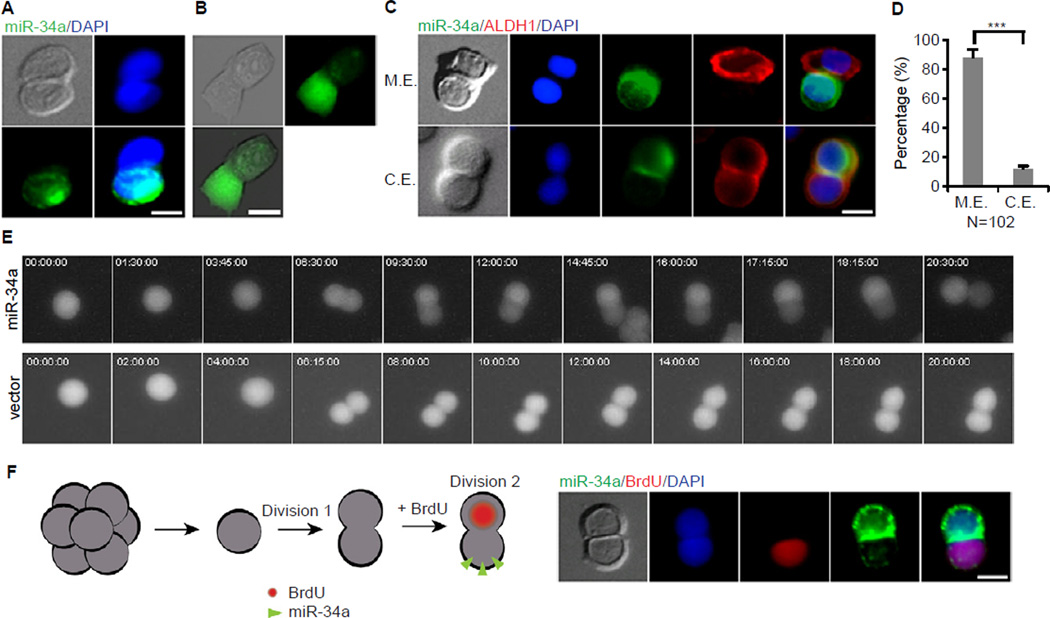
We next evaluated whether miR-34a levels are regulated differentially depending on whether a daughter cell adopts a CCSC or non-CCSC identity. We observed asymmetric distribution of miR-34a in pair-cell assays of early stage CCSCs using RNA fluorescent in-situ hybridization (FISH) with a miR-34a probe (Figure 3A and Figure S4A). In contrast, expression of miR-34c, another member of the miR-34 family, always remained low (data not shown). Asymmetric distribution of miR-34a in dividing pairs was further confirmed using the miR-34a D2EGFP reporter lines described previously (Figure 3B). Time-lapse movies of CCSCs from the reporter line showed that during division, one daughter cell started to express miR-34a, which inhibited GFP expression. In contrast, CCSCs infected with the D2GFP control vector (i.e. no 3′UTR miR-34a binding sites) always expressed GFP symmetrically during division (Figure 3E and Movie S1).
Figure 3. Symmetric and Asymmetric Distribution of miR-34a during division. Also see Figure S4.
(A) Representative image of symmetric and asymmetric distribution of miR-34a (green) during division shown by RNA FISH. (B) Representative image of asymmetric expression of miR-34a shown by a miR-34a GFP reporter in living cells. A high GFP fluorescent signal indicates a low miR-34a expression level. (C) Representative images showing miR-34a (RNA FISH) and ALDH1 are mutually exclusive (M.E., top row) or are co-expressed (C.E., bottom row). (D) Percentages of CCSC divisions wherein miR-34a and ALDH1 are M.E. or C.E.. (E) Representative image of time-lapse images of asymmetric miR-34a expression during CCSC division with a miR-34a GFP reporter (top). CCSCs infected with a D2GFP control vector divided GFP symmetrically (bottom) (also see Movie S2). (F) miR-34alow daughter cell has more proliferative potential. Left, schematic representation of the experimental approach. Single cells were allowed to divide once in 24 hours (division 1). Cells were then treated with BrdU for 3 hours to label cells that were entering the 2nd division (division 2) before co-staining for BrdU/miR-34a. Right, representative image showing that the miR-34alow daughter was more proliferative and incorporated BrdU. Blue is DAPI staining of the nucleus. Scale bar, 8 µm. Error bars denote the s.d. between triplicates. ***, p<0.001.
Co-immunofluorescence showed that miR-34a and ALDH1 expression were mutually exclusive in daughter cells during 88% of CCSC1 divisions (Figures 3C and 3D) and 83% of CCSC2 division (Figure S4B and S4C), consistent with miR-34alow daughters being mostly CCSCs and miR-34ahigh daughters mostly non-CCSCs. Furthermore, pair-cell assays with CCSC1 and CCSC2 spheres incubated with BrdU showed that miR-34alow daughter cells have higher proliferative rates than miR-34ahigh daughter cells (Figure 3F and Figure S4D). This observation is consistent with the previous finding that CCSC (ALDH1+) daughter cells have higher proliferative rates than non-CCSC (CK20+) daughter cells after asymmetric division (Figure 2D and Figure S3K).
miR-34a Suppresses Notch Signaling to Promote Daughter Differentiation
miR-34a has been reported to suppress Notch1 protein levels (Li et al., 2009). RT-qPCR and Western blot confirmed that miR-34a downregulates Notch1 expression in early stage CCSCs (Figures 4A and 4B). Notch signaling has been shown to promote CCSC self-renewal (Sikandar, 2010). CCSCs have high levels of Notch activity, expressing high levels of Notch intercellular domain (NICD) and the Notch target gene Hes1 (Figures 4C and 4D). High Notch activity is specific to CCSCs, as depletion of CCSCs by either FACs sorting or FBS-induced differentiation significantly reduced Notch activity (Figures 4C and 4D). Inhibition of Notch by the γ-secretase inhibitor DAPT reduced the CCSC population and increased the non-CCSC population (Figures 4E–4G), confirming that Notch promotes CCSC self-renewal. We then constitutively expressed shRNAs that targeted against the canonical Notch transcription factor RBPJk, which efficiently inhibited Notch signaling as shown by Western blot (Figure S5A). RBPJκ knockdown and downstream Notch signaling inhibition significantly reduced the ability of CCSC to form xenograft tumors in vivo (Figures 4H and 4I). Given that miR-34a suppresses Notch, these findings are consistent with previous observations that ectopic expression of miR-34a inhibited CCSC self-renewal and tumor formation (Figures 1B, 1C, 1L, 1M and 1N).
