Abstract
Objective
DBA/2 apoE−/− mice have ~10-fold larger lesions than AKR apoE−/− mice. The objective of this study was to determine if macrophages from these two strains had altered cholesterol metabolism that might play a role in their divergent atherosclerosis susceptibility.
Approach and results
AKR and DBA/2 macrophages incubated with acetylated LDL (AcLDL) resulted in higher cholesterol ester (CE) and lower free cholesterol (FC) accumulation in the DBA/2 cells. However, these strains had equivalent AcLDL uptake and cholesterol esterification activity. Cholesterol efflux from unloaded cells to apoAI or HDL was similar in the two strains. However, upon AcLDL loading, cholesterol efflux was impaired in the DBA/2 cells, but this impairment was corrected by loading in the presence of an inhibitor of cholesterol esterification. Thus, the cholesterol efflux capabilities are similar in these strains, but there appeared to be a defect in lipid droplet (LD)-stored CE mobilization in DBA/2 cells. Lalistat-1, a specific inhibitor of lysosomal acid lipase, completely blocked the hydrolysis of LD-stored CE, implying that LD autophagy is responsible for CE turnover in these cells. CE turnover was 2-fold slower in DBA/2 vs. AKR cells. Autophagic flux, estimated by a fluorescent LC3-II reporter and the increase in p62 levels after chloroquine treatment, was higher in AKR vs. DBA/2 macrophages, which had an apparent decrease in autophagosome fusion with lysosomes. When autophagy was activated by amino acid starvation, CE levels decreased in DBA/2 cells.
Conclusions
Physiological regulation of autophagy in macrophages controls CE accumulation and may modify atherosclerosis susceptibility.
Keywords: cholesterol ester, foam cell, atherosclerosis, autophagy
Atherosclerosis, the primary cause of cardiovascular disease and the leading cause of death worldwide1, is characterized by the progressive build up of cholesterol-rich plaques in the arteries. Apolipoprotein E (apoE)-deficient mice develop aortic lesions that progress from fatty streaks to fibrous plaques with a predilection to form at regions with disturbed flow2, and are a commonly used model for this disease. Atherosclerosis in various mouse models is sensitive to the genetic background, and we have previously shown that apoE-deficient DBA/2 mice have 10-fold larger lesion areas than apoE-deficient AKR mice when fed a chow diet for 16 weeks.3 As an early step in the formation of atherosclerotic lesions is the transformation of arterial wall macrophages into lipid-loaded foam cells4, we studied cholesterol metabolism after cholesterol loading with acetylated LDL (AcLDL) in bone-marrow derived macrophages from apoE-deficient AKR and DBA/2 mice in vitro.
We found that DBA/2 vs. AKR macrophages accumulated more cholesterol esters (CE) in lipid droplets (LD) due to a decreased rate of LD-stored CE hydrolysis. Recently, Ouimet et al. characterized that macrophage foam cell LD are delivered to lysosomes via autophagy where LD-stored CE is hydrolyzed into free cholesterol (FC) by lysosomal acid lipase.5 We found that the same pathway is responsible for the majority of LD-stored CE hydrolysis in apoE-deficient AKR and DBA/2 macrophages, but that autophagic flux was slower in DBA/2 cells resulting in more CE accumulation in LD. These studies imply that the regulation of autophagy may play an important physiological role in foam cell formation and atherogenesis.
