Abstract
Of the four native nucleosides, 2′-deoxyguanosine (dGuo) is most easily oxidized. Two lesions derived from dGuo are 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy)·dGuo. Furthermore, while steady-state levels of 8-oxodGuo can be detected in genomic DNA, it is also known that 8-oxodGuo is more easily oxidized than dGuo. Thus, 8-oxodGuo is susceptible to further oxidation to form several hyperoxidized dGuo products. This review addresses the structural impact, the mutagenic and genotoxic potential, and biological implications of oxidatively damaged DNA, in particular 8-oxodGuo, Fapy·dGuo, and the hyperoxidized dGuo products.
Keywords: 8-oxo-7,8-dihydro-2′-deoxyguanosine; hyperoxidized dG lesions; guanine quadruplexes; trinucleotide repeat expansion; oxidatively damaged DNA as regulator of gene expression
Introduction
Cellular DNA is subjected to a multitude of damaging agents. In particular, oxidation of DNA can occur following exposure to a wide variety of endogenous and exogenous species. Endogenous oxidizing agents are produced during normal cellular processes such as respiration and as part of an inflammatory response [1, 2]. These oxidizing agents include reactive oxygen species (ROS) such as superoxide (O2•–), hydrogen peroxide (H2O2, singlet oxygen (1O2), hydroxyl radical (•OH), and peroxynitrite (ONOO–). Under physiological conditions, ONOO– reacts with CO2 to form nitrosoperoxycarbonate (ONOOCO2–) which homolyzes to form •NO2 and CO3•–, and the latter oxidizes the guanine (Gua) nucleobase [3]. 1O2 is also able to oxidize Gua whereas •OH reacts with all four nucleobases and also the 2′-deoxyribose sugar ring [4]. O2•– and H2O2 do not react with DNA directly [4]. However, O2•– can react with the neutral Gua radical that arises from one electron oxidation and deprotonation of G [5]. Furthermore, in the presence of Fe2+ or Cu2+, H2O2 can be converted to •OH via the Fenton reaction. Exogenous sources of DNA damaging agents include ionizing radiation [6-9], ultraviolet light [10], and chemotherapeutics such as bleomycin [6].
Types of oxidatively damaged DNA
Both the sugar-phosphate backbone and the nucleobases of DNA represent targets for oxidizing agents. This review will focus on chemistry occurring at nucleobases, in particular at Gua, and will examine the influences of the oxidatively damaged nucleobases at the molecular, genetic, and cellular levels. As a note on nomenclature, we will follow recent recommendations [11] in which the guanine nucleobase is referred to as Gua, the 2′-deoxnucleoside as dGuo, and the guanine nucleobase in DNA is referred to with the single letter G. We will use the same system when referring to oxidatively damaged nucleobases, oxidatively damaged 2′-deoxnucleosides, and oxidatively damaged nucleobases in DNA.
Of the four 2′-deoxynucleosides, dGuo has the lowest reduction potential (E0 = 1.3, 1.4, 1.6, and 1.7 V versus NHE for dGuo, dAdo, dCyt, and dThy, respectively) and is most susceptible to oxidation [12]. It has been shown that solution-borne ROS show nonspecific reaction at all guanines in a sequence [13]. In contrast, selective oxidation of the 5′-G of 5′-GG-3′ guanine doublets or 5′-GGG-3′ guanine triplets is observed in DNA-mediated charge transfer chemistry, in which the pi orbitals of the DNA nucleobases mediate charge transfer between a donor and an acceptor [14]. Ab initio molecular orbital calculations predict that in a guanine doublet or triplet, the bulk of the HOMO lies on the 5′-G, which has a lower reduction potential than a single G [15, 16].
8-oxo-7,8-dihydroguanine (8-oxoG)
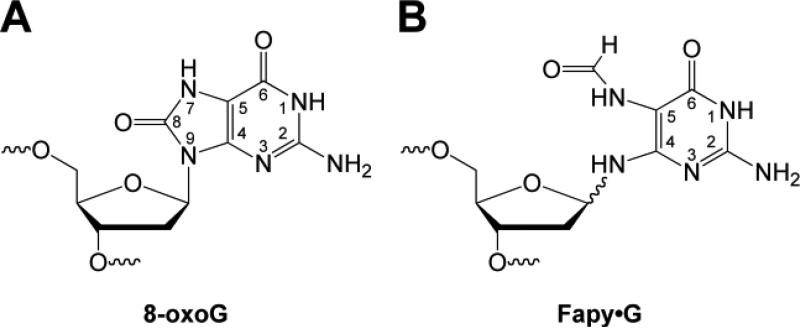
A prototypic oxidized form of G found in DNA is 8-oxoG (Figure 1A). This lesion serves as a biomarker of oxidative stress since it is formed by reaction of G with a variety of oxidants including, but not limited to, •OH, 1O2, and CO3•– [17]. Older methods for determining the amount of 8-oxoG in DNA generated artifacts during sample work up, but newer methodology makes it possible to use this biomarker for tissue and urine analysis [18, 19]. Due to the concerted efforts of laboratories involved in the European Standard Committee on Oxidative DNA Damage (ESCODD), recommended protocols for the extraction of DNA from cells and tissues now exist, along with methodologies for the digestion of the DNA, and analysis of the digestion products [20-24]. Using these protocols, which include the addition of antioxidants, metal chelators, and/or free radical trapping agents during sample preparation, it is now estimated that 8-oxoG is present at steady-state levels of 0.3-4.2 per 106 G and those levels are elevated under conditions of oxidative stress [24]. For more details regarding the quantitation of 8-oxoG in genomic DNA the reader is referred to another review in this special issue on oxidatively damaged DNA [25].
Figure 1.
Structure of (A) 8-oxoG and (B) Fapy·G.
2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy)·G
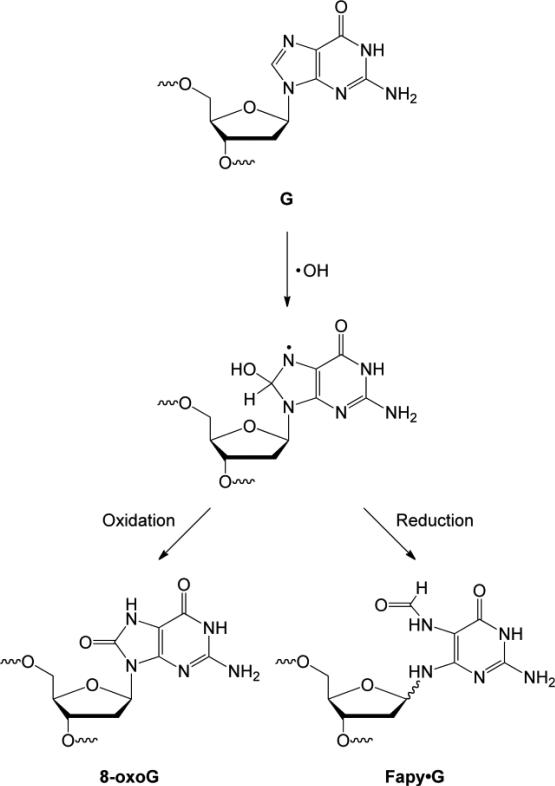
Fapy·G (Figure 1B) is believed to form from the C-8 hydroxyl radical adduct of G [26]. Notably, the hydroxyl radical adduct can be converted to 8-oxoG or Fapy·G, depending on reaction conditions. Formation of 8-oxoG occurs via an oxidative pathway whereas Fapy·G is generated by a reductive pathway (Figure 2). When chromatin is exposed to γ-radiolysis in the presence of oxygen, 8-oxoG is formed preferentially whereas under anoxic conditions Fapy·G is the major product [27]. 8-oxoG and Fapy·G are formed in comparable amounts when DNA is irradiated with ultraviolet light [28]. Fapy·G is found in ~3-fold higher amount than 8-oxoG in human leukemia cells exposed to ionizing radiation [9]. While a greater number of studies have focused on 8-oxoG, the development of methods to chemically synthesize oligonucleotides containing Fapy·G [29-33] has significantly facilitated research on this DNA lesion and the results obtained suggest that Fapy·G may also serve as a biomarker of oxidative stress.
Figure 2.
Formation of 8-oxoG and Fapy·G from a common precursor.
Hyperoxidized guanine products
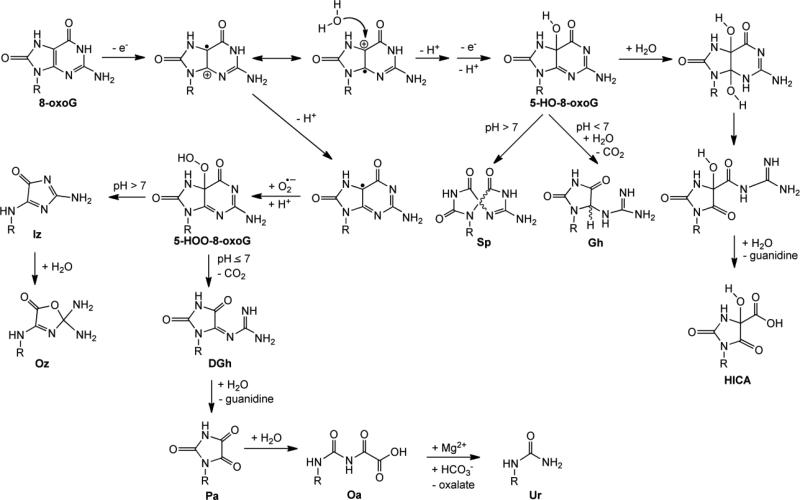
8-oxodGuo has a reduction potential that is significantly lower than dGuo, 0.7 V versus NHE [34, 35]. Thus, 8-oxodG is susceptible to further oxidation and this secondary oxidation event gives rise to the hyperoxidized dG products [2, 36]. Indeed, the oxidation of 8-oxoG yields intermediates that decompose to form several hyperoxidized products as shown in Figure 3. The first step is a one-electron oxidation to form the 8-oxoG radical cation. Subsequent addition of water and deprotonation, and a second one-electron oxidation and deprotonation results in the formation of 5-HO-8-oxoG. At pH > 7, diastereomers of the hyperoxidized product spiroiminodihydantoin (Sp) are formed from 5-HO-8-oxoG by a 1,2 shift [37]. At pH < 7, hydrolysis and decarboxylation of 5-HO-8-oxoG occur to form the diastereomers of the hyperoxidized product guanidinohydantoin (Gh) [37, 38]. The pH-dependence of the formation of Sp and Gh has also been examined using computational methods [39]. Furthermore, once formed, Sp can be converted to Gh in acidic conditions [40]. Addition of H2O to 5-HO-8-oxoG can lead to formation of 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid (HICA) [41]. Notably, O2•– can add to the neutral 8-oxoG radical to form 5-HOO-8-oxoG, which at pH > 7 is converted to imidazolone (Iz) and its hydrolysis product oxazolone (Oz) [42]. Alternatively, at pH ≤ 7, 5-HOO-8-oxoG is decarboxylated to form dehydroguanidinohydantoin (DGh) [43]. DGh can undergo hydrolysis to generate parabanic acid (Pa), and addition of H2O yields oxaluric acid (Oa). Finally, in the presence of physiologically relevant concentrations of Mg2+ and bicarbonate, Oa undergoes facile hydrolysis to urea (Ur) [44].
Figure 3.
Summary of the 8-oxoG-derived hyperoxidized G products.
Interestingly, the formation of hyperoxidized G products is dependent on the dose rate of oxidant delivered to the system. For example, at a low dose rate of ONOO–, Sp, Gh, and HICA are formed, while at a high dost rate Ca, Iz, and Oa are generated [37]. These two classes of 8-oxoG-derived products, seen with high and low dose rates of ONOO–, can be rationalized mechanistically [2]. At high dose rates, ONOO– itself acts as a nucleophile after the initial oxidation of 8-oxoG whereas at lower dose rates water can also compete and act as a nucleophile.
Of the hyperoxidized G products, Sp and Gh have garnered particular attention in the literature because synthetic methods to generate these hyperoxidized G products in olignonucleotides have been developed [45]. Furthermore, while Sp, Gh, Iz, Oz, DGh, Pa, Oa, Ur and HICA have been identified in vitro using nucleoside and/or oligonucleotide model systems, Sp has been detected in vivo. Using LC-MS, Hailer and co-workers observed that Sp was generated in E. coli treated with K2CrO4 [46].