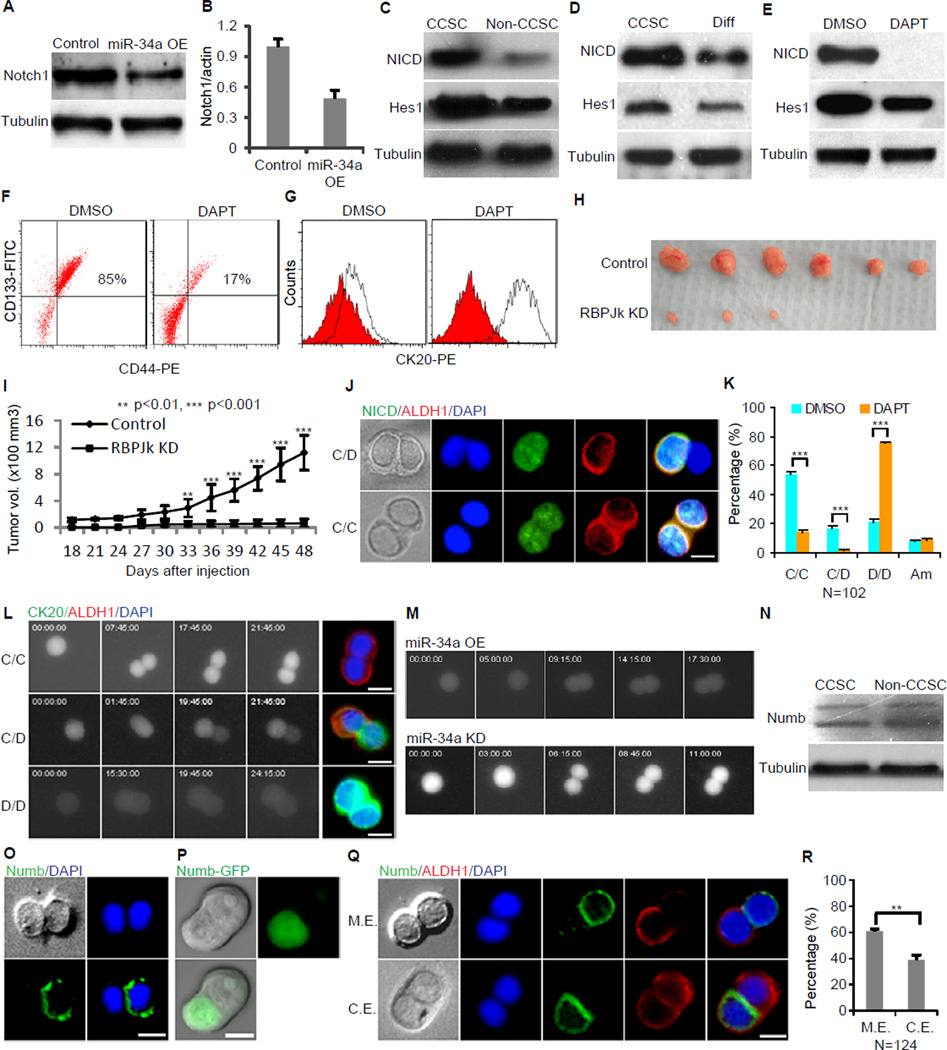
Figure 4. miR-34a Targets Notch to Determine Cell Fate. Also see Figure S5.
(A and B) Western blot (A) and RT-qPCR (B) showing that ectopic miR-34a expression (miR-34a OE) downregulates Notch1 expression. Error bars denote the s.d. between triplicates. (C) Notch signaling (NICD and Hes1) is upregulated in CCSCs and downregulated in non-CCSCs isolated by FACS. (D) Notch signaling (NICD and Hes1) is upregulated in CCSCs and downregulated in differentiation medium-induced differentiated cells. Diff: differentiation medium-induced differentiated cells. (E) The γ-secretase inhibitor DAPT inhibits Notch signaling (NICD, HES1) in CCSCs. (F) Notch inhibition by DAPT depletes CCSCs (CD133+CD44+) from spheres compared to the control (DMSO). (G) Notch inhibition by DAPT induces differentiation. The red histograms represent isotype controls, and the blank histograms represent CK20+ cells. (H and I) Inhibition of Notch signaling by anti-RBPJK shRNA reduced tumorigenicity of the sphere cells, shown by tumor images (H) and growth curves (I). Error bars denote the s.d. derived from 6 mice per group. (J) Representative images of immunofluorescence for NICD and ALDH1. Notch signaling (NICD, green) is only expressed in ALDH1+ (red) cells. (K) DAPT inhibits asymmetric division (C/D) and increases differentiation (D/D). C/C, CCSC/CCSC daughter pair; C/D, CCSC/non-CCSC daughter pair; D/D, non-CCSC/non-CCSC daughter pair ; Am, ambiguous. Error bars denote the s.d. between triplicates. (L) Representative time-lapse images of a Notch GFP reporter cell line showing the three types of division. Immunofluorescence of the same daughter pairs immediately after the movies confirmed that the Notch+ daughters were ALDH1+ and the Notch− daughters were CK20+ (also see Movie S2). (M) Representative time-lapse images showing that ectopic miR-34a expression increases symmetric Notchlow/Notchlow cell division (top) whereas miR-34a knockdown increases symmetric Notchhigh/Notchhigh cell division (bottom). (N) Western blot showing Numb levels in CCSCs and non-CCSCs. (O) Representative image of symmetric and asymmetric segregation of endogenous Numb (green) shown by immunofluorescence with Numb antibodies. (P) Representative image of asymmetric segregation of the Numb-GFP fusion protein in living cells. (Q) Representative image showing Numb and ALDH1 are mutually exclusive (M.E., top row) or are co-expressed (C.E) in at least one of the daughter cells (bottom row) during division. (R) Percentages of CCSC divisions wherein Numb and ALDH1 are M.E. or C.E. in daughter cells. Error bars denote the s.d. between triplicates. Blue is DAPI staining of the nucleus. Scale bar, 8 µm. **, p<0.01; ***, p<0.001.