Results
Macrophage cholesterol accumulation and uptake
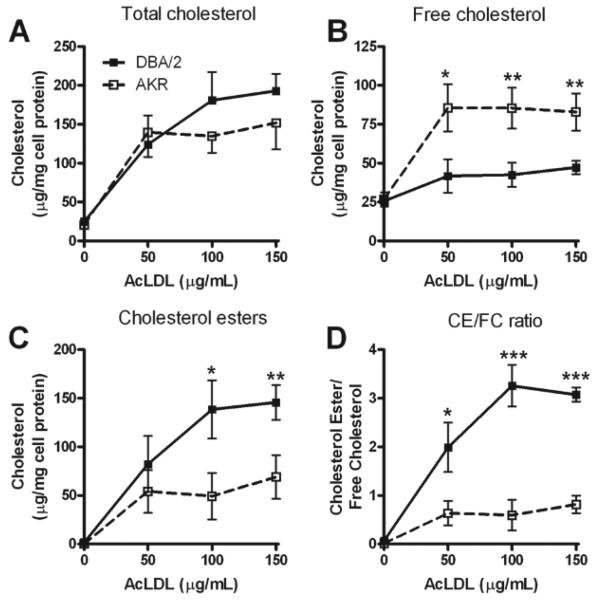
In order to examine cholesterol metabolism in apoE-deficient bone marrow derived macrophages (here after described as macrophages) from the atherosclerosis resistant AKR and atherosclerosis sensitive DBA/2 backgrounds, we incubated these cells for 24h in presence of increasing concentrations of AcLDL and measured total cholesterol (TC), FC and CE levels. TC levels increased upon AcLDL loading in both strains and this loading appears to be saturable, as expected since it is receptor mediated (Figure 1A). The amount of loading during 24h was variable using different batches of AcLDL, but in general unloaded cells contained ~30 to 50 μg/ml cholesterol/mg cell protein, while loaded cells contained from 100 to 300 μg/ml cholesterol/mg cell protein. However, using different batches of AcLDL we consistently observed that the distribution of FC and CE was significantly different between the two strains at all doses of AcLDL. DBA/2 macrophages had lower levels of FC (Figure 1B) and higher levels of CE (Figure 1C) than AKR macrophages, resulting in CE/FC ratios of ~ 2 to 3 in DBA/2 loaded macrophages and <1 in AKR loaded macrophages (Figure 1D).
Figure 1. AcLDL loading of macrophages from apoE-deficient AKR and DBA/2 mice.
Macrophages from AKR (open square and dash line) or DBA/2 (black square and solid line) mice were loaded for 24h at 37°C with 0, 50, 100 or 150 μg/mL AcLDL. TC (A), FC (B), CE (C) and CE/FC ratio (D) are expressed as the mean±SD of triplicates. *, p<0.05; **, p<0.01; ***, p<0.001 by two-tailed t-test comparing the two strains at each dose.
We hypothesized that this strain-effect on cholesterol loading and CE storage might be due to differences in: 1) lipoprotein uptake; 2) AcLDL-bound CE hydrolysis to FC, transport of FC to the endoplasmic reticulum or subsequent cholesterol esterification by ACAT; 3) FC efflux to extracellular acceptors; or 4) CE hydrolysis from LD.
To test our first hypothesis, we measured the uptake of DiI-labeled AcLDL by macrophages from both strains using flow cytometry after a 30min incubation. We found similar levels of AcLDL uptake by AKR and DBA/2 macrophages (Supplemental Figure IA). We confirmed this result with an alternate method measuring [3H]-AcLDL uptake (Supplemental Figure IB). Thus, we ruled out AcLDL uptake as the mechanism responsible for the differences observed in cholesterol loading at 50 μg/mL AcLDL, and in general for the differences in CE/FC distribution in AKR and DBA/2 macrophages.