Structural and thermodynamic properties of oxidatively damaged DNA
Structural impact of 8-oxoG in DNA oligonucleotides
Studies of the structural impact of 8-oxoG were greatly facilitated by the development of synthetic methods for oligonucleotides containing the lesion [47]. Several structural studies of duplexes containing 8-oxoG have examined the effect of the lesion on both the local and global duplex organization. Circular dichroism (CD) spectropolarimetry was used to examine the impact on global DNA structure of 8-oxoG [48]. In these experiments, the 8-oxoG was base paired to C, G, A, or T within a 13-mer duplex. Regardless of the nucleobase to which it was paired, 8-oxoG was judged not to change the global structure of the DNA; all four 8-oxoG-containing duplexes exhibited spectral shapes characteristic of B-form DNA.
While CD can be informative regarding changes to DNA structure at the global level, methods such as NMR and X-ray crystallography can provide details regarding structural changes that occur not only globally, but also locally. It is of note that many experimental systems for NMR and crystallography use self-complementary oligonucleotides in order to facilitate data analysis. In such cases the final duplex product contains two 8-oxoG:X base pairs where X can be C, G, A, or T.
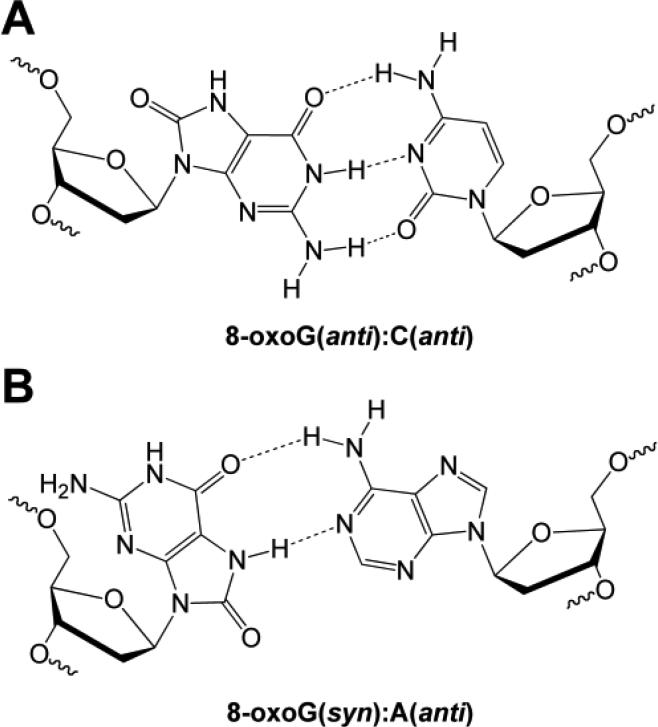
Consistent with the results obtained by CD, using a self-complementary 12-mer, NMR studies showed little change in the global duplex structure for an 8-oxoG:C base pair relative to the corresponding unmodified duplex with a G:C base pair [49]. The base stacking, base pairing, and helical twist are not perturbed. However, small perturbations in the local structure surrounding the lesion were revealed by comparison of the imino proton chemical shifts for the modified and unmodified duplexes. Imino proton resonances also indicate that 8-oxoG is in the keto form and, furthermore, a lack of nuclear Overhauser effect (NOE) connectivity to other protons revealed that the N7H of 8-oxoG is not involved in base pairing [49]. Indeed, NOE connectivity indicates that 8-oxoG is paired to C in a canonical Watson-Crick base pair with both bases in the anti conformation (Figure 4A). Furthermore, 31P-NMR spectroscopy revealed that the phosphorus resonances from the sugar-phosphate backbone of the 8-oxoG-containing duplex were comparable to those of undamaged DNA [49]. An X-ray structure of a self-complementary 10-mer confirmed that both nucleosides in an 8-oxoG:C base pair are in the anti conformation [50]. While confirming the global structure is not altered relative to a duplex containing a G:C base pair, the crystal structure did reveal minor perturbations, namely that the plane of the base pair is slightly offset compared to the unmodified duplex.
Figure 4.
Structure of (A) 8-oxoG(anti):C(anti) and (B) 8-oxoG(syn):A(anti) base pair.
More recent NMR studies have again confirmed the lack of change to the global structure for a duplex containing an 8-oxoG:C base pair, but by monitoring line broadening of the exchangeable protons as a function of temperature, showed that the 8-oxoG:C base pair has increased ability to exchange with solvent, and thus opens to solution more readily than its unmodified counterpart [51]. Additionally, the base pair located 5′ to 8-oxoG also showed increased exchange. However, this technique represents an underestimation of the rate of base pair opening, as it does not account for the fact that a base pair may open several times without exchanging a proton with the solvent. In different work, when a glycine catalyst was added to increase the rate of imino proton exchange with solvent, the apparent equilibrium constant for base pair opening of the 8-oxoG:C pair was the same as a G:C base pair [52]. Thus, the overall time that an 8-oxoG:C base pair spends open to solution is the same as a G:C base pair. However, it should be noted that these experiments do not separately account for the rate of base pair opening and closing. Since the apparent equilibrium constant is equal to the ratio of the opening and closing rates, it is possible that the rate of 8-oxoG:C base pair opening and the rate of closing are both faster compared to G:C, or that both rates are slower relative to G:C. Both of these scenarios could have implications for detection of 8-oxodG within the genome by DNA repair proteins.
In addition to the 8-oxoG:C base pair, NMR and X-ray crystallography have been used to investigate the effect on duplex structure of an 8-oxoG:A base pair. DNA polymerase is known to perform translesion synthesis past 8-oxoG and insert C or A [53]. Thus, the 8-oxoG:A base pair is the result of a misincorporation event by polymerase. The first structural investigation employed NMR to elucidate the impact of the 8-oxoG:A base pair in a non self-complementary 12-mer duplex [54]. Based on NOE connectivities, it was determined that 8-oxoG rotates about its glycosidic linkage and adopts the syn conformation, which allows for Hoogsteen base pairing with A; in this base pair the A remains in the anti conformation (Figure 4B). Surprisingly, this non-canonical base pairing had little effect on the global structure of the duplex. Further investigation of the 8-oxoG:A base pair by X-ray crystallography using a self-complementary 12-mer confirmed the minimal structural distortion imposed by the 8-oxoG:A base pair, indicating that much like the 8-oxoG:C base pair, the overall B-form duplex is maintained [55].
8-oxoG alters both the thermal and thermodynamic stability of DNA duplexes
The melting temperature (Tm) of a duplex provides a measure of the thermal stability of the DNA. Two techniques that are commonly used to obtain the Tm of a duplex are optical spectroscopy and differential scanning calorimetry (DSC). In optical spectroscopy the absorbance of the DNA is measured as a function of temperature. In DSC, changes in heat capacity are measured as a function of temperature. Regardless of the technique used to determine thermal stability, the presence of an 8-oxoG:C base pair decreases the Tm relative to a control duplex which contains an unmodified G:C base pair; these results indicate that an 8-oxoG:C base pair is thermally destabilizing to a duplex [48, 49, 51, 55-57]. As seen in Table I, the sequence context of 8-oxoG, size of the duplex, and location of the damaged nucleobase within the duplex can all affect the magnitude of the thermal destabilization. Nevertheless, while the extent to which an 8-oxoG:C base pair is thermally destabilizing can range from small to quite large, in all cases the presence of an 8-oxoG:C base pair in a duplex causes a decrease in the Tm. Indeed, this thermal destabilization caused by 8-oxoG was recently exploited in nanopore ion channel recordings in order to detect the oxidatively damaged nucleobase in DNA [58].
Table I.
Thermal and thermodynamic impact of 8-oxoG in DNA.
| DNAa | Base Pairing | Tm (°C) | ΔTm (°C) | ΔH (kcal/mol) | ΔG (kcal/mol) | Reference |
|---|---|---|---|---|---|---|
| 5'-CGCGAATTCGCG-3' | G:C | 47 | - | - | [49] | |
| 5'-CGCGoxAATTCGCG-3' | 8-oxoG:C | 44 | 3 | - | - | [49] |
| 5'-CGCCAATTGGCG-3' | G:C | 53.9 | - | - | [55] | |
| 5'-CGCCAATTTGoxCG-3' | 8-oxoG:C | 40.5 | 13.4 | - | - | [55] |
| 5'-CGCAAATTTGCG-3' | G:A | 19.3 | - | - | [55] | |
| 5'-CGCAAATTTGoxCG-3' | 8-oxoG:A | 48.4 | –29.1 | - | - | [55] |
| 5'-GACCGGGTCAGTGCTACT-3' | G:C | 65.2 ± 0.3 | - | –16.6 ± 1.1 | [56] | |
| 5'-GACCGoxGGTCAGTGCTACT-3' | 8-oxoG:C | 64.0 ± 0.7 | 1.2 ± 0.8 | - | –15.6 ± 0.6 | [56] |
| 5'-GACCGGGTCAGTGCTACT-3' | G:A | 58.0 ± 0.8 | - | –13.8 ± 0.7 | [56] | |
| 5'-GACCGoxGGTCAGTGCTACT-3' | 8-oxoG:A | 61.1 ± 0.3 | –3.1 ± 0.9 | - | –14.3 ± 0.4 | [56] |
| 5'-GATTGAATCAGTGCTACT-3' | G:C | 54.8 ± 0.4 | - | –12.8 ± 0.3 | [56] | |
| 5'-GATTGoxAATCAGTGCTACT-3' | 8-oxoG:C | 53.3 ± 0.2 | 1.5 ± 0.4 | - | –11.9 ± 0.3 | [56] |
| 5'-GATTGAATCAGTGCTACT-3' | G:A | 46.1 ± 0.5 | - | –9.6 ± 0.1 | [56] | |
| 5'-GATTGoxAATCAGTGCTACT-3' | 8-oxoG:A | 49.1 ± 0.5 | –3.0 ± 0.7 | - | –10.8 ± 0.2 | [56] |
| 5'-GACCGGGTCAGTGCTACT-3' | G:C | 75.8 ± 0.2 | 96.9 ± 1.9 | 19.3 ± 0.3 | [48] | |
| 5'-GACCGoxGGTCAGTGCTACT-3' | 8-oxoG:C | 73.7 ± 0.2 | 2.1 ± 0.3 | 89.6 ± 1.8 | 17.3 ± 0.4 | [48] |
| 5'-GACCGGGTCAGTGCTACT-3' | G:A | 64.5 ± 0.2 | 71.6 ± 2.9 | 12.5 ± 0.4 | [48] | |
| 5'-GACCGoxGGTCAGTGCTACT-3' | 8-oxoG:A | 70.7 ± 0.2 | –6.2 ± 0.3 | 88.1 ± 1.8 | 15.9 ± 0.3 | [48] |
| 5'-ACTGATAGACGCACT-3' | G:C | - | 107 ± 2 | 15.4 ± 0.3 | [57] | |
| 5'-ACTGATAGoxACGCACT-3' | 8-oxoG:C | - | - | 84 ± 2 | 13.0 ± 0.3 | [57] |
| 5'-GCGAATTCGC-3' | G:C | 53.0 | 72.3 ± 3 | 8.2 ± 0.2 | [51] | |
| 5'-GCGAATTCGoxC-3' | 8-oxoG:C | 39.2 | 13.8 | 33.2 ± 3 | 3.0 ± 0.2 | [51] |
| 5'-GAGAGCGCTCTC-3' | G:C | 59.5 | 92.0 ± 3 | 12.5 ± 0.2 | [51] | |
| 5'-GAGAGoxCGCTCTC-3' | 8-oxoG:C | 51.2 | 8.3 | 39.5 ± 3 | 4.6 ± 0.2 | [51] |
For clarity, the sequence of only one strand of the duplex is provided. The pairing partner of the underlined base is indicated in the column labeled ‘Base Pairing’. Gox represents 8-oxoG.
Interestingly, when 8-oxoG is base paired with A, the Tm is higher relative to the unmodified parent duplex containing a G:A base pair [48, 55]. This increase in thermal stability is likely due to the presence of hydrogen bonding and favorable base stacking in the 8-oxoG:A base pair, compared to the G:A base pair. However, when the Tm of duplexes containing an 8-oxoG:A base pair is compared to that of duplexes containing a canonical G:C base pair, the effect of 8-oxoG on thermal stability becomes apparent; the Tm of the modified duplex is lower than the unmodified parent duplex [48, 55].
These data illustrate the importance of not relying on melting temperature data to deduce the structural impact of a DNA lesion. Indeed, while 8-oxoG:C and 8-oxoG:A base pairs do decrease thermal stability relative to G:C, albeit modest in some cases, neither base pair causes a change in the global DNA structure.