Because differential Notch signaling levels enable asymmetric division of certain normal stem cells, we asked whether Notch signaling levels are differentially regulated during CCSC asymmetric division, and whether they are correlated with daughter cell fate outcomes. Co-immunofluorescence staining of CCSCs in pair-cell assays showed that NICD was distributed asymmetrically, appearing in the CCSC (ALDH1+) daughter only (Figure 4J). Inhibition of Notch signaling by DAPT suppressed asymmetric division and reduced symmetric CCSC-CCSC division significantly (Figure 4K), similar to the effect of ectopic miR-34a expression on CCSC division (Figure 2E).
We then integrated a lentiviral Notch pathway EGFP reporter into CCSC spheres and performed time-lapse microscopy to visualize directly the distribution of Notch signaling during cell division. This reporter contained multiple RBPJκ response elements upstream of a basal promoter driving expression of EGFP to measure endogenous Notch signaling activity. In these movies, Notch(EGFP)+ cells either divided symmetrically into two Notch(EGFP)+ daughter cells or divided asymmetrically into a Notch(EGFP)+ daughter cell and a Notch(EGFP)− daughter cell, whereas Notch-EGFP- cells mostly divided into two Notch(EGFP)− daughter cells (Figure 4L and Movie S2). Co-immunofluorescence staining of the same daughter pairs immediately after time-lapse imaging verified that Notch(EGFP)+ daughter cells were CCSCs (ALDH1+CK20−) and Notch(EGFP)− daughter cells were non-CCSCs (ALDH1−CK20+) (Figure 4L).
Because differential miR-34a levels (Figures 3A, 3B and 3E) can potentially contribute to differential Notch signaling levels through suppression of Notch1 expression, we examined whether disruption of differential miR-34a levels interferes with differential Notch signaling levels. Indeed, ectopic expression of miR-34a increased Notch(EGFP) −/Notch(EGFP)− pairs while knockdown of miR-34a increased Notch(EGFP)+/Notch(EGFP)+ pairs during cell division. In both cases, asymmetric distribution of Notch signaling was significantly reduced (Figures 4M). Overall, these time-lapse movies indicated that, in miR-34ahigh daughter cells, miR-34a suppresses Notch signaling to promote differentiation.
Numb Localization Correlates Less Closely with Daughter Cell Fate Outcomes than miR-34a
A well-known mechanism for enabling asymmetric division is the cell fate determinant Numb, which, like miR-34a, also suppresses Notch signaling (Neumuller and Knoblich, 2009). During asymmetric division, Numb localizes to one side of the dividing cell and causes degradation of membrane-bound Notch receptors and NICD (McGill and McGlade, 2003; Schweisguth, 2004). Numb protein levels are slightly upregulated in non-CCSCs relative to CCSCs (Figure 4N). Immunofluorescence staining of dividing pairs shows that endogenous Numb localized asymmetrically during ~20% of CCSC divisions (Figure 4O). Analysis of a CCSC line stably expressing a Numb-GFP fusion protein from a weak UBC promoter confirmed that even before the completion of cell division, Numb-GFP localizes to one side of the dividing cell (Figure 4P). Interestingly, despite being a better-characterized cell fate determinant, Numb and ALDH1 were mutually exclusive in only 61% of CCSC1 divisions and 55% of CCSC2 divisions whereas they were co-expressed in at least one daughter cell (symmetric Numb vs. asymmetric ALDH1 or vice versa) in 39% of CCSC1 divisions and 45% of CCSC2 divisions (Figure 4Q, 4R, Figures S5B and S5C). miR-34a and ALDH1 expression, on the other hand, were mutually exclusive in daughter cells during 88% of CCSC1 divisions and 83% of CCSC2 divisions as previously described (Figure 3C, 3D, S4B and S4C). Therefore, miR-34a correlates more closely with CCSC daughter cell differentiation than Numb.