AcLDL-bound CE metabolism and re-esterification
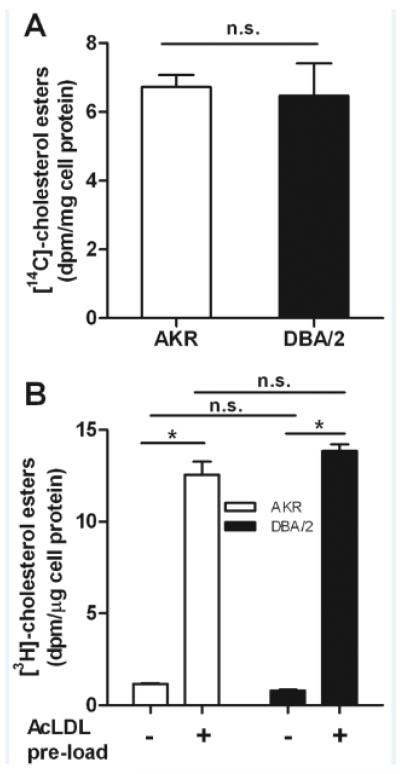
We next addressed the second hypothesis that the strain difference in cholesterol loading might be due to altered levels of AcLDL-bound CE catabolism, delivery of FC to the endoplasmic reticulum, or activity of ACAT, the endoplasmic reticulum enzyme in macrophages that esterifies FC to CE for storage in lipid droplets. In order to determine if there was a strain effect on lysosomal hydrolysis of endocytosed CE in AcLDL, we measured the % CE remaining in cells after AcLDL loading (50 μg/ml) for 16h in the presence of ACAT inhibitor (ACATi). Both AKR and DBA/2 macrophages contained <6% of their TC in CE (not significant vs. each other), demonstrating that lysosomal hydrolysis of AcLDL-derived CE was similar in these strains and ruling out a lysosomal storage disorder in DBA/2 cells. We then assessed ACAT activity in two assays using either broken cell lysates or living cells. This was important since there is a well characterized ACAT protein polymorphism in the AKR strain, although this N-terminal 33 residue truncation is not associated with altered ACAT activity.6, 7 In broken cell lysates made from cholesterol loaded cells, we found no difference in ACAT activity between the two strains (6.7±0.3 and 6.5±0.9 dpm CE/mg cell protein in AKR and DBA/2 macrophages, respectively, p=0.70, Figure 2A). In order to confirm these results and to verify that the CE/FC ratio difference we observed in loaded cells was not be due to altered delivery of FC to the endoplasmic reticulum, we measured the initial ACAT activity in live unloaded or pre-loaded AKR and DBA/2 macrophages by incubation with [3H]cholesterol-AcLDL for 1h. In both AKR and DBA macrophages, CE formation was induced in pre-loaded cells vs. unloaded cells by over 10-fold (p<0.0001, Figure 2B). However, there were no significant differences in ACAT activity between the two strains in either unloaded or pre-loaded cells. These results confirm the data in the broken cell lysates and imply that the delivery of FC to the site of ACAT in the endoplasmic reticulum is comparable in both strains. Thus, AcLDL CE metabolism and re-esterification does not appear to be responsible for the observed strain effects on CE/FC ratio.
Figure 2. Comparison of ACAT activity in macrophages from apoE-deficient AKR and DBA/2 mice.
(A) AKR (open bar) and DBA/2 (solid bar) cells were loaded with AcLDL, and cell lysates were used to measure ACAT activity as described in the Methods section. Results were normalized to cell protein and expressed as mean±SD of triplicates. (B) AKR (open bars) and DBA/2 (solid bars) ACAT activity was measured in live cells, as described in the Methods section, with or without preloading with 50 μg/mL AcLDL. Results were normalized to cell protein and expressed as mean±SD of triplicates, *, p<0.0001 by ANOVA posttest.