While it is common practice in the literature to interpret Tm data in terms of free energy changes, thermal melting temperature is not a thermodynamic parameter. To determine the thermodynamic impact of a DNA lesion, the enthalpy of the melting transition must be obtained. The enthalpy can be extracted from data obtained by optical spectroscopy using van't Hoff analysis or obtained directly from heat capacity measurements [59]. Using a variety of duplexes of differing lengths, and in which the 8-oxoG is in different sequence contexts and in different positions in the duplex, it has been demonstrated that an 8-oxoG:C base pair is thermodynamically destabilizing (Table I) [48, 51, 56, 57]. Interestingly, while an 8-oxoG:C base pair provides an enthalpic destabilization, it also provides an entropic stabilization. This phenomenon, called ‘enthalpy-entropy compensation’, is often observed for melting transitions of duplexes containing a DNA lesion. As a result of this compensation, favorable entropic effects offset some of the free energy penalty for the enthalpic destabilization. Nevertheless, duplexes containing an 8-oxoG:C pair are thermodynamically destabilized relative to the unmodified duplex; this is reflected in Table 1 by the more positive values for ΔG of the damage-containing duplexes.
Similar to the results obtained in studies of thermal stability, duplexes containing an 8-oxoG:A base pair are thermodynamically more stable than the corresponding duplex containing a G:A base pair [48, 56]. This result is reflected in the values for ΔG that are more negative than duplexes containing a G:A base pair. However, the 8-oxoG:A base pair is thermodynamically less stable than the duplex with a canonical G:C base pair.
Structural, thermal, and thermodynamic impact of Fapy·G in DNA oligonucleotides
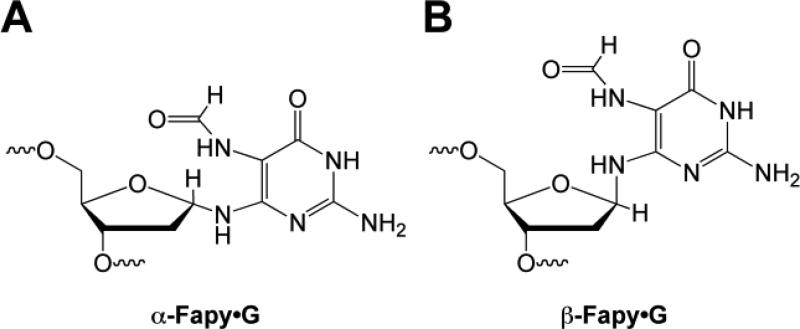
The ability to incorporate Fapy·G into oligonucleotides enabled studies of the lesion within the context of DNA [29-33]. An important consideration with respect to Fapy·G is the availability of a lone pair on the glycosidic nitrogen. This lone pair facilitates epimerization at the formamidopyrimidine anomeric center with the α and β anomers shown in Figure 5. In order to probe the distribution of anomers in duplex DNA, the selective cleavage of α-nucleotides by endonuclease IV (endo IV) was exploited [60]. Endo IV incised less than 5% of Fapy·G DNA, suggesting that the lesion exists predominately as the β-anomer in DNA.
Figure 5.
Structure of (A) α-Fapy·G and (B) β-Fapy·G.
Recently developed synthetic and separation strategies have allowed for the preparation of oligonucleotides containing individual Fapy·G anomers [33]. Duplexes containing the individual anomers exhibited distinct thermal and thermodynamic properties. The α-Fapy·G: C base pair was considerably more thermally destabilizing to the duplex, causing a ~15 degree decrease in the Tm relative to the corresponding duplex containing a G:C base pair; the β-Fapy·G:C base pair decreased the Tm by 6 degrees [33]. Using van't Hoff analysis the ΔΔG for the α-Fapy·G:C and β-Fapy·G:C base pairs was –5.3 and –1.5 kcal/mol, respectively, as compared to the G:C containing duplex. In previous work a destabilization of –3.3 kcal/mol was observed for the Fapy·G:C base pair in DNA [61], which was likely > 95% β-anomer based on incision by endo IV[60]. The differences observed for destabilization of duplexes by the β-Fapy·G:C base pair may be due to sequence context of the base pair and/or length of the duplex.
As observed for 8-oxoG, the Tm of Fapy·G base paired with A is higher relative to the unmodified parent duplex containing a G:A base pair [61]. A similar result was observed when Fapy·G was paired with G. These results suggest that the Fapy·G lesion is more tolerant of mispairing than G. However, when the Tm of duplexes containing a Fapy·G:A or Fapy·G:G base pair is compared to that of duplexes containing a canonical G:C base pair, the effect of Fapy·G on thermal stability is apparent; the Tm of the modified duplex is lower than the unmodified parent duplex.
Structural, thermal, and thermodynamic impact of hyperoxidized guanine products
Hyperoxidized guanine products are structurally diverse with many being ring-opened, bulky, and/or non-planar structures. As a result, one might predict that these oxidatively damaged nucleobases would significantly perturb the structure and stability of a duplex. While many of the hyperoxidized guanine products remain uncharacterized with respect to their impact on a duplex, there are several reports which address the impact of Sp. While there are some differences in the CD spectra for a duplex containing the Sp lesion as compared to 8-oxoG or G, the spectral shapes for all three duplexes are consistent with B-form DNA [57]. Using computational methods, the two diastereomers of Sp were shown to favor positioning in the major groove, with both anti and syn conformations possible depending on the base pairing partner [62]. Both diastereomers adversely impacted base pair stacking and hydrogen bonding and resulted in a widening of the grooves. Studies using NMR revealed that strand slippage can occur in DNA primer/template models and that Sp is in a single base bulge instead of forming a base pair [63]. Similar primer slippage has also been proposed to occur during primer extension of templates containing hyperoxidized G lesions [56].
Melting temperature data reveals that Sp, Gh, and Ca dramatically decrease the thermal stability of duplexes and primer/template pairs containing these lesions and can lower the Tm of a duplex by up to 20 degrees [56, 57, 64]. The presence of Sp or Gh also thermodynamically destabilizes a duplex regardless of the base pairing partner and sequence context, with Sp being more severe [56, 57].
Mutational and genotoxic properties of oxidatively damaged DNA
Two important parameters relating to the genetic effects of oxidatively damaged DNA are the mutagenic and genotoxic potentials of the lesion. If a lesion is a block to replication by DNA polymerases, with both partial and complete blockage of a polymerase possible, the lesion is referred to as toxic. If replication occurs but the incorrect base is incorporated by polymerase opposite the lesion, the lesion is considered mutagenic. Since 8-oxoG and the hyperoxidized G lesions are derived from G, the ‘correct’ incorporation is C; if A, G, or T are incorporated, the lesion would code for a mutation.
Origin of the mutational properties of 8-oxoG
When in the anti conformation, 8-oxoG is replicated by DNA polymerases as if it was G, and C is paired with the lesion. This replication event would be non-mutagenic. However, when in the anti conformation there is electronic repulsion between the C8 carbonyl oxygen of 8-oxoG and the oxygen of the 2′-deoxyribose ring (Figure 4A). Rotation about the glycosidic bond to adopt the syn conformation can relieve this repulsion. When in the syn conformation the Hoogsteen face of 8-oxoG is available for base pairing and DNA polymerases incorporate A opposite the oxidized nucleobase (Figure 4B). Following a subsequent round of replication this misincorporation event results in a G → T transversion, which is the characteristic mutation caused by 8-oxoG.
Mutational and genotoxic properties of 8-oxoG in vitro
The incorporation of dNTPs opposite 8-oxoG by several classical DNA polymerases has been examined [65, 66]. Steady-state kinetic data for the E. coli Klenow fragment of polymerase I exo– (KF–), polymerase II exo– (Pol II–), and bacteriophage T7 exo– (T7–) are provided in Table II. Insertion efficiencies, which provide a measure of the specificity for competing substrates, apparent misinsertion frequencies (f), and the corresponding percent of misinsertion are shown. KF–, Pol II–, and T7– all show a preference for incorporating dCTP over dATP opposite 8-oxoG. Misinsertion frequencies range from 0.3-1 which correspond to 23-50% misinsertion of dATP opposite 8-oxoG. Furthermore, the proofreading ability of Pol I and KF did not recognize 8-oxoG:A or 8-oxoG:C base pairs as substrates [53]. Interestingly, with HIV-1 reverse transcriptase (RT) the preference for incorporation is reversed and dATP incorporation is favored with a misinsertion frequency of 14; this misinsertion frequency corresponds to 93% misinsertion of dATP opposite 8-oxoG [66].
Table II.
Steady-state kinetic parameters for the incorporation of dNTPs opposite 8-oxoG.
| dNTP: template base | Insertion Efficiency kcat/Km (μM-1sec-1)a | fb | Percent Misinsertionc | Reference | |
|---|---|---|---|---|---|
| KF– | dCTP:G | 0.340 | [65] | ||
| dCTP:8-oxoG | 0.012 | [65] | |||
| dATP:8-oxoG | 0.003 | 0.3 | 23 | [65] | |
| Pol II– | dCTP:G | 0.110 | [65] | ||
| dCTP:8-oxoG | 0.0004 | [65] | |||
| dATP:8-oxoG | 0.0004 | 1 | 50 | [65] | |
| T7– | dCTP:G | 0.050 | [66] | ||
| dCTP:8-oxoG | 0.00035 | [66] | |||
| dATP:8-oxoG | 0.00020 | 0.6 | 37 | [66] | |
| HIV-1 RT | dCTP:G | 0.330 | [66] | ||
| dCTP:8-oxoG | 0.00003 | [66] | |||
| dATP:8-oxoG | 0.00040 | 14 | 93 | [66] |
In vitro experiments using eukaryotic DNA polymerases have shown that dNTP incorporation opposite of 8-oxoG is polymerase dependent. The ratio of incorporation of dCTP to dATP opposite 8-oxoG was 1:200 and 1:5 for the replicative DNA polymerases α (human) and δ (calf thymus), respectively [53]. Indeed, these replicative polymerases preferentially incorporate dATP opposite of 8-oxoG. However, a preference for dCTP incorporation was observed for the yeast replicative polymerase δ [67]. The human repair polymerase β (pol β) has a ratio of incorporation of 4:1 and preferentially pairs dC with 8-oxoG [53]. Evolutionarily it would be advantageous for repair polymerases to be less promiscuous when incorporating nucleotides. Additionally, the translesion polymerases η and Dpo4 also preferentially incorporate dCTP [67, 68].
In vitro models have shown that 8-oxoG, while potentially mutagenic, does not represent a block to most polymerases and the lesion itself lacks significant toxicity. However, extension past an 8-oxoG:X base pair, where X = A or C, is polymerase dependent. KF–, KF+, and E. coli Pol II– efficiently extended an 8-oxoG:A base pair, but extension of an 8-oxoG:C base pair was greatly impaired [53, 65]. In contrast, pol η efficiently extended an 8-oxoG:C base pair, but not an 8-oxoG:A base pair [67]. T7– and HIV-1 RT extended both base pairs, although faster rates were observed for 8-oxoG:A [66]. In the cases where the 8-oxoG:A base pair does not block further replication, the polymerase will continue to replicate the DNA, leading to a G → T mutation if the mismatch is not repaired.
Mutational and genotoxic properties of 8-oxoG in vivo
Studies performed in vitro demonstrated that 8-oxoG has the ability to miscode and cause mutations. However, 8-oxoG is only mildly mutagenic when replicated in vivo owing to the presence of an extensive repair system that has evolved to counter its genetic effects [69]. In mammalian cells, the glycosylase/AP lyase OGG1 (MutM in E. coli) excises 8-oxoG from duplex DNA when it is paired with C. A second glycosylase, MYH (MutY in E. coli), removes A from an 8-oxoG:A mispair. By removing the A from this mispair, the cell is given a second attempt to incorporate the correct base C opposite 8-oxoG. Finally, MTH1 (MutT in E. coli), is a phosphatase that converts 8-oxodGTP to 8-oxodGMP; this action removes 8-oxodGTP from the nucleotide pool and prevents incorporation of 8-oxodGTP during replication. The repair of oxidatively damaged DNA is reviewed elsewhere in this special issue and the reader is directed there for a more detailed discussion [70].
Determining the genetic properties of 8-oxoG in vivo was greatly facilitated by the development of methods to incorporate the lesion into a vector that could be replicated in bacterial and/or mammalian cells. In the first studies to address the impact of 8-oxoG in vivo, the number of transformants or antibiotic resistant colonies recovered relative to an unmodified vector was used to assess toxicity of the lesion. A lacZ reporter assay or nucleotide sequencing analysis in the region corresponding to the lesion site described the mutagenicity of the lesion. When replicated in wild-type E. coli, vectors containing 8-oxoG displayed mutation frequencies of less than 2% [71-74]. In simian kidney (COS-7) cells the mutation frequencies were less than 5% [73]. In both systems the primary mutation observed at the site of replication was G → T transversions. Furthermore, in both bacterial and mammalian cells 8-oxoG was not toxic and was replicated as readily as an unmodified vector.