miR-34a Generates a Bimodal Notch Distribution
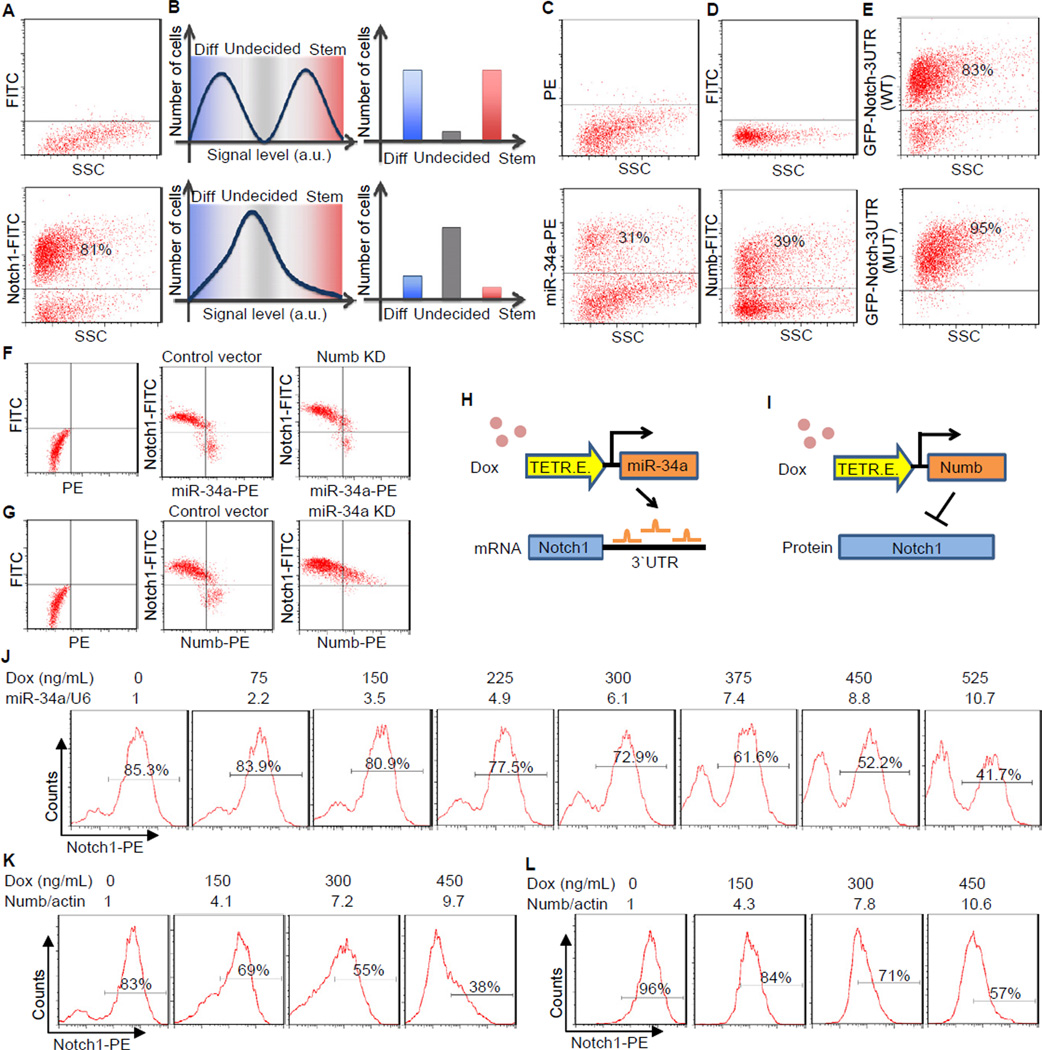
To understand why miR-34a is more closely correlated with non-CCSC daughter cells than Numb after asymmetric division, we measured the distribution of endogenous Notch1, a direct target of both miR-34a and Numb. FACS of CCSC1 sphere cells showed that Notch1 displayed a bimodal distribution with well-separated peaks (Figure 5A, bottom). The bimodal distribution of Notch was also confirmed in other early stage CCSC lines, including CCSCs freshly isolated from CRC tumors (Figure S6A). Notch1 bimodality is important for robust cell fate decisions, because bimodal signals enable the majority of daughter cells to specify their CCSC vs. non-CCSC identity unequivocally, while non-bimodal signals leave a substantial portion of the population undecided and subject to stochastic variations (Figure 5B).
Figure 5. miR-34a Generates Bimodal Notch levels. Also see Figure S6.
(A) FACS plots of sphere cells showing bimodal Notch in CCSC sphere cells. The cutoff threshold was determined by the negative control in the top panel with isotype-matched IgG followed by FITC or PE conjugated secondary antibodies. Cutoff thresholds for the remaining FACs plots in Figure 5 were determined in a similar way. (B) Schematic representation showing signaling bimodality is important for robust cell fate decision. Bimodal signals enables the majority of cells to determine their fate unequivocally, while unimodal signals leave a big portion of the population undecided and subject to stochastic variations. (C and D) FACS plots showing miR-34a (C) and Numb (D) distribution in CCSC sphere cells. (E) FACS plot showing GFP levels from Notch1 3′UTR reporters with native (top) and mutated (bottom) miR-34a binding sites. (F) FACS plots showing the distribution of miR-34a and Notch levels in Numb knockdown (KD) and control CCSC sphere cells. Numb was knocked down by an shRNA vector. (G) FACS plots showing the distribution of Numb and Notch levels in miR-34a KD and control CCSC sphere cells. miR-34a was knocked down by microRNA sponges. (H) Schematic illustrating the inducible miR-34a construct used in the experiments shown in (J). (I) Schematic illustrating the inducible Numb construct used in the experiments shown in (K) and (L). (J) Notch1 displayed a bimodal, on-off response when miR-34a expression was incrementally induced by doxycycline, as shown by FACS. The miR-34a levels were measured by RT-qPCR and shown on top of the FACs plots. (K and L) FACS plots showing Notch1 distribution in wild-type CCSC sphere cells (K) and miR-34a KD ]CCSC sphere cells (L) when Numb expression was incrementally induced by doxycycline. The Numb levels were measured by RT-qPCR and shown on top of the FACs plots.
We next addressed whether miR-34a or Numb was responsible for Notch1 bimodality in sphere cells. FACS of sphere cells with both miR-34a FISH probes and Numb antibodies showed that both miR-34a and Numb distribution in sphere cells were semi-bimodal, though not to the same extent as Notch1 (Figures 5C and 5D). Since miR-34a suppresses Notch1 post-transcriptionally while Numb suppresses Notch1 post-translationally, to delineate their respective effects we constructed a cell line that stably expresses a lentiviral reporter in which the 3′UTR of Notch1 was fused to the EGFP gene. In this system, Notch 3′UTR-EGFP expression displayed a similar bimodal distribution as Notch1, confirming that a post-transcriptional mechanism acting on the Notch1 3′UTR is sufficient to generate bimodality (Figure 5E, top panel). Mutation of the miR-34a binding sites in the 3′UTR abolished EGFP bimodality (Figure 5E, bottom panel). Altogether, these data indicate that miR-34a contributes to Notch1 bimodality.