Macrophage cholesterol efflux
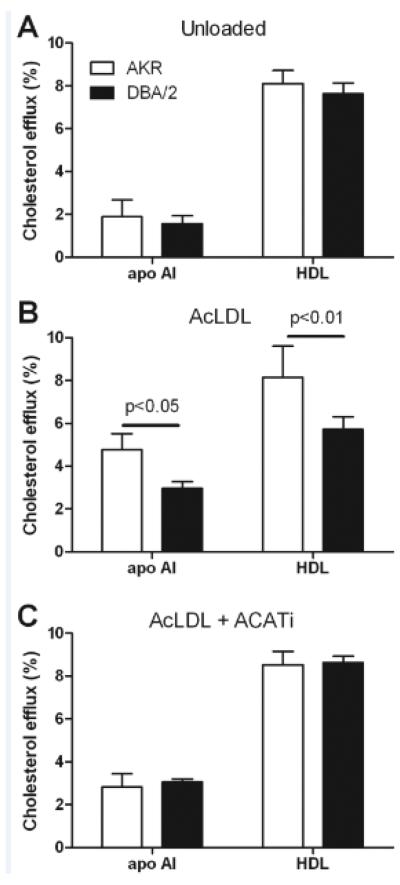
We evaluated efflux of cholesterol to apoAI and HDL in unloaded cells, where most cholesterol is in FC, and in pre-loaded cells, where CE stores accumulate. In the unloaded cells there was no significant difference between AKR and DBA/2 macrophages in terms of efflux to 10 μg/mL apoAI (1.9±0.8% vs. 1.6±0.4%, respectively) or 100 μg/mL HDL (8.1±0.6% vs. 7.6±0.5%, respectively, Figure 3A). However, when loaded cells were incubated with these acceptors, AKR macrophages, vs. DBA/2 macrophages, had increased efflux to both apoAI (4.8±0.8% vs. 3.0±0.3%, p<0.05) and HDL (8.1±1.5% vs. 5.7±0.6%, p<0.01, Figure 3B). To determine if this strain-effect on the difference in efflux between the unloaded and loaded cells was due to CE stores, we repeated the efflux study in cells in which AcLDL loading was performed in the presence of an ACATi to prevent CE formation. As in the unloaded cells, the macrophages loaded in the presence of ACATi had no significant strain differences in their efflux to either apoAI or HDL (Figure 3C). Thus, there is no inherent difference in efflux capacity between AKR and DBA/2 macrophages when the cells contain mostly FC; however, in conditions where CE stores are present, the DBA/2 macrophages have impaired efflux activity. Taken together, the results suggest that the strain effect on CE/FC distribution may be due to different efficiencies in CE hydrolysis.
Figure 3. Cholesterol efflux from macrophages of apoE-deficient AKR and DBA/2 mice.
AKR (open bars) and DBA/2 (solid bars) cells were incubated overnight in presence of FBS 1% (A), 50 μg/mL AcLDL (B) or 50 μg/mL AcLDL and 2 μg/mL ACATi (C) and then chased for 4h to apoAI or HDL. Results are represented as the mean±SD of triplicates. Strain effect significant p-values were only observed in AcLDL loaded cells (B), as indicated (two-tailed t-test).
Macrophage cholesterol ester hydrolysis
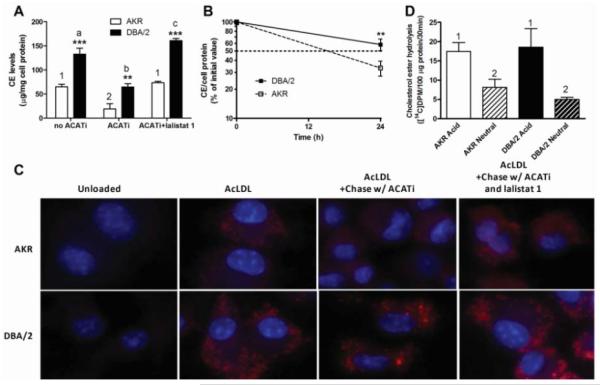
Esterified cholesterol in cholesterol-loaded macrophages is stored in LD, and in order for this CE to be mobilized for efflux it must first undergo hydrolysis to FC. A recent report by Ouimet et al. highlights the involvement of autophagy in regulating LD-stored CE hydrolysis and cholesterol efflux from cholesterol loaded macrophages.5 This study demonstrated the engulfment of LD by autophagosomes delivering LD-stored CE to lysosomal acid lipase via the formation of autolysosomes. In order to test the involvement of lysosomal acid lipase in the hydrolysis of LD-stored CE in AKR and DBA/2 macrophages, we followed a protocol similar to Ouimet et al.5, in which AcLDL-loaded macrophages, with concomitant CE stores, were chased for 24h with apoAI in absence or presence of ACATi, or in the presence ACATi plus lalistat 1, a specific inhibitor of lysosomal acid lipase.8, 9 For the chase in the absence of ACATi, CE levels were 2-fold higher in DBA/2 vs. AKR cells (133.0±12.2 vs. 64.8±5.6 μg/mg cell protein, p<0.001, respectively, Figure 4A), representative of the higher initial CE storage of DBA/2 macrophages (compare with Figure 1C). When ACATi was added to the chase media, in order to prevent the re-esterification of hydrolyzed LD-stored CE, the CE levels dropped in both strains, but still resulted in significantly more CE in DBA/2 vs. AKR macrophages (64.8±6.6 vs. 29.9±10.1 μg/mg cell protein, p<0.01, Figure 4A). ACATi led to a 63% decrease in CE in the AKR cells vs. a 51% in DBA/2 cells (p<0.001 for both strains by ANOVA posttest). In the presence of both ACATi and lalistat 1 during the chase, LD-stored CE hydrolysis was inhibited and the CE levels in both strains were similar to those observed in the absence of ACATi. These results suggest that lysosomal acid lipase is responsible for the hydrolysis of LD-stored CE in these foam cells and that LD-stored CE hydrolysis via lysosomal acid lipase may be slower in DBA/2 macrophages.