Recently developed techniques have allowed for a more high-throughput analysis of the mutagenicity and toxicity of a particular DNA lesion when replicated in E.coli. The competitive replication of adduct bypass (CRAB) assay and restriction endonuclease and postlabeling (REAP) analysis of mutation frequency and specificity, described in more detail in [75], have been used to study the genetic effects of not only 8-oxoG, but also Fapy·G and several hyperoxidized G lesions. Similar to the results obtained in earlier work, 8-oxoG was found to be readily replicated in E.coli and to be weakly mutagenic. The results obtained with Fapy·G and the hyperoxidized G lesions are summarized in subsequent sections of this review.
Incorporation of 8-oxodGTP from the nucleotide pool
In addition to considering the consequences of 8-oxoG when the lesion forms in DNA, one must also consider the consequences of oxidation of dGTP to 8-oxodGTP in the nucleotide pool. Namely, is 8-oxodGTP recognized as a substrate by polymerases, resulting in the incorporation of 8-oxoG into the nascent strand? Indeed, 8-oxodGTP is incorporated opposite A and C by the α subunit of E. coli DNA polymerase III [76]. Furthermore, the relative efficiency of the potentially mutagenic event where 8-oxodGTP is incorporated opposite A is nearly the same as pairing with C.
Subsequent work examined the incorporation of 8-oxodGTP into DNA by several other polymerases and is reviewed in [77]. Shown in Table III are steady-state kinetic parameters describing the insertion efficiencies, misinsertion frequencies, and percent misinsertion of 8-oxodGTP by Pol II–, KF–, HIV-1 RT, and T7– [78]. For all the polymerases examined, the insertion of dGTP opposite C is more efficient than insertion of 8-oxodGTP. However, the preference of 8-oxodGTP insertion opposite A or C varies between the polymerases with T7– showing the highest preference for incorporating 8-oxodGTP opposite A with 97% misinsertion.
Table III.
Steady-state kinetic parameters for the incorporation of 8-oxodGTP.
| Polymerase | dNTP: template base | Insertion Efficiency kcat/Km (mM-1min-1) | fa | Percent Misinsertionb | Reference |
|---|---|---|---|---|---|
| Pol II– | dGTP:C | 4800 | [78] | ||
| 8-oxodGTP:C | 0.015 | [78] | |||
| 8-oxodGTP:A | 0.00067 | 0.045 | 4 | [78] | |
| KF– | dGTP:C | 17000 | [78] | ||
| 8-oxodGTP:C | 0.11 | [78] | |||
| 8-oxodGTP:A | 0.048 | 0.44 | 31 | [78] | |
| HIV-1 RT | dGTP:C | 6000 | [78] | ||
| 8-oxodGTP:C | 0.26 | [78] | |||
| 8-oxodGTP:A | 0.13 | 0.50 | 33 | [78] | |
| T7– | dGTP:C | 2300 | [78] | ||
| 8-oxodGTP:C | 0.00067 | [78] | |||
| 8-oxodGTP:A | 0.021 | 31 | 97 | [78] |
Apparent misinsertion frequency, defined as f = [(kcat/Km)A/(kcat/Km)C].
Percent misinsertion = (f/(1+f)) × 100.
In addition to steady-state experiments, single-turnover methods have been used to study nucleotide incorporation reactions, and are reviewed in [79]. In a steady-state experiment, the steady-state rate kcat is dominated by the slowest step in the reaction cycle. For DNA polymerases this is often the dissociation of the polymerase-DNA product. Thus, kcat does not represent the chemistry step describing the addition of a dNTP to the nascent strand. By performing experiments under single-turnover conditions in which [polymerase]>>[DNA], the observed rate reports the rate of the slowest step up to and including the chemistry step in which the dNTP is covalently attached to the nascent strand [79]. Using single-turnover methods and the mitochondrial DNA polymerase γ, 8-oxodGTP was a 12.5-times better substrate when incorporated opposite A than opposite C [80]. Release of the pyrophosphate product was slow in the case of incorporation opposite C and allowed for reversal of the chemistry step. The reversibility of the chemistry step was proposed to result in 8-oxodGTP being a poorer substrate when incorporated opposite C. However, once formed the 8-oxoG:C base pair was extended 96% of the time whereas 8-oxoG:A was extended only 70% of the time.
The consequences of the incorporation of 8-oxodGTP from the nucleotide pool are highlighted by the mutation rates observed in strains of E. coli. Strains lacking MutM or MutY, the repair enzymes that counter 8-oxoG when base paired in genomic DNA, have ~10-fold higher rate of mutation than wild type E. coli [81, 82]. However, strains lacking MutT, which removes 8-oxodGTP from the nucleotide pool, have a 100-10,000-fold higher mutation rate than wild type strains [83]. It is noteworthy that the extent to which 8-oxodGTP contributes to this higher mutation rate has been questioned [84]. A detailed discussion of the prevention of misincorporation of oxidatively damaged DNA from the nucleotide pool is provided elsewhere in this issue [85].
Mutational and genotoxic properties of Fapy·G in vitro and in vivo
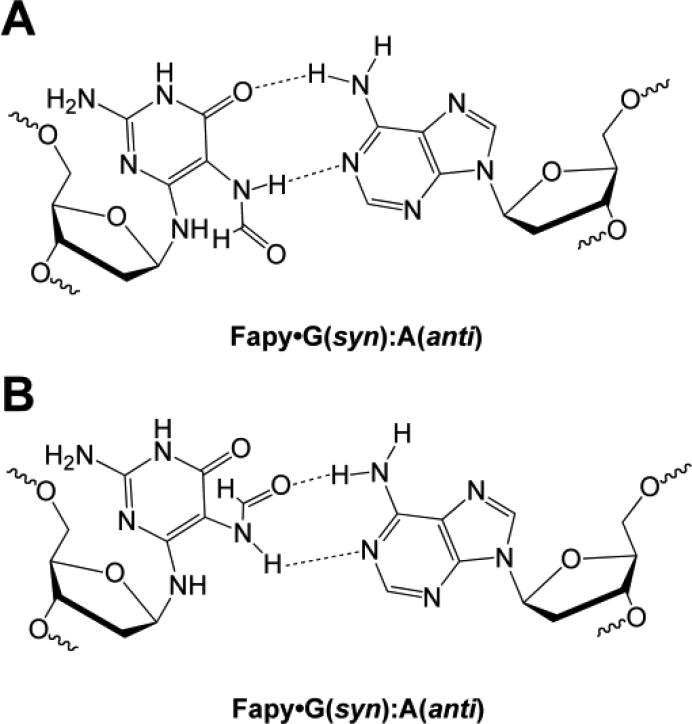
The incorporation of dNTPs opposite Fapy·G has been examined in vitro and, similar to 8-oxoG, dCTP and dATP are incorporated opposite the lesion. Incorporation of dATP can be rationalized when the lesion is in the syn conformation (Figure 6). Two rotamers of the formamide group allow Fapy·G (syn) to present thymidine-like hydrogen bonding groups and pair with A. Steady-state kinetic data for KF– and E. coli Klenow fragment of polymerase I with active proofreading activity (KF+) are provided in Table IV [86]. Insertion efficiencies, which provide a measure of the specificity for competing substrates, and apparent misinsertion frequencies (f) are shown. Both KF– and KF+ show a preference for incorporating dCTP over dATP opposite Fapy·G. While this preference is maintained in different sequence contexts, variations in insertion efficiency were observed depending on the 3′-flanking nucleotide [61, 86]. Comparing the insertion efficiency for dCTP and dATP incorporation opposite G reveals that the fidelity of KF+ is ~7-times greater than KF– [86]. However, the misinsertion frequency for dATP insertion opposite Fapy·G is ~3.5-fold higher with KF+ than with KF–. Indeed, Fapy·G has a misinsertion frequency that is 500-times higher than G, even when the polymerase has proofreading ability.
Figure 6.
Structure of Fapy·G(syn):A(anti) base pair with two rotamers of the formamide group of Fapy·G shown (top and bottom).
Table IV.
Steady-state kinetic parameters for the incorporation of dNTPs opposite Fapy·G.
| dNTP: template base | Insertion Efficiency Vmax/Km (%·min-1·M-1)a | fa | Reference | |
|---|---|---|---|---|
| KF– | dCTP:G | 3.5 ± 2.5 × 108 | [86] | |
| dATP:G | 1.5 ± 0.9 × 104 | 4.3 × 10–5 | [86] | |
| dCTP:Fapy·G | 7.5 ± 2.6 × 107 | [86] | ||
| dATP:Fapy·G | 6.1 ± 2.0 × 104 | 8.1 × 10–4 | [86] | |
| KF+ | dCTP:G | 4.7 ± 0.1 × 108 | [86] | |
| dATP:G | 2.8 ± 1.7 × 103 | 6.0 × 10–6 | [86] | |
| dCTP:Fapy·G | 2.1 ± 0.1 × 107 | [86] | ||
| dATP:Fapy·G | 5.9 ± 0.9 × 104 | 2.8 × 10–3 | [86] |
Apparent misinsertion frequency, defined as f = [(Vmax/Km)A/(Vmax/Km)C].
With respect to extension past a Fapy·G-containing base pair, experiments using KF– showed that extension past a Fapy·G:A or Fapy·G:C base pair is ~100-fold slower than a G:C base pair [61]. For KF+, extension past a Fapy·G:C base pair is 3-times more efficient than excision by the proofreading activity; however, extension past a Fapy·G:A base pair was not observed, indicating that proofreading is more efficient than extension.
The nucleotide pool may also serve as a source of Fapy·G lesions in DNA. Indeed, Fapy·dGTP is incorporated opposite C and A by KF+, although much less efficiently than dGTP is incorporated [87]. Steady-state kinetic parameters reveal that Fapy·dGTP is incorporated opposite C ~1000-times less efficiently than dGTP, and opposite A ~40,000-times less efficiently than dTTP. Notably, Fapy·dGTP is a poor substrate for MutT which enhances the kinetic viability of incorporation [87].
The mutagenicity and genotoxicity of Fapy·G in vivo have also been examined. Replication of a single-stranded vector containing Fapy·G in COS-7 cells results in 8-30% G → T mutations, depending on the sequence context of the lesion [88]. Notably, in these experiments Fapy·G was slightly more mutagenic than 8-oxoG, which also exhibited a similar sequence context effect. However, the mutation frequency of Fapy·G in E. coli, as determined by the REAP assay (vida supra) is significantly lower than in COS-7 cells, with less than 2% G → T mutations observed [86]. The reason for this differential mutagenicity in E. coli and COS-7 cells is unknown. In E. coli Fapy·G was also replicated less efficiently than 8-oxoG [86].
Mutational and genotoxic properties of hyperoxidized products when replicated in vitro
In general, the hyperoxidized G lesions serve as sources of both mutations and toxicity when replicated in vitro. In many cases the severity of the toxicity, and the extent to which the lesions serve as a block to replication, is polymerase-dependent.
The products generated by oxidation of an 8-oxoG-containing oligonucleotide with the oxidant [IrCl6]2– were shown to be both mutagenic and a partial block to replication by KF–, calf thymus DNA polymerase α (pol α), and human pol β [89, 90]. Incorporation of dATP and dGTP opposite the oxidized form(s) of 8-oxoG were observed. It is now known that the lesions associated with this observed mutagenicity and toxicity were the diastereomers of Sp and/or Gh.
When oligonucleotides containing authentic Sp (as a mixture of diastereomers) or Gh were used in primer extension experiments with KF–, dATP and dGTP were incorporated opposite the hyperoxidized guanine products instead of the correct base dCTP, suggesting that the lesions would cause G → T and G → C mutations, respectively [56]. Although the lesions were a significant block to replication by KF–, running start experiments showed enhanced replication relative to a standing start replication. Furthermore, sequence context affected replication efficiency as templates containing two guanines immediately 3′ to the hyperoxidized lesion were replicated more readily than templates containing two adenines. It was proposed that the additional stability provided to the duplex by the adjacent G:C base pairs provides a better template for polymerase than the A:T base pairs [56]. In other work, Gh was also a strong block to replication by calf thymus pol α, human pol β [56, 90, 91], and exonuclease deficient RB69 polymerase [92] where a purine was incorporated opposite Gh when replication did occur.