We then tested whether knockdown of miR-34a or Numb affects Notch1 bimodality in CCSC1 and CCSC2 spheres. Two-color FACS using miR-34a FISH probes and Notch1 antibodies revealed that Numb knockdown (Numb KD) by a lentiviral shRNA vector did not completely abolish Notch1 bimodality (Figure 5F and Figure S6B) (Numb knockdown efficiency was validated by western blot (Figure S6C)). In contrast, miR-34a knockdown completely abolished Notch bimodality even though the distribution of Numb in the population remained similar (Figure 5G and Figure S6D). Combined, these results indicate that in CCSC miR-34a plays a more important role than Numb in causing Notch1 bimodality.
To characterize quantitatively the contribution of miR-34a and Numb to Notch bimodality, we induced miR-34a and Numb expression and monitored how they affected the distribution of Notch1 in CCSC1 and CCSC2 spheres. First, we constructed a CCSC line integrated with a Tet-inducible lentiviral vector that can incrementally increase miR-34a expression levels (Figure 5H). Instead of gradually reducing Notch1 levels in all cells, incremental increase of miR-34a levels (confirmed by RT-qPCR) sharply switched off Notch1 expression in individual cells to maintain Notch1 bimodality (Figure 5J and Figure S6E). FACS with FISH probes further showed that the induced miR-34a levels do not need to be bimodal to cause Notch bimodality (Figure 5J and Figure S6F). Next, we generated CCSC lines that stably express Tet-inducible Numb (Figure 5I). In contrast to the effect of miR-34a overexpression, increasing Numb levels reduce Notch1 levels in all cells gradually and shift the entire Notch1 distribution in a continuously graded manner without creating two separate populations (Figure 5K and Figure S6G). Furthermore, after we knocked down miR-34a to remove potential crosstalk between Numb and miR-34a, Notch1 remained unimodal throughout the induction of Numb (Figure 5L and Figure S6H).
Collectively, these experiments support a model in which both Numb and miR-34a regulate Notch1, but miR-34a has an intrinsic ability to cause Notch1 bimodality. This role is consistent with our previous findings that miR-34a correlates with cell fate asymmetry more strongly than Numb.
miR-34a:Notch1 mRNA Mutual Sequestration Creates a Sharp Threshold Response
To understand how miR-34a generates Notch1 bimodality, we constructed a mathematical model of miR-34a regulation based on published measurements and equations of microRNA regulation (Osella et al., 2011; Vohradsky et al., 2010) (see Supplemental Information). This analysis revealed a potential mechanism for microRNAs to generate bimodality without feedback.
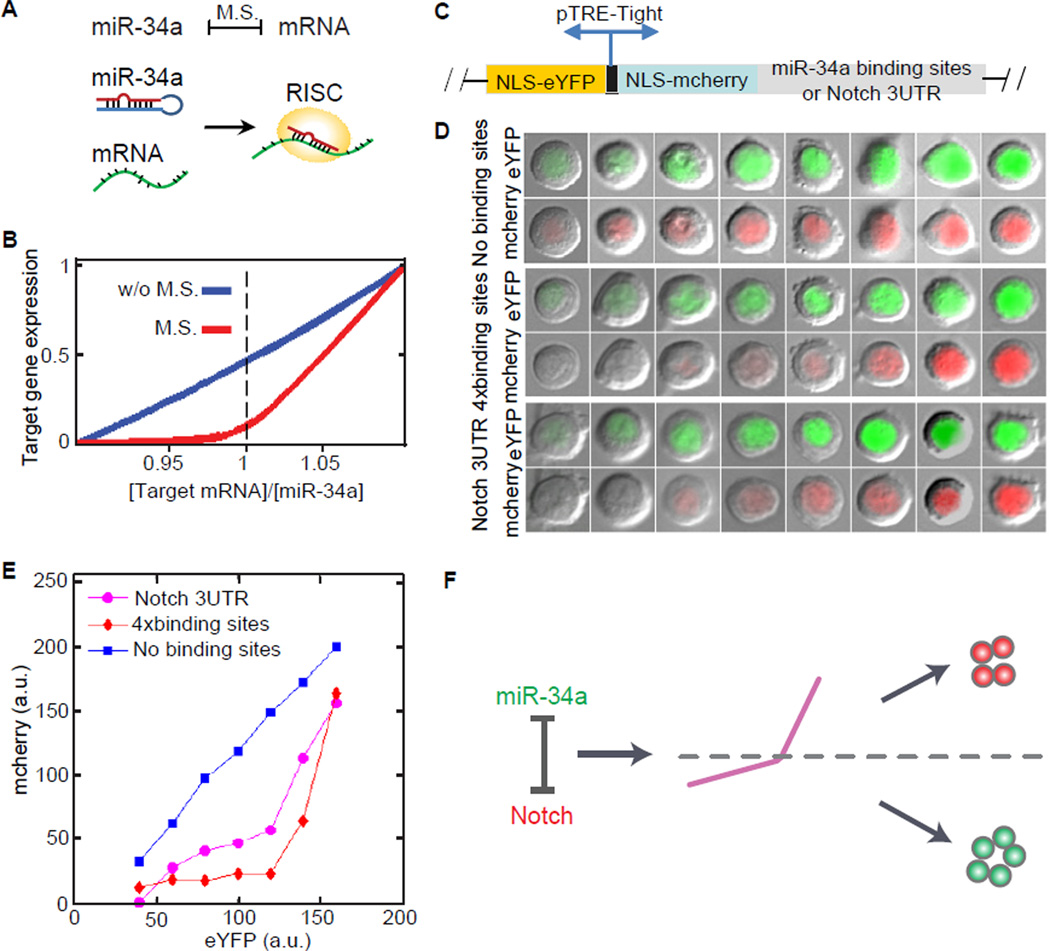
Incorporated in the RNA-induced silencing complex (RISC), a given microRNA and its target mRNA sequester each other when they bind together (Levine et al., 2007; Liu et al., 2005) (Figure 6A). The strength of this mutual sequestration is dependent on kinetic factors including binding and disassociation rates, degradation rates, the number and matching sequences of microRNA binding sites, and the recycling time of the microRNA. If the mutual sequestration is sufficiently strong, microRNAs will quickly turn off target genes when the activity level of microRNAs exceeds the target mRNA level (Figure 6B). This leads to a threshold response of target gene expression to the microRNA level, which has been demonstrated using synthetic constructs in Hela cells (Mukherji et al., 2011).