Figure 4. Cholesterol esters turnover in macrophages from apoE-deficient AKR and DBA/2 mice.
(A) CE mass in AcLDL loaded AKR (open bars) or DBA/2 (solid bars) cells chased for 24h with apoAI in presence or absence of 2 μg/mL ACATi or 10 μM lalistat 1. Results are expressed as mean±SD of triplicates. **, p<0.001; and ***, p<0.0001, DBA/2 compared to AKR for each condition by two-tailed t-test. For the three AKR conditions, 1 vs. 2, p<0.001. For the three DBA/2 conditions, a or c vs. b, p<0.001; a vs. c, p<0.05 by ANOVA posttest (B) CE turnover in AcLDL loaded AKR (open square and dash line) or DBA/2 (black square and solid line) cells chased to apoAI in presence of ACATi. Results represent the mean±SD of 3 independent experiments and were normalized to the initial values for each strain and experiment. (C) Nile red-stained lipid droplets and DAPI-stained nuclei (blue) in unloaded and AcLDL loaded cells before and after a 24h chase in presence of ACATi with or without lalistat 1. (D) Acidic and neutral CE hydrolase activity normalized to protein content. AKR and DBA/2 lysates from AcLDL loaded cells were assayed as described in Methods. Results represent the mean±SD of 6 wells. 1 vs. 2, p<0.001 by ANOVA posttest.
To more precisely measure LD-stored CE hydrolysis rates we measured cellular CE levels after AcLDL loading (0h chase) or 24h after chasing with apoAI in the presence of an ACATi to block FC re-esterification. We varied the AcLDL loading dose in order to load the AKR and DBA/2 macrophages with similar levels of CE, nevertheless, we normalized our data to the CE content of the cells at 0h. Combining the data from three independent experiments, we observed 41±8% and 66±14% reductions in CE content after the 24h chase in the AKR and DBA/2 cells, respectively (p=0.01 by t-test, Figure 4B). This corresponds to a CE half life of 13.9h in AKR cells, which was doubled in DBA/2 macrophages to 30.4h, thus confirming that LD-stored CE hydrolysis is slower in DBA/2 cells.