Steady-state kinetic experiments were used to determine insertion efficiencies for the incorporation of dATP and dGTP opposite Sp and Gh [56]. Shown in Table V, when Sp is present in the template as a mixture of diastereomers, dATP and dGTP are incorporated opposite of Sp with similar efficiencies, while dGTP is incorporated opposite Gh ~10-times more efficiently than dATP. When the two diastereomers of Sp were separated and studied individually, the diastereomer that eluted first from an anion exchange HPLC column (Sp1) had an insertion efficiency comparable to that observed for the mixture of diastereomers [56]. However, the insertion efficiency of the slower-eluting diastereomer (Sp2) was 3-fold lower. These in vitro results establish that the R and S diastereomers of Sp may have different coding properties.
Table V.
Steady-state kinetic parameters for the incorporation of dNTPs opposite Sp and Gh.
| dNTP: template base | Insertion Efficiency kcat/Km (μM-1sec-1) | Reference |
|---|---|---|
| dCTP:G | 0.70 | [56] |
| dCTP:8-oxoG | 0.04 | [56] |
| dATP:8-oxoG | 0.019 | [56] |
| dGTP:8-oxoG | 18 × 10-5 | [56] |
| dATP:Spa | 8.3 × 10-5 | [56] |
| dGTP:Spa | 10 × 10-5 | [56] |
| dATP:Gha | 8.2 × 10-5 | [56] |
| dGTP:Gha | 79 × 10-5 | [56] |
Present as a mixture of diastereomers.
A crystal structure of the RB69 DNA polymerase in complex with DNA containing a Gh lesion provides insight into the source of toxicity and mutagenicity of the lesion [92]. In the crystal, the R configuration of Gh was captured and the lesion was in the high syn conformation. Additionally, in the crystal structure the Gh is extrahelical and is not positioned to serve as a templating base; this positioning of the lesion suggests that it would be toxic and would serve as a block to replication. However, when in the S configuration, which was not observed in the crystal, it was proposed that Gh would be in a better position to serve as a templating base and this configuration may allow for replication of the lesion. Indeed, when Gh is in the high syn conformation the hydantoin group displays functional groups similar to a pyrimidine and explains why purines are incorporated opposite Gh when replication does occur.
In addition to the mutagenicity and toxicity of Sp and Gh, Iz is mutagenic as well as toxic, causing G → C transversions with frequency of 4.2% compared to a non-mutagenic replication event; notably, Pol I continued to extend only 27% of the Iz:G pair [93]. Additionally, dIzTP can be incorporated from the nucleotide pool and is paired exclusively with G.
The lesion Oz, which is generated by the hydrolysis of Iz, is paired mainly with dATP when replicated by KF– or Taq polymerase, suggesting that formation of Oz in DNA would lead to G → T mutations [94]. Oz is also a block to replication by both polymerases with inhibition of replication past Oz:A being more severe with Taq polymerase. Oz does not serve as a substrate for the repair polymerase pol β and was a complete block to replication with this polymerase.
In the case of Oa, different mutational properties are observed depending on the polymerase [95]. Incorporation of dATP is observed with KF–, while both dGTP and dATP are incorporated by Taq polymerase; these results suggest that Oa could be the source of G → T and/or G → C mutations. While KF– could fully replicate the template in the presence of all 4 dNTPs, DNA synthesis by Taq was dramatically inhibited with only small amounts of the fully extended primer observed. As observed for Oz, the Oa lesion was potently toxic to pol β and no nucleotide incorporation was observed.
The lesion Ua is a block to replication by KF–, pol β, AMV reverse transcriptase, and Sequenase with the polymerases stalling one nucleotide before the lesion, and at the lesion site [96]. However, with prolonged incubation times the full length product was observed indicating that these polymerases can replicate past Ua. When replication does occur, both dATP and dGTP are inserted opposite Ua suggesting that this lesion is a source of G → T and G → C mutations. Steady-state kinetic experiments with Ua in the template revealed an order of magnitude difference in the insertion efficiency by KF–, favoring the incorporation of dATP opposite Ua.
In contrast to most of the other hyperoxidized guanine products, Ca does not block replication by KF; no termination of the primer was observed opposite Ca [64]. dATP was predominately incorporated opposite Ca and, to a much lesser extent dGTP. These results suggest that Ca would cause mostly G → T transversions, with small amounts of G → C transversions.
Mutational and genotoxic properties of hyperoxidized products when replicated in vivo
Primer extension experiments using purified polymerases have shown that most of the hyperoxidized G lesions are partial, and in some cases severe, blocks to replication. When replication does occur, all of the hyperoxidized lesions examined have the ability to miscode and cause point mutations. These in vitro experiments provide a measure of the inherent mutagenicity and toxicity of the hyperoxidized lesions when replicated by a given polymerase. However, both prokaryotic and eukaryotic cells possess several polymerases that can be recruited to participate in replication. Furthermore, there is the possibility that DNA repair enzymes may remove the lesion prior to replication. Thus, replication of a lesion-containing vector in vivo, in the presence of the full replication and repair capacity of the cell, provides a measure of the mutagenicity and toxicity in a biological context.
Using the CRAB and REAP assays (vida supra) the toxicity, mutation type, and frequency of mutation for several hyperoxidized dG lesions has been determined when replicated in E. coli [75]. Both assays use an M13 single-stranded vector containing a single hyperoxidized lesion in a well-defined location. The CRAB assay measures the bypass of a lesion relative to a G-containing control vector. In this case, bypass of a lesion refers to replication past the lesion and is an indication of toxicity. The REAP assay takes advantage of the fact that the hyperoxidized lesion is positioned at a known site in the M13 vector. Therefore, following replication of the vector to produce progeny, the site that originally contained the lesion can be interrogated to reveal the type and frequency of mutations. Presence of a G at the site that originally contained the hyperoxidized guanine lesion indicates the lesion is non-mutagenic. In contrast, if this nucleobase is A, C, or T, the lesion codes for a mutation.
A summary of the bypass efficiencies, mutation type, and mutation frequencies for 8-oxoG, the two diastereomers of Sp, Gh, Iz, Oz, Ca, Oa, and Ur when the lesions are replicated in wild type E. coli is provided in Table VI [44, 97-102]. As described previously, 8-oxoG is readily bypassed in both bacterial and mammalian cells and is weakly mutagenic causing ~5% G → T transversions [71-74]. The bypass efficiencies of the hyperoxidized lesions range from 10-100%. Ur is the most significant block to replication with only 10% bypass efficiency relative to dG [44]. In contrast, Oa could be replicated just as readily as the G control [100, 102]. Notably, REAP and CRAB experiments for some hyperoxidized lesions have been performed in two different sequence contexts: 5′-dTXG-3′ and 5′-dGXA-3′. In these two sequence contexts most lesions are bypassed with comparable efficiencies, however, significant sequence context effects are observed for Gh and Oa [97, 98, 100, 102].
Table VI.
Bypass efficiencies, mutation type, and mutation frequency induced by hyperoxidized G lesions when replicated in E. coli.
| Lesion | Bypass Efficiency (%) | Mutation Type and Frequency (%) | Total Mutation Frequency (%) | References | |
|---|---|---|---|---|---|
| 5'-dTXG-3' | 5'-dGXA-3' | ||||
| 8-oxoG | 100 | 90 | G → T (5)a | 5 | [44,97,98,100,101,102] |
| Sp1b | 20 | 10 | G → T (80)a | 100 | [98,100,101,102] |
| G → C (20) | |||||
| Sp2c | 20 | 10 | G → T (50)a | 100 | [98,100,101,102] |
| G → C (50) | |||||
| Gh | 20 | 80 | 5'-TXG-3' | 100 | [100,102] |
| G →T (40) | |||||
| G → C (60) | |||||
| 5'-GXA-3' | 100 | [97] | |||
| G → T (2) | |||||
| G → C (98) | |||||
| Iz | nd | 60 | G → T (90)d | 90 | [99] |
| Oz | nd | 57 | G → T (95)d | 95 | [97] |
| Ca | nd | 65 | G → T (90)d | 90 | [97] |
| Oa | 100 | 52 | G → T (99)a | 99 | [44,97,100,102] |
| Ur | 10 | 11 | 5'-TXG-3' | 100 | [100,102] |
| G → T (55) | |||||
| G → C (35) | |||||
| G → A (10) | |||||
| 5'-GXA-3' | 99 | [44] | |||
| G → T (99) | |||||
Mutation type and frequency are similar in the sequence contexts of 5'-TXG-3' and 5'-GXA-3'.
Diastereomer of Sp that elutes first from an anion exchange HPLC column.
Diastereomer of Sp that elutes second from an anion exchange HPLC column.
Values reported are for the 5'-GXA-3' sequence context.
nd: not determined.
In contrast to the weakly mutagenic 8-oxoG, the hyperoxidized G lesions are potently mutagenic with mutation frequencies of 90-100% [44, 97-102]. The hyperoxidized lesions induce predominately G → T and G → C transversions although the frequency of each mutation is lesion dependent. As observed for bypass efficiency, the type of mutations caused by some of the lesions, in particular Gh and Ur, is dependent on the sequence context of the lesion [97, 98, 100, 102].
Using strains of E.coli deficient in a particular polymerase or DNA repair enzyme has provided insight into the processing of the hyperoxidized lesions in vivo. With the exception of Oa, which was readily bypassed, the bypass efficiencies of Sp, Gh, and Ur were dramatically reduced in polymerase V (Pol V) deficient cells [102]. These results indicate a critical role for Pol V in replicating these hyperoxidized dG lesions. When replicated in cells lacking MutY, no change in the mutation type or frequency was observed for Sp, Gh, Oa, or Ur [100]. These results are in contrast to those obtained with 8-oxoG, which is a known substrate for MutY when mispaired with A, where an increase in mutation frequency from 3 to 44% G → T mutations was observed in cells lacking MutY. These results indicate that MutY does not have glycosylase activity on Sp, Gh, Oa, or Ur. However, the Sp lesions are bypassed less efficiently in cells that possess MutY; thus, MutY may play a protective role by limiting the survival of cells that contain these lesions [100]. It is noteworthy that while MutY does not appear to have glycosylase activity on the hyperoxidized lesions studied, the glycosylase Nei does play a role in reducing the level of Sp in E. coli exposed to [Cr2O7]2- [46]. The level of Sp was 20-fold higher in E. coli lacking Nei than in wild-type cells.
The ability of hyperoxidized G lesions to be incorporated into the genome via the nucleotide pool has also been examined. Interestingly, in contrast to the potent mutagenicity of Gh and Sp when the lesions are replicated by polymerases in vitro and in vivo, when dGhTP and dSpTP were introduced to the nucleotide pool of wild-type E. coli, no increase in mutation frequency was observed [103]. Furthermore, MutT was not able to hydrolyze the hyperoxidized triphosphates in vitro suggesting that the absence of mutagenicity of dGhTP and dSpTP might be due to the inability of E. coli polymerases to incorporate the lesions in vivo, or that the lesions are incorporated opposite G to form the ‘correct’ base pair [103]. Further studies are needed to understand the lack of mutagenicity of hyperoxidized lesions when the lesions are part of the nucleotide pool.
To date the mutational and genotoxic properties of hyperoxidized products in mammalian cells have not been examined and, with respect to carcinogenicity, it will be important to determine if the potent mutagenicity and toxicity observed when the lesions are replicated in E. coli are also observed in mammalian cells.
Biological implications of oxidatively damaged DNA
Similar to experiments which explored the biochemistry and mutagenicity of Fapy·G and the hyperoxidized lesions, studies to address the biological implications of these oxidatively damaged DNA lesions have lagged behind those for 8-oxoG and remain future work. For this reason, this section highlights the biological implications of 8-oxoG. Selected topics are described below, and the reader is directed to other reviews in this special issue that focus on the contributions of oxidatively damaged DNA to cancer [104], aging [105], cardiovascular health [106], neurological disease [107], and epigenetic regulation [108].
The role of 8-oxoG in Huntington's disease
In general, the repair of oxidatively damaged DNA nucleobases minimizes their mutagenicity and/or toxicity and is beneficial to an organism [109]. Indeed, removal of 8-oxoGua from an 8-oxoG:C base pair by OGG1 minimizes the G → T transversion mutations caused by this DNA lesion. However, the repair of 8-oxoG by OGG1 within a cytosine-adenine-guanine (CAG) trinucleotide repeat (TNR) sequence has been linked to expansion of the repetitive sequence [110, 111]. This TNR expansion is the pathogenic signature of several neurodegenerative disorders including Huntington's disease (HD). In healthy individuals the number of CAG TNR repeats falls in the range of 5-35; in patients afflicted by HD the TNR sequence is expanded and includes greater than 40 CAG repeats [112].