Figure 6. miR-34a generates Notch1 threshold response. Also see Figure S7.
(A) A cartoon illustration of the mutual sequestration between miR-34a and Notch1 mRNA. (B) Mutual sequestration leads to a sharp, threshold response in simulation. M.S., mutual sequestration. (C) Schematic of a two-color fluorescent reporter. The reporter contains a bidirectional Tet-inducible promoter driving the expression of nuclear localization sequences (NLS) tagged mCherry and enhanced yellow fluorescent protein (eYFP). Notch 3′UTR or 4 repeats of bugled miR-34a binding sequence were cloned into the 3′UTR of mCherry. (D) Representative images of single cells expressing YFP and mCherry. Their 2 color reporters contain Notch 3′UTR (bottom), miR-34a binding sequence (middle) or neither (top). The reporters containing Notch 3′UTR or miR-34a binding sites show a sharper turn-on with a threshold-like response. (E) Transfer function relating eYFP to mCherry generated by binning the imaged cells according to eYFP intensity and plotting the mean mCherry level in each bin (a.u: arbitrary units). (F) A schematic illustration of the model. Mutual sequestration generates a threshold response that separates bimodal populations.
Using a similar experimental approach as Mukherji et al, we tested specifically whether miR-34a generates a threshold response in Notch1 expression in CCSC1 and CCSC2 spheres. Briefly, we generated CCSC lines that stably express a two-color fluorescent reporter. This reporter contains a bidirectional Tet-inducible promoter driving the expression of two genes encoding the fluorescent proteins mCherry and enhanced yellow fluorescent protein (eYFP) (Figure 6C). The 3′UTR of mCherry contains either the Notch1 3′UTR sequence or 4 repeats of miR-34a binding sites. The eYFP fluorescence indicates baseline transcriptional activity and the mCherry fluorescence reflects the level of a miR-34a target gene (such as Notch1 or a miR-34a reporter). By normalizing the mCherry fluorescence with the eYFP fluorescence, the effect of miR-34a regulation in single cells can be compared.
After induction by Doxycycline, we measured the levels of eYFP and mCherry in individual cells using fluorescence microscopy. Without miR-34a binding sites in its 3′UTR, the expression level of mCherry was proportional to the level of eYFP expression. When the mCherry 3′UTR contained either the Notch1 3′UTR or tandem miR-34a binding sites, the mCherry level initially showed no significant increase compared to increasing eYFP levels until a threshold was reached. After this point, the mCherry level increased rapidly (Figure 6D and Figure S7A). This threshold behavior was quantitatively characterized by plotting the transfer function between the mCherry and eYFP levels (Mukherji et al., 2011) (Figure 6E). The transfer function confirmed the threshold response of miR-34a target genes, because the level of mCherry, which represents the target gene expression, does not significantly rise until a threshold level of eYFP is reached.
Stochastic simulations confirmed that the threshold response can generate the observed Notch1 bimodality from the measured miR-34a distribution (Figures S7B and S7C). Hence the data collectively support a model that mutual sequestration between miR-34a and Notch1 mRNA generates a sharp response with a threshold, which separates the bimodal Notch+/miR-34a− CCSC and Notch−/miR-34a+ non-CCSC subpopulations (Figure 6F).
A caveat is that microRNA regulation does not always generate such thresholds. As mentioned previously, the strength of mutual inhibition depends on multiple factors. Mukherji et al showed that the existence of a threshold depends on both the number and affinity of the microRNA binding sites in the target mRNA (Mukherji et al., 2011). For example, Mycn, another target of miR-34a (Choi et al., 2011), does not display a bimodal distribution (Figure S6I). Therefore, miR-34a may selectively target a subset of genes for bimodal outputs.
DISCUSSION
By analyzing the properties of cultured human CRC cells, we determined that the microRNA miR-34a acts as a bimodal switch to target Notch in early stage CCSCs. This switch controls the choice of daughter cells to self-renew or to differentiate during division. The ability of miR-34a to generate robust binary signals may contribute to its stronger correlation with cell fate markers during CCSC division than Numb. However, the extent of correlation between Numb and cell fate markers may be confounded by other factors. First, the phosphorylation state of Numb plays a major role in its localization during asymmetric cell division (Neumuller and Knoblich, 2009), so the level of active Numb rather than the total level of Numb may be a better indicator of its function. Second, our characterization of CCSC differentiation may be simplistic. Normal ISCs first differentiate into trans-amplifying (TA) progenitor cells, which then become more terminally differentiated cells. It is possible that there is a progenitor cell population in the CCSC spheres as well, although to date there is no identified marker that can distinguish this progenitor subgroup. Alternatively, CRC cells may have “lost” this intermediate state compared to normal ISCs. If distinct CRC TA/progenitors do exist, it is conceivable that these cells might have distinct Numb levels that affect the correlation between Numb and CCSC markers.