We further addressed the role of lysosomal acid lipase in LD-stored CE turnover qualitatively using fluorescent microscopy. Unloaded AKR and DBA/2 macrophages, or cells loaded with 50 μg/mL AcLDL in the absence or presence of a chase period were stained with Nile red to visualize LD and co-stained with DAPI (nuclear stain). The results, presented in Figure 4C, show that loaded cells accumulate LD in both AKR and DBA/2 cells but to a greater extent in DBA/2, consistent with the CE biochemical measurement in Figure 1C. After a 24h chase in presence of ACATi, the number of LD is dramatically reduced in AKR cells and their number appears moderately decreased in DBA/2 macrophages, but still higher than in the AKR cells, again consistent with the biochemical CE measures in Figure 4A. When lalistat 1 and ACATi were added to the chase media, there was no discernable decrease in the number of LD vs. the loaded cells in both strains. These results confirm the role of lysosomal acid lipase in CE lipid droplet turnover in both strains, and suggest that either decreased activity of lysosomal acid lipase or decreased autophagy of LD may be responsible for decreased LD-stored CE turnover in DBA/2 macrophages. We also examined small early foam cell lesions in the aortic root of chow diet fed AKR and DBA/2 mice using Bodipy to stain cholesterol ester laden LD. In a preliminary quantitative study, we observed that the lipid droplet % of the foam cell area was >2-fold larger in DBA/2 vs. AKR lesions (p<0.01, Supplemental Figure II). This finding is consistent with our cell culture observation of more CE content in DBA/2 vs. AKR AcLDL-loaded macrophages (Figure 4).
Cholesterol esterase activity was measured in cell lysates from AKR and DBA/2 macrophages. In both strains there was >2-fold more acidic than neutral cholesterol esterase activity (p<0.01 by ANOVA posttest), but there was no significant strain difference between the acidic or neutral cholesterol esterase activities (Figure 4D). Thus, the activity of lysosomal acid lipase does not appear to be limiting in the DBA/2 macrophages. These results are also consistent with the absence of defect in AcLDL-bound CE hydrolysis in DBA/2 cells.
Autophagy role in strain difference in CE ester hydrolysis
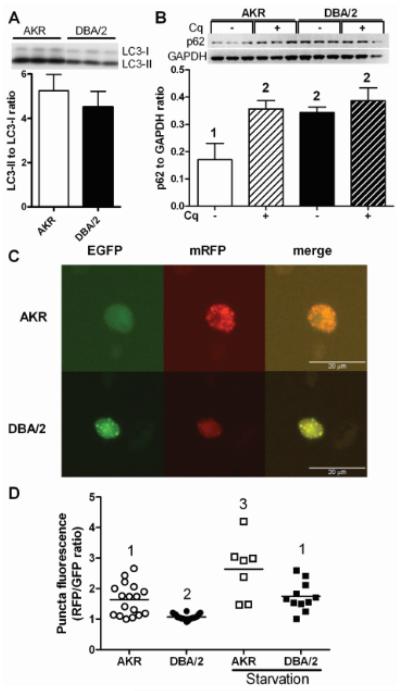
To investigate whether autophagy may play a role in the observed impairment of LD-stored CE hydrolysis in DBA/2 cells, the key autophagy proteins microtubule-associated protein 1 light chain 3 (LC3) and p62 were assessed using western blot, normalized to GAPDH used as a loading control. LC3 is synthesized as a precursor and is cleaved to form the cytosolic form LC3-I which is then covalently modified with phosphatidylethanolamine to generate LC3-II, which specifically binds to the elongating autophagosome and remains bound through to fusion with the lysosome where it is degraded.10, 11 Low levels of LC3-II can result from low conversion from LC3-I, which occurs when autophagy is blocked at an early stage leading to LC3-I accumulation.12 We compared the LC3-II to LC3-I ratio in AcLDL loaded macrophages, and found in both cell types that there was ~5-fold higher levels of LC3-II than LC3-I (Figure 5A), indicating that both cell types have sufficient LC3-II in order to initiate autophagy. The levels of p62 can be used as an indication of autophagic flux, as p62 targets protein aggregates to the autophagosome and is degraded along with the aggregate; thus, low levels of p62 that are increased by chloroquine treatment, which blocks its lysosomal degradation, are associated with high levels of autophagic flux.10, 11, 13 We assessed p62 levels in AcLDL loaded macrophages in the absence or presence of a 4h treatment with 30 μM chloroquine. Chloroquine treatment induced p62 levels 2.1-fold in AKR macrophages (p<0.01), while this treatment led a non-significant 13% increase in p62 levels in DBA/2 macrophages (Figure 