Using an R6/1 mouse model of HD, the degree of TNR expansion correlated with the level of 8-oxoG [110]. Furthermore, TNR expansion was abrogated in somatic cells of OGG1-/- HD mice [110]. Thus 8-oxoG, and its repair by OGG1, is implicated in HD by facilitating TNR expansion. This result is counterintuitive as repair of DNA damage is usually advantageous, but in this case the OGG1 repair event is implicated in a disease-initiating TNR expansion.
OGG1 is a glycosylase that is known to initiate the BER process in mammalian cells by excising 8-oxoGua from the sugar-phosphate backbone via cleavage of the N-glycosydic bond [109]. The product generated by OGG1 is DNA containing an abasic site, and the subsequent actions of several enzymes including apurinic/apyrimidinic endonuclease 1 (APE1), pol β, and DNA ligase complete the repair event to replace the damaged nucleobase with an unmodified Gua [113, 114]. It is noteworthy that while OGG1 is typically associated with short-patch BER, in which pol β incorporates a single nucleotide as part of the repair event, it has been shown that CAG TNR DNA can also undergo long-patch BER (LP-BER) [111]. In LP-BER, instead of incorporating a single nucleotide, pol β performs strand displacement synthesis and incorporates two or more nucleotides. Flap endonuclease (FEN1) removes the displaced single strand of DNA and ligase completes the repair event. LP-BER is typically utilized when the abasic site is chemically modified by alkylation or oxidation [115, 116]. However, it has been shown that LP-BER occurs on CAG TNR DNA even in the presence of an unmodified abasic site [111].
It has been proposed that LP-BER on these repetitive sequences may be facilitated by the ability of TNR DNA to adopt non-canonical structures [111]. For example, single-stranded DNA containing CAG repeats has been shown to adopt a stem-loop hairpin both in vitro[117-122] and in vivo [123]. Furthermore, formation of a stem-loop hairpin by the displaced strand generated in LP-BER has been shown to render the strand refractory to cleavage by FEN1 [111]. Thus, following ligation, the entire length of the stem-loop hairpin is incorporated into the DNA to cause an expansion of the CAG repeat sequence.
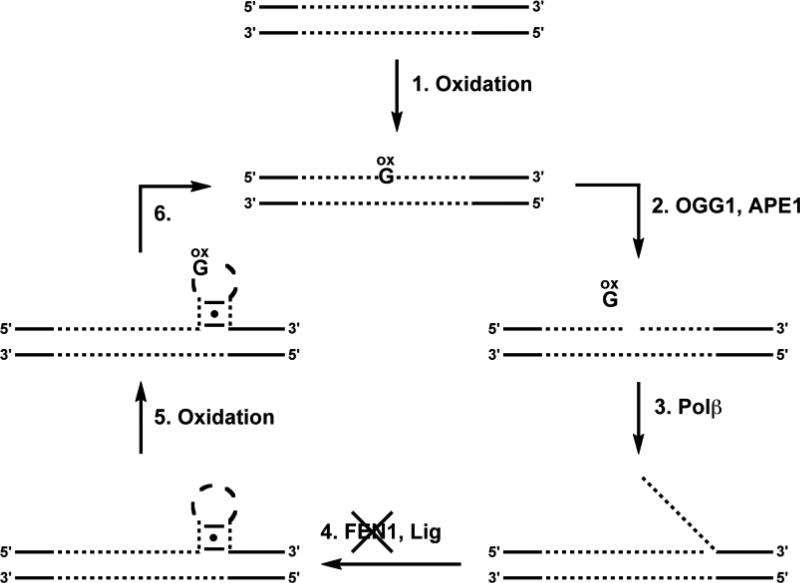
Additionally, we have shown recently that the stem-loop hairpin formed by CAG repeats is hyper-susceptible to oxidative DNA damage, yet is inefficiently repaired by OGG1 [124, 125]. Thus, damage is expected to accumulate in the stem-loop hairpin. If this damage-containing hairpin is ligated into DNA and reincorporated into duplex, a bone fide substrate for OGG1 is regenerated, and the cycle of repair and expansion can be restarted. Indeed, a ‘toxic oxidation cycle’ has been proposed in which a reiterative cycle of repair and expansion works to incrementally expand the region of TNR DNA [110, 125] (Figure 7). Considering this proposed cycle, the role of OGG1 in TNR expansion is to remove 8-oxoGua and initiate a LP-BER repair event; due to the propensity for the repetitive DNA to form non-canonical structures, the displaced single strand is not removed as it should be and, instead, is incorporated into the DNA to cause the TNR expansion.
Figure 7.
Toxic oxidation cycle. (1) Genomic DNA is oxidized to generate 8-oxoG and (2) OGG1 and APE1 initiate BER by removing the oxidatively damaged nucleobase from duplex TNR DNA (shown as dotted lines) and cleaving the backbone at the abasic site. (3) The resulting gap is repaired via LP-BER, producing a TNR flap that (4) is refractory to FEN1 and ligation occurs, trapping a stem-loop hairpin in the duplex. (5) The resulting stem-loop hairpin is highly susceptible to DNA damage in the loop region and the lack of efficient repair of this damage by OGG1 results in (6) its being incorporated into the expanded duplex. The final product of the cycle now contains DNA damage that will be recognized by OGG1. Therefore, the cycle is repeated and the DNA can be processed again, incrementally expanding the TNR tract of DNA.
Telomeres may protect the genome from oxidatively damaged DNA
The linear chromosomes of eukaryotic organisms have at their termini repetitive DNA sequences called telomeres. These are non-coding regions that serve to protect the genome from the ‘end replication problem’ that arises during lagging strand DNA synthesis [126, 127]. Human telomeric DNA is composed of ~10 kilobases of double stranded (5′-TTAGGG-3′/5′-CCCTAA-3′)n repeats [128]. The extreme 3′-end is a 200-300 base single-stranded overhang of the G-containing strand [129]. It is known that in vitro this single-stranded G-rich sequence has the ability to form highly-ordered structures called guanine quadruplexes [130]. Quadruplexes consist of stacked tetrads where each tetrad is a planar array of four Hoogsteen-bonded guanines [131]. Support for the formation of these structures in vivo has been demonstrated using an antibody that is specific for guanine quadruplexes [132].
Given the high G content of telomeric DNA, and the relative ease of oxidation of dGuo as compared to the other 2′-deoxynucleosides, it has been proposed that telomeric DNA may serve as a hot spot for oxidation within the genome. Several reports have shown that oxidative stress and UVA/UVB irradiation induces oxidatively damaged DNA in oligonucleotides with telomeric repeats in vitro [133, 134]. Recently, it has also been shown that the telomeric regions of DNA isolated from mouse kidney cells are more susceptible to the oxidative stress induced by H2O2/Cu2+ treatment than non-telomeric DNA [135]. However, when U2OS osteosarcoma cells were treated with menadione, which generates ROS, the frequency of MutM-sensitive sites in telomeric DNA was the same as in non-telomeric DNA immediately following the menadione exposure [135]. MutM is the bacterial homolog of OGG1 and removes 8-oxoGua from DNA; thus, the number of MutM-sensitive sites directly correlates with the number of oxidatively damaged guanines. In contrast to the comparable amount of DNA damage immediately following exposure to menadione, 6 h after the treatment no MutM-sensitive sites were detected in the non-telomeric DNA, indicating that they had been repaired, whereas MutM-sensitive sites were detected in the telomeric DNA. These results suggest that while telomeric DNA may not be more susceptible to oxidative damage in vivo, repair may be less efficient in telomeres and can lead to an accumulation of oxidative damage over time in telomeric DNA in vivo.
Considering the ability of DNA to mediate long-range charge transfer reactions [14], telomeric DNA or other G-rich regions of the genome might provide regions to which oxidizing equivalents are funneled. Indeed, the number of 5′-GGG-3′ triple guanine sites was shown statistically to be elevated in the regions flanking protein-coding exons, and thus it was suggested that oxidizing charges injected into DNA might be funneled to these sacrificial G-rich introns or to the non-coding sequences of telomeric DNA. Heller has proposed a means of cathodic DNA protection similar to the way in which Zn2+ protects steel [136]. In support of this proposal, using an in vitro model of human telomeric DNA which contains both a duplex region and single-stranded overhang capable of forming a quadruplex, it was shown that long-range charge transfer to quadruplex structures could trap oxidative DNA damage, such as 8-oxoG [137].
8-oxoG and colorectal cancer
Recently, mutations in the gene that encodes MYH have been linked to an individual's predisposition for colorectal cancer [138-141]. This genetic disposition is referred to as MYH-associated polyposis (MAP) and is illustrated by biallelic-inherited mutations that result in defective activity of MYH [142]. MYH is an adenine-specific DNA glycosylases that removes adenine from an 8-oxoG:A base pair [143]. Removal of 8-oxoGua from this base pair is not beneficial, as it would result in a C → A mutation. MYH activity gives the cell a chance to place the proper base, C, opposite 8-oxoG, which would then allow OGG1 another opportunity to remove the lesion prior to replication.
The two most prevalent mutations observed in MAP-affected individuals are Y165C and G382D and these two altered amino acids are highly conserved in MutY enzymes across phylogeny [140, 144]. These MYH variants fail to complement the activity of the E. coli MutY protein in MutY– E. coli [145]. Furthermore, in vitro analysis of the E. coli [145] and mouse [146, 147] enzymes corresponding to the MYH variants observed in humans revealed a reduction in the adenine glycosylase activity of the mutant enzymes. Using a non-cleavable substrate analog, 2′-deoxy-2′-fluoroadenosine, both substrate affinity and the ability to differentiate between 8-oxoG and G were greatly reduced for the mutant enzymes [148].
Interestingly, analysis of the adenomatous polyposis coli (APC) gene of individuals producing the Y165C and G382D MYH variants identified a specific sequence context, 5′-GAA-3′, in which there was a significant increase in the number of C → A mutations [140]. In vitro kinetic experiments using two DNA duplex substrates, one containing the 5′-GAA-3′ sequence context and one with 5′-GAG-3′, found that the reduced activity observed for the mutant MYH enzymes relative to the wild type enzyme was magnified in the ‘hotspot’ 5′-GAA-3′ sequence [145, 146].
8-oxoG as a regulator of gene expression
Altering of the state of chromatin organization at a gene promoter provides a means to regulate gene expression. Epigenetic modification of the DNA and/or histone proteins has been linked to reorganization of chromatin from tightly packaged and transcriptionally-silenced heterochromatin to the more loosely packaged and transcriptionally-active euchromatin and vice versa [149]. Interestingly, it has also been shown that oxidatively damaged DNA can regulate gene expression and, in particular, estrogen-induced gene expression.
Following hormone activation, estrogen receptors interact with transcription co-activators, which modify histones [149] and contribute to the control of gene expression [150]. Specifically, demethylation of lysine 9 of the histone 3 (H3K9) nucleosomal histone is required for efficient transcription in vivo. Perillo and co-workers investigated the molecular mechanism by which demethylation of lysine affects estrogen-induced transcription [151]. The demethylation is known to be performed by lysine-specific demethylase 1 (LSD1) which is a flavin-dependent oxidase [152]. In order to complete the catalytic cycle the flavin cofactor is re-oxidized using oxygen as the electron acceptor, resulting in the production of H2O2. This localized production of H2O2 leads to formation of 8-oxoG, presumably by conversion of H2O2 to ROS that can react directly with G. Additionally, chromatin immunoprecipitation (ChIP) analysis revealed that OGG1 and topoisomerase IIβ accumulated at these sites of DNA damage in an estrogen-dependent manner [151]. Perillo and co-workers proposed that removal of 8-oxoGua by OGG1 and the initiation of the BER pathway would generate a nick that could be utilized by topoisomerase IIβ and cause the chromatin relaxation necessary to accommodate the transcription initiation complex. This proposed mechanism highlights how a cell may use controlled production of oxidatively damaged DNA to regulate gene expression and suggests that 8-oxoG should not be regarded solely as an undesired damage product.
A prebiotic role for 8-oxoG in the repair of thymine dimers
It has recently been proposed that prior to the evolution of more sophisticated enzyme cofactors, 8-oxoG may have served a role in catalyzing reactions [153]. In particular, the ability of 8-oxoG to act as a primordial flavin cofactor and as a substitute for the DNA repair enzyme photolyase was examined. Photolyase is known to repair cyclobutane pyrimidine dimers via a one electron reduction, with a photoexcited FADH– cofactor serving as the source of the electron [154]. The redox potential of 8-oxoG is closer to that of the flavin than the canonical DNA bases and suggested that 8-oxoG may be able to mimic the behavior of the flavin.