Our studies demonstrate that the ability to generate a threshold response in its target genes allows miR-34a to regulate Notch as a bimodal switch. This switch determines cell fate asymmetry in a robust and precise way during CCSC division. In addition, miR-34a likely targets multiple targets besides Notch to further enforce cell fate determination. Given that various microRNAs are expressed in different types of stem cells, it will not be surprising if some of those microRNAs also act as bimodal switches like miR-34a. Important parameters to generate these switches will include the number and sequence matching of the microRNA binding sites in target mRNAs. In electrical circuit design, switches are widely used because they can function in the presence of environmental and intrinsic noises. That cells and electrical circuits share a similar design principle even though regulatory networks and electronic devices are vastly different illustrates the fundamental importance of mechanisms that convert noisy signals into unambiguous signals for robust decision-making.
The miR-34a switch is necessary and sufficient for Notch bimodality. However, the bimodality of miR-34a and Numb levels may further contribute to Notch bimodality. In fact, their mutual correlation with Notch suggest that miR-34a and Numb are not independent regulators. Instead, they likely share common upstream regulators or crosstalk with each other to determine cell fate synergistically. Hence, the miR-34a switch is probably part of a complex mechanism to ensure robust cell fate decisions.
Pair-cell assays with early stage CCSC showed that (a) high miR-34a levels decreased both symmetric CCSC-CCSC division and asymmetric division, resulting in fewer CCSC daughter cells and more non-CCSC daughter cells and (b) low miR-34a levels increased symmetric CCSC-CCSC division but still decreased asymmetric division, resulting in more CCSC daughter cells and fewer non-CCSC daughter cells. Since CCSC daughter cells have higher proliferation rates than non-CCSC daughter cells, low miR-34a levels promote proliferation as well as self-renewal and symmetric division. Interestingly, asymmetric division in this system requires miR-34a levels to reside in a “sweet spot” in the middle: either too much or too little miR-34a abolishes asymmetric division. These data support a model where a single microRNA can regulate three distinct cell division outcome “states”: symmetric self-renewal, asymmetric division and non-self-renewal. Given this ability of microRNAs to enable highly precise and nuanced regulation of the relative proportions of different cell types in a population, we anticipate that future studies will show important roles for microRNAs to regulate tissue homeostasis and pattern formation for many normal stem cell systems that use microRNAs’ ability to “fine tune” the balance between asymmetric and symmetric stem cell division. Restoration of such roles for microRNAs in cancer cells may represent an important therapeutic strategy for future cancer treatment.
EXPERIMENTAL PROCEDURES
Isolation and Culture of CCSCs
CCSCs were isolated as described previously (Sikandar, 2010). For this study, CCSCs were derived from three early stage and two late stage CRC patient tumors (Table 1). Briefly, after washing with PBS, fresh human CRC tumors were dissociated using collagenase and were strained with a 40 µm filter. The tumor cells were initially sorted using anti-CD133 (clone C24B9, 1:50, Cell Signaling) and anti-CD44 (clone156-3C11, 1:100, Cell Signaling) antibodies and later switched to ALDH1 using the Aldeflour kit (Stem Cell Technologies). CCSCs were cultured as spheres in ultralow-attachment flasks (Corning) in DMEM/F12 (Invitrogen), supplemented with nonessential amino acids (Fisher), sodium pyruvate (Fisher), Penicillin-streptomycin (Fisher), N2 supplement (Invitrogen), B27 supplement (Invitrogen), 4 µg/mL heparin (Sigma), 40 ng/mL epidermal growth factor (Invitrogen), and 20 ng/mL basic fibroblast growth factor (Invitrogen) at 37 °C and 5% CO2. To propagate in vitro, spheres were collected by gentle centrifugation, dissociated into single cells, and then cultured to form next generation spheres.
Immunofluorescence
CCSCs were first plated on an uncoated glass culture slide (BD Biosciences). After fixed in cold methanol, the cells were blocked in 10% normal goat serum for 1 hour and then incubated with anti-ALDH1 (clone H-4, 1:100, Santa Cruz), anti-CD44 (clone156-3C11, 1:400, Cell Signaling), anti-CD133 (1:200, Abcam), anti-CK20 (clone H-70, 1:100, Santa Cruz), anti-CEA (1:200, Abcam), anti-Numb (clone C44B4, 1:100, Cell Signaling, or 1:100, Abcam) and anti-NICD (1:100, R&D Systems) antibody overnight at 4 °C. The cells were then incubated with Rhodamine Red or Alexa Fluor 488 labeled secondary antibody (Invitrogen) for 1 hour at room temperature. After counterstained with DAPI (Invitrogen), the slide was observed under a fluorescent microscope (Olympus).
RNA FISH
RNA FISH was performed as described by Jing Lu et al (Lu and Tsourkas, 2009). The procedures are described in detail in the Supplemental Experimental Procedures.
miR-34 Threshold Assay
The threshold assay was performed as described by Mukherji et al (Mukherji et al., 2011). The procedures are described in detail in the Supplemental Experimental Procedures.
Statistical Analysis of Xenograft Tumors
Data were expressed as mean ± standard deviation of no smaller than three biological repeats, and analyzed for statistical significance using the GraphPad Prism 5 software. Two-way ANOVA was used to compare the mean responses of different tumor sizes at different time points, followed by Bonferroni post-hoc test to determine statistical significance.
Supplementary Material
Highlights.
miR-34a regulates colon cancer stem cell asymmetric division.
miR-34a generates a sharp threshold response.
miR-34a converts Notch signaling into a toggle switch.
Binary Notch levels specify self-renewal vs. differentiation.