Using DNA duplexes that contain 8-oxoG and a cyclobutane pyrimdine dimer, it was shown that 8-oxoG acts in a mechanism consistent with photolyase; photoexcited 8-oxoG donates an electron to the pyrimidine dimer to catalyze bond cleavage [153]. A subsequent back electron transfer regenerates 8-oxoG and two pyrimidine monomers in DNA. Notably, 8-oxoG was effective in catalyzing the photorepair of both a thymine dimer and a uracil dimer. These results suggest that the oxidatively damaged guanine product 8-oxoG may have played a role in catalyzing the repair of another type of damage, the pyrimidine dimer.
Conclusions
The ease of oxidation of G gives rise to several types of oxidatively damaged DNA. The primary oxidation product 8-oxoG has been studied extensively. While 8-oxoG does not affect the global structure of DNA, it is mildly mutagenic in cells and the source of this mutagenicity is well understood. Despite our understanding of the mutagenic potential of 8-oxoG, the full biological consequences and roles of 8-oxoG are still being unraveled. Indeed, while 8-oxoG is known to play detrimental roles in cells and to contribute to onset of several diseases, this oxidatively damaged DNA lesion has also been shown to play a specialized role in cellular processes such as gene expression and DNA repair.
In addition to 8-oxoG, the Fapy·G lesion has begun to emerge as an important component of oxidatively damaged DNA that under some conditions is formed in amounts comparable to 8-oxoG. Furthermore, initial studies in mammalian cells have revealed that Fapy·G is slightly more mutagenic than 8-oxoG. Further studies are needed to elucidate the full biological consequences of Fapy·G.
Similar to Fapy·G, the hyperoxidized G products have been less extensively characterized than 8-oxoG, however it is known that some of these oxidation products have a more dramatic structural impact than 8-oxoG. Additionally, in contrast to 8-oxoG, many of these DNA lesions are genotoxic and potently mutagenic in cells. Nevertheless, our understanding of the consequences of the hyperoxidized G products is in its infancy. Elucidating the levels of these hyperoxidized G products in cells will be a first step towards revealing their true cellular and biological consequences.
Abbreviations
- APC
adenomatous polyposis coli
- APE1
apurinic/apyrimidinic endonuclease 1
- T7–
bacteriophage T7 exo–
- ChIP
chromatin immunoprecipitation
- CD
circular dichroism
- CRAB
competitive replication of adduct bypass assay
- CAG
cytosine-adenine-guanine
- DGh
dehydroguanidinohydantoin
- dGuo
2′-deoxyguanosine
- pol α
DNA polymerase α
- FEN1
flap endonuclease
- 8-oxodGuo
8-oxo-7,8-dihydro-2′-deoxyguanosine
- DSC
differential scanning calorimetry
- ESCODD
European Standard Committee on Oxidative DNA Damage
- Fapy
formamidopyrimidine
- Gh
guanidinohydantoin
- HICA
4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid
- HD
Huntington's disease
- Iz
imidazolone
- KF–
E. coli Klenow fragment of polymerase I exo–
- LP-BER
long-patch base excision repair
- LSD1
lysine-specific demethylase 1
- MAP
MYH-associated polyposis
- NOE
nuclear Overhauser effect
- Oa
oxaluric acid
- Oz
oxazolone
- Pa
parabanic acid
- Pol II–
polymerase II exo–
- pol β
polymerase β
- ROS
reactive oxygen species
- REAP
restriction endonuclease and postlabeling assay
- RT
reverse transcriptase
- Sp
spiroiminodihydantoin
- TNR
trinucleotide repeat
- Ur
urea
Footnotes
Declaration of Interest
Work in the authors’ laboratory was supported by the NIH grant ES019296.
References
- 1.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 2.Niles JC, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: Structures and mechanisms of product formation. Nitric Oxide. 2006;14:109–121. doi: 10.1016/j.niox.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Yermilov V, Yoshie Y, Rubio J, Ohshima H. Effects of carbon dioxide/bicarbonate on induction of DNA single-strand breaks and formation of 8-nitroguanine, 8-oxoguanine and base-propenal mediated by peroxynitrite. FEBS Lett. 1996;399:67–70. doi: 10.1016/s0014-5793(96)01288-4. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: Formation, measurement, and biological significance. Rev Physiol Biochem Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 5.Misiaszek R, Crean C, Joffe A, Geacintov NE, Shafirovich V. Oxidative DNA damage associated with combination of guanine and superoxide radicals and repair mechanisms via radical trapping. J Biol Chem. 2004;279:32106–32115. doi: 10.1074/jbc.M313904200. [DOI] [PubMed] [Google Scholar]
- 6.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc Natl Acad Sci U S A. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regulus P, Spessotto S, Gateau M, Cadet J, Favier A, Ravanat JL. Detection of new radiation-induced DNA lesions by liquid chromatography coupled to tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2223–2228. doi: 10.1002/rcm.1612. [DOI] [PubMed] [Google Scholar]
- 8.Candeias LP, O'Neill P, Jones GD, Steenken S. Ionization of polynucleotides and DNA in aqueous solution by 193 nm pulsed laser light: Identification of base-derived radicals. Int J Radiat Biol. 1992;61:15–20. doi: 10.1080/09553009214550571. [DOI] [PubMed] [Google Scholar]
- 9.Pouget JP, Douki T, Richard MJ, Cadet J. DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and comet assay. Chem Res Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 10.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Cooke MS, Loft S, Olinski R, Evans MD, Bialkowski K, Wagner JR, Dedon PC, Moller P, Greenberg MM, Cadet J. Recommendations for standardized description of and nomenclature concerning oxidatively damaged nucleobases in DNA. Chem Res Toxicol. 2010;23:705–707. doi: 10.1021/tx1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 13.Margolin Y, Cloutier JF, Shafirovich V, Geacintov NE, Dedon PC. Paradoxical hotspots for guanine oxidation by a chemical mediator of inflammation. Nat Chem Biol. 2006;2:365–366. doi: 10.1038/nchembio796. [DOI] [PubMed] [Google Scholar]
- 14.Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito I, Takayama M, Sugiyama H, Nakatani K. Photoinduced DNA cleavage via electron-transfer - demonstration that guanine residues located 5′ to guanine are the most electron-donating sites. J Am Chem Soc. 1995;117:6406–6407. [Google Scholar]
- 16.Sugiyama H, Saito I. Theoretical studies of GC-specific photocleavage of DNA via electron transfer: Significant lowering of ionization potential and 5′-localization of homo of stacked GG bases in B-form DNA. J Am Chem Soc. 1996;118:7063–7068. [Google Scholar]
- 17.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 18.European Standards Committee on Urinary (DNA) Lesion Analysis. Evans MD, Olinski R, Loft S, Cooke MS. Toward consensus in the analysis of urinary 8-oxo-7,8-dihydro-2′-deoxyguanosine as a noninvasive biomarker of oxidative stress. FASEB J. 2010;24:1249–1260. doi: 10.1096/fj.09-147124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mistry V, Teichert F, Sandhu JK, Singh R, Evans MD, Farmer PB, Cooke MS. Noninvasive assessment of oxidatively damaged DNA: Liquid chromatography-tandem mass spectrometry analysis of urinary 8-oxo-7,8-dihydro-2′-deoxyguanosine. Methods Mol Biol. 2011;682:279–289. doi: 10.1007/978-1-60327-409-8_20. [DOI] [PubMed] [Google Scholar]
- 20.European Standards Committee on Oxidative DNA Damage Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: An approach to consensus. Carcinogenesis. 2002;23:2129–2133. doi: 10.1093/carcin/23.12.2129. [DOI] [PubMed] [Google Scholar]
- 21.European Standards Committee on Oxidative DNA Damage Comparison of results from different laboratories in measuring 8-oxo-2′-deoxyguanosine in synthetic oligonucleotides. Free Radical Res. 2002;36:649–659. doi: 10.1080/10715760290029047. [DOI] [PubMed] [Google Scholar]
- 22.European Standards Committee on Oxidative DNA Damage Inter-laboratory validation of procedures for measuring 8-oxo-7,8-dihydroguanine/8-oxo-7,8-dihydro-2′-deoxyguanosine in DNA. Free Radical Res. 2002;36:239–245. doi: 10.1080/10715760290019246. [DOI] [PubMed] [Google Scholar]
- 23.European Standards Committee on Oxidative DNA Damage Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radical Bio Med. 2003;34:1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 24.European Standards Committee on Oxidative DNA Damage Establishing the background level of base oxidation in human lymphocyte DNA: Results of an interlaboratory validation study. FASEB J. 2004;18:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 25.Moller P, Loft S, Olinski R, Collins A, Cooke M. Free Rad Res. 2012 doi: 10.3109/10715762.2011.644241. [DOI] [PubMed] [Google Scholar]
- 26.Munk BH, Burrows CJ, Schlegel HB. Exploration of mechanisms for the transformation of 8-hydroxy guanine radical to FapyG by density functional theory. Chem Res Toxicol. 2007;20:432–444. doi: 10.1021/tx060187t. [DOI] [PubMed] [Google Scholar]
- 27.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free-radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 28.Doetsch PW, Zastawny TH, Martin AM, Dizdaroglu M. Monomeric base damage products from adenine, guanine, and thymine induced by exposure of DNA to ultraviolet-radiation. Biochemistry. 1995;34:737–742. doi: 10.1021/bi00003a005. [DOI] [PubMed] [Google Scholar]
- 29.Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing FapydG (N6-(2-deoxy-α,β-d-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine). J Am Chem Soc. 2001;123:8636–8637. doi: 10.1021/ja0160952. [DOI] [PubMed] [Google Scholar]
- 30.Haraguchi K, Delaney MO, Wiederholt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and characterization of oligodeoxynucleotides containing formamidopyrimidine lesions and nonhydrolyzable analogues. J Am Chem Soc. 2002;124:3263–3269. doi: 10.1021/ja012135q. [DOI] [PubMed] [Google Scholar]
- 31.Jiang YL, Wiederholt CJ, Patro JN, Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing FapydG (N-6-(2-deoxy-α,β-d-erythropentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine) using a 5′-dimethoxytrityl dinucleotide phosphoramidite. J Org Chem. 2005;70:141–149. doi: 10.1021/jo048253o. [DOI] [PubMed] [Google Scholar]
- 32.Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, Rizzo CJ. Site-specific synthesis and characterization of oligonucleotides containing an N(6)-(2-deoxy-d-erythropentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-n-methylformamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem Res Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukin M, Minetti CA, Remeta DP, Attaluri S, Johnson F, Breslauer KJ, de Los Santos C. Novel post-synthetic generation, isomeric resolution, and characterization of Fapy-dG within oligodeoxynucleotides: Differential anomeric impacts on DNA duplex properties. Nucleic Acids Res. 2011;39:5776–5789. doi: 10.1093/nar/gkr082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger M, Anselmino C, Mouret JF, Cadet J. High-performance liquid-chromatography electrochemical assay for monitoring the formation of 8-oxo-7,8-dihydroadenine and its related 2'-deoxyribonucleoside. J Liq Chromatogr. 1990;13:929–940. [Google Scholar]
- 35.Yanagawa H, Ogawa Y, Ueno M. Redox ribonucleosides. Isolation and characterization of 5-hydroxyuridine, 8-hydroxyguanosine, and 8-hydroxyadenosine from torula yeast RNA. J Biol Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]
- 36.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 37.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: Mechanistic studies with 18O. Chem Res Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 38.Ye Y, Muller JG, Luo WC, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of C-13-, N-15-, and O-18-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 39.Munk BH, Burrows CJ, Schlegel HB. An exploration of mechanisms for the transformation of 8-oxoguanine to guanidinohydantoin and spiroiminodihydantoin by density functional theory. J Am Chem Soc. 2008;130:5245–5256. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- 40.Ye Y, Munk BH, Muller JG, Cogbill A, Burrows CJ, Schlegel HB. Mechanistic aspects of the formation of guanidinohydantoin from spiroiminodihydantoin under acidic conditions. Chem Res Toxicol. 2009;22:526–535. doi: 10.1021/tx800402y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niles JC, Wishnok JS, Tannenbaum SR. Mass spectrometric identification of 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid during oxidation of 8-oxoguanosine by peroxynitrite and KHSO5/CoCl2. Chem Res Toxicol. 2004;17:1501–1509. doi: 10.1021/tx040003f. [DOI] [PubMed] [Google Scholar]
- 42.Cadet J, Berger M, Buchko GW, Joshi PC, Raoul S, Ravanat JL. 2,2-diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-d-erythropentofuranosyl) amino]-5-(2H)-oxazolone. A novel and predominant radical oxidation-product of 3′,5′-di-O-acetyl-2′-deoxyguanosine. J Am Chem Soc. 1994;116:7403–7404. [Google Scholar]
- 43.Chworos A, Coppel Y, Dubey I, Pratviel G, Meunier B. Guanine oxidation: NMR characterization of a dehydro-guanidinohydantoin residue generated by a 2e- oxidation of d(GpT). J Am Chem Soc. 2001;123:5867–5877. doi: 10.1021/ja003945p. [DOI] [PubMed] [Google Scholar]
- 44.Henderson PT, Neeley WL, Delaney JC, Gu F, Niles JC, Hah SS, Tannenbaum SR, Essigmann JM. Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations. Chem Res Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- 45.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 46.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodepudi V, Shibutani S, Johnson F. Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem Res Toxicol. 1992;5:608–617. doi: 10.1021/tx00029a004. [DOI] [PubMed] [Google Scholar]
- 48.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry. 1995;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 49.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. Nmr studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc Natl Acad Sci U S A. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh SK, Szulik MW, Ganguly M, Khutsishvili I, Stone MP, Marky LA, Gold B. Characterization of DNA with an 8-oxoguanine modification. Nucleic Acids Res. 2011;39:6789–6801. doi: 10.1093/nar/gkr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crenshaw CM, Wade JE, Arthanari H, Frueh D, Lane BF, Nunez ME. Hidden in plain sight: Subtle effects of the 8-oxoguanine lesion on the structure, dynamics, and thermodynamics of a 15-base pair oligodeoxynucleotide duplex. Biochemistry. 2011;50:8463–8477. doi: 10.1021/bi201007t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 54.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 55.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 56.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (klenow fragment). Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 57.Chinyengetere F, Jamieson ER. Impact of the oxidized guanine lesion spiroiminodihydantoin on the conformation and thermodynamic stability of a 15-mer DNA duplex. Biochemistry. 2008;47:2584–2591. doi: 10.1021/bi701502t. [DOI] [PubMed] [Google Scholar]
- 58.Schibel AE, Fleming AM, Jin Q, An N, Liu J, Blakemore CP, White HS, Burrows CJ. Sequence-specific single-molecule analysis of 8-oxo-7,8-dihydroguanine lesions in DNA based on unzipping kinetics of complementary probes in ion channel recordings. J Am Chem Soc. 2011;133:14778–14784. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 60.Patro JN, Haraguchi K, Delaney MO, Greenberg MM. Probing the configurations of formamidopyrimidine lesions FapydA and FapydG in DNA using endonuclease IV. Biochemistry. 2004;43:13397–13403. doi: 10.1021/bi049035s. [DOI] [PubMed] [Google Scholar]
- 61.Wiederholt CJ, Greenberg MM. Fapy-dG instructs klenow exo(-) to misincorporate deoxyadenosine. J Am Chem Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 62.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 63.Fenn D, Chi LM, Lam SL. Effect of hyperoxidized guanine on DNA primer-template structures: Spiroiminodihydantoin leads to strand slippage. FEBS Lett. 2008;582:4169–4175. doi: 10.1016/j.febslet.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 64.Gasparutto D, Da Cruz S, Bourdat AG, Jaquinod M, Cadet J. Synthesis and biochemical properties of cyanuric acid nucleoside-containing DNA oligomers. Chem Res Toxicol. 1999;12:630–638. doi: 10.1021/tx980255e. [DOI] [PubMed] [Google Scholar]
- 65.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo- and II exo. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 66.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 67.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 68.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 70.Sampath H, McCullough AK, Lloyd RS. Free Rad Res. 2012 doi: 10.3109/10715762.2012.655730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 72.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman AP. Site-specific mutagenesis using a gapped duplex vector: A study of translesion synthesis past 8-oxodeoxyguanosine in E. Coli. Mutat Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 73.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C→T.A transversions in simian kidney cells. Proc Natl Acad Sci U S A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 75.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 76.Maki H, Sekiguchi M. Mutt protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 77.Katafuchi A, Nohmi T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: Their efficiency and template base preference. Mutat Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 78.Einolf HJ, Schnetz-Boutaud N, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry. 1998;37:13300–13312. doi: 10.1021/bi981346d. [DOI] [PubMed] [Google Scholar]
- 79.Joyce CM. Techniques used to study the DNA polymerase reaction pathway. Biochim Biophys Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxodeoxyguanosine by the human mitochondrial DNA polymerase. J Biol Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 81.Cabrera M, Nghiem Y, Miller JH. MutM, a 2nd mutator locus in Escherichia coli that generates G.C→T.A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michaels ML, Pham L, Cruz C, Miller JH. MutM, a protein that prevents G.C→T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yanofsky C, Cox EC, Horn V. Unusual mutagenic specificity of an E coli mutator gene. Proc Natl Acad Sci USA. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tassotto ML, Mathews CK. Assessing the metabolic function of the MutT 8-oxodeoxyguanosine triphosphatase in Escherichia coli by nucleotide pool analysis. J Biol Chem. 2002;277:15807–15812. doi: 10.1074/jbc.M200965200. [DOI] [PubMed] [Google Scholar]
- 85.Nakabeppu Y. Free Rad Res. 2012 [Google Scholar]
- 86.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, FapydG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 87.Imoto S, Patro JN, Jiang YL, Oka N, Greenberg MM. Synthesis, DNA polymerase incorporation, and enzymatic phosphate hydrolysis of formamidopyrimidine nucleoside triphosphates. J Am Chem Soc. 2006;128:14606–14611. doi: 10.1021/ja065525r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muller JG, Duarte V, Hickerson RP, Burrows CJ. Gel electrophoretic detection of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine via oxidation by Ir(IV). Nucleic Acids Res. 1998;26:2247–2249. doi: 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 92.Aller P, Ye Y, Wallace SS, Burrows CJ, Doublie S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kino K, Sugiyama H. Possible cause of G-C→C-G transversion mutation by guanine oxidation product, imidazolone. Chem Biol. 2001;8:369–378. doi: 10.1016/s1074-5521(01)00019-9. [DOI] [PubMed] [Google Scholar]
- 94.Duarte V, Gasparutto D, Jaquinod M, Cadet J. In vitro DNA synthesis opposite oxazolone and repair of this DNA damage using modified oligonucleotides. Nucleic Acids Res. 2000;28:1555–1563. doi: 10.1093/nar/28.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duarte V, Gasparutto D, Jaquinod M, Ravanat J, Cadet J. Repair and mutagenic potential of oxaluric acid, a major product of singlet oxygen-mediated oxidation of 8-oxo-7,8-dihydroguanine. Chem Res Toxicol. 2001;14:46–53. doi: 10.1021/tx0001629. [DOI] [PubMed] [Google Scholar]
- 96.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem Res Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 97.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 98.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 99.Neeley WL, Delaney JC, Henderson PT, Essigmann JM. In vivo bypass efficiencies and mutational signatures of the guanine oxidation products 2-aminoimidazolone and 5-guanidino-4-nitroimidazole. J Biol Chem. 2004;279:43568–43573. doi: 10.1074/jbc.M407117200. [DOI] [PubMed] [Google Scholar]
- 100.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 101.Delaney S, Delaney JC, Essigmann JM. Chemical-biological fingerprinting: Probing the properties of DNA lesions formed by peroxynitrite. Chem Res Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 103.Hori M, Suzuki T, Minakawa N, Matsuda A, Harashima H, Kamiya H. Mutagenicity of secondary oxidation products of 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate (8-hydroxy-2′- deoxyguanosine 5′-triphosphate). Mutat Res. 2011;714:11–16. doi: 10.1016/j.mrfmmm.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 104.TBD cancer review. Free Rad Res. 2012 [Google Scholar]
- 105.TBD aging review. Free Rad Res. 2012 [Google Scholar]
- 106.Herbert K. Free Rad Res. 2012 [Google Scholar]
- 107.Perry G. Free Rad Res. 2012 [Google Scholar]
- 108.Olinski R. Free Rad Res. 2012 [Google Scholar]
- 109.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between polymerase β and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 113.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 114.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: Passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase beta-dependent long patch base excision repair. J Biol Chem. 2000;275:2211–2218. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- 116.Sung JS, DeMott MS, Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. J Biol Chem. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- 117.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2254. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paiva AM, Sheardy RD. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–14227. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- 119.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mitas M, Yu A, Dill J, Kamp TJ, Chambers EJ, Haworth IS. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith GK, Jie J, Fox GE, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 1995;23:4303–4311. doi: 10.1093/nar/23.21.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gacy AM, Goellner G, Juranic N, Macura S, Mcmurray CT. Trinucleotide repeats that expand in human-disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 123.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG)•(CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jarem DA, Wilson NR, Delaney S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 125.Jarem DA, Wilson NR, Schermerhorn KM, Delaney S. Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion. DNA Repair. 2011;10:887–896. doi: 10.1016/j.dnarep.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singer MS, Gottschling DE. TLC1: Template RNA component of saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 127.McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 128.Collins K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–383. doi: 10.1016/s0955-0674(00)00103-4. [DOI] [PubMed] [Google Scholar]
- 129.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gilbert DE, Feigon J. Multistranded DNA structures. Curr Opin Struct Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 131.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 132.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kruk PA, Rampino NJ, Bohr VA. DNA damage and repair in telomeres: Relation to aging. Proc Natl Acad Sci U S A. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kawai K, Fujitsuka M, Majima T. Selective guanine oxidation by UVB-irradiation in telomeric DNA. Chem Comm. 2005:1476–1477. doi: 10.1039/b418000c. [DOI] [PubMed] [Google Scholar]
- 135.Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair. 2011;10:34–44. doi: 10.1016/j.dnarep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Friedman KA, Heller A. On the non-uniform distribution of guanine in introns of human genes: Possible protection of exons against oxidation by proximal intron poly-G sequences. J Phys Chem B. 2001;105:11859–11865. [Google Scholar]
- 137.Delaney S, Barton JK. Charge transport in DNA duplex/quadruplex conjugates. Biochemistry. 2003;42:14159–14165. doi: 10.1021/bi0351965. [DOI] [PubMed] [Google Scholar]
- 138.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in myh predispose to multiple colorectal adenoma and somatic G:C→T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 139.Cheadle JP, Sampson JR. Exposing the myth about base excision repair and human inherited disease. Hum Mol Genet. 2003;12:R159–R165. doi: 10.1093/hmg/ddg259. [DOI] [PubMed] [Google Scholar]
- 140.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 141.Lipton L, Sieber OM, Crabtree M, Heinimann K, Fidalgo P, Phillips RKS, Bisgaard ML, Ornoft TF, Aaltonen LA, Hodgson SV, Tomlinson IPM, Thomas HJW. Multiple colorectal adenomas, familial adenomatous polyposis and germline mutations in MYH. Gastroenterology. 2003;124:A45–A45. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 142.Sampson JR, Jones S, Dolwani S, Cheadle JP. MutYH (MYH) and colorectal cancer. Biochem Soc T. 2005;33:679–683. doi: 10.1042/BST0330679. [DOI] [PubMed] [Google Scholar]
- 143.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 144.Zhou XL, Djureinovic T, Werelius B, Lindmark G, Sun XF, Lindblom A. Germline mutations in the MYH gene in swedish familial and sporadic colorectal cancer. Genetic Testing. 2005;9:147–151. doi: 10.1089/gte.2005.9.147. [DOI] [PubMed] [Google Scholar]
- 145.Chmiel NH, Livingston AL, David SS. Insight into the functional consequences of inherited variants of the hmyh adenine glycosylase associated with colorectal cancer: Complementation assays with hMYH variants and pre-steady-state kinetics of the corresponding mutated E. coli enzymes. J Mol Biol. 2003;327:431–443. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 146.Pope MA, Chmiel NH, David SS. Insight into the functional consequences of hMYH variants associated with colorectal cancer: Distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair. 2005;4:315–325. doi: 10.1016/j.dnarep.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 147.Pope MA, David SS. DNA damage recognition and repair by the murine MutY homologue. DNA Repair. 2005;4:91–102. doi: 10.1016/j.dnarep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 148.Livingston AL, Kundu S, Henderson Pozzi M, Anderson DW, David SS. Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 2005;44:14179–14190. doi: 10.1021/bi050976u. [DOI] [PubMed] [Google Scholar]
- 149.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 150.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 151.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9Me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 152.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS letters. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 153.Nguyen KV, Burrows CJ. A prebiotic role for 8-oxoguanosine as a flavin mimic in pyrimidine dimer photorepair. J Am Chem Soc. 2011;133:14586–14589. doi: 10.1021/ja2072252. [DOI] [PubMed] [Google Scholar]
- 154.Heil K, Pearson D, Carell T. Chemical investigation of light induced DNA bipyrimidine damage and repair. Chem Soc Rev. 2011;40:4271–4278. doi: 10.1039/c000407n. [DOI] [PubMed] [Google Scholar]