ACKNOWLEDGEMENTS
We thank Michael Elowitz, members of the Shen, Lipkin, Elowitz, and Jin laboratories for discussions and advice. We also thank Harley McAdams, Kenneth Kemphues, Tudorita Tumbar and Robert Weiss for their comments on the manuscript. This work was supported by NIGMS R01GM95990, NSF 1137269, DARPA 19-1091726, Cornell Nanobiotechnology Center, Cornell Stem Cell Program, NCI R21CA162483, NCI R21CA153049, and a generous gift from Matthew Bell.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information for this article includes seven figures, two movies, Supplemental Movie Legends, Supplemental Experimental Procedures and Supplemental References can be found with this article online.
REFERENCES
- Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer stem cells: in the line of fire. Cancer treatment reviews. 2012;38:589–598. doi: 10.1016/j.ctrv.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Arrowsmith J. Trial watch: Phase II failures: 2008–2010. Nature reviews Drug discovery. 2011a;10:328–329. doi: 10.1038/nrd3439. [DOI] [PubMed] [Google Scholar]
- Arrowsmith J. Trial watch: phase III and submission failures: 2007–2010. Nature reviews Drug discovery. 2011b;10:87. doi: 10.1038/nrd3375. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nature cell biology. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa EMF, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell stem cell. 2011;9:476–485. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Dey-Guha I, Wolfer A, Yeh AC, Albeck JG, Darp R, Leon E, Wulfkuhle J, Petricoin EF, 3rd, Wittner BS, Ramaswamy S. Asymmetric cancer cell division regulated by AKT; Proceedings of the National Academy of Sciences of the United States of America; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmink BL, Van Houdt WJ, Vries RG, Hoogwater FJ, Govaert KM, Verheem A, Nijkamp MW, Steller EJ, Jimenez CR, Clevers H, et al. Differentiated human colorectal cancer cells protect tumor-initiating cells from irinotecan. Gastroenterology. 2011;141:269–278. doi: 10.1053/j.gastro.2011.03.052. [DOI] [PubMed] [Google Scholar]
- Goulas S, Conder R, Knoblich JA. The par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell stem cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Clevers H. Wnt/beta-catenin and MAPK signaling: allies and enemies in different battlefields. Sci Signal. 2012;5:e15. doi: 10.1126/scisignal.2002921. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas H, Palacios J, Moreno-Bueno G. Molecular profiling of docetaxel cytotoxicity in breast cancer cells: uncoupling of aberrant mitosis and apoptosis. Oncogene. 2007;26:2902–2913. doi: 10.1038/sj.onc.1210102. [DOI] [PubMed] [Google Scholar]
- Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer research. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Blat IC, Jacks T, Clevers H, van Oudenaarden A. Optimality in the development of intestinal crypts. Cell. 2012;148:608–619. doi: 10.1016/j.cell.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell stem cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, Sikandar SS, Gumus ZH, Lipkin SM. miR-23a Promotes the Transition from Indolent to Invasive Colorectal Cancer. Cancer discovery. 2012 doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HY, Hwang do W, Lee DS, Kim S. A reporter gene imaging system for monitoring microRNA biogenesis. Nature protocols. 2009;4:1663–1669. doi: 10.1038/nprot.2009.119. [DOI] [PubMed] [Google Scholar]
- LaPointe LC, Dunne R, Brown GS, Worthley DL, Molloy PL, Wattchow D, Young GP. Map of differential transcript expression in the normal human large intestine. Physiol Genomics. 2008;33:50–64. doi: 10.1152/physiolgenomics.00185.2006. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E, McHale P, Levine H. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput Biol. 2007;3:e233. doi: 10.1371/journal.pcbi.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Madison BB, Zacharias W, Kolterud A, States D, Gumucio DL. Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol Genomics. 2007;29:290–301. doi: 10.1152/physiolgenomics.00269.2006. [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer research. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature cell biology. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, Van’t Veer L, Tollenaar RA, Jackson DB, Agrawal D, et al. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tsourkas A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011 doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, Tsatsanis A, Gallinger S, Dick JE. ID1 and ID3 Regulate the Self-Renewal Capacity of Human Colon Cancer-Initiating Cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Osella M, Bosia C, Cora D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1001101. e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, Ridgway RA, Sansom OJ, Nathke IS. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell stem cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22:187–193. doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–R138. [PubMed] [Google Scholar]
- Sikandar S, Dizon D, Shen X, Li Z, Besterman J, Lipkin SM. The class I HDAC inhibitor MGCD0103 induces cell cycle arrest and apoptosis in colon cancer initiating cells by upregulating Dickkopf-1 and non-canonical Wnt signaling. Oncotarget. 2010;1:596–605. doi: 10.18632/oncotarget.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikandar S, Edwards R, Lyles K, Waterman M, Lipkin SM. NOTCH signaling is required for colon cancer initiating cell tumor formation, self-renewal and repression of secretory cell lineage differentiation. Cancer research. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo MM. Reflections on the spread of metastasis to cancer prevention. Cancer Prev Res (Phila) 2011;4:324–328. doi: 10.1158/1940-6207.CAPR-11-0046. [DOI] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell stem cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/[gamma]-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vohradsky J, Panek J, Vomastek T. Numerical modelling of microRNA-mediated mRNA decay identifies novel mechanism of microRNA controlled mRNA downregulation. Nucleic Acids Res. 2010;38:4579–4585. doi: 10.1093/nar/gkq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell stem cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbel MA, McIntire MG, Dwyer P, Fiebiger E. CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PLoS ONE. 2011;6:e16442. doi: 10.1371/journal.pone.0016442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn BS, Kim YJ, Mantel C, Yu KY, Broxmeyer HE. Blocking of c-FLIP(L)--independent cycloheximide-induced apoptosis or Fas-mediated apoptosis by the CC chemokine receptor 9/TECK interaction. Blood. 2001;98:925–933. doi: 10.1182/blood.v98.4.925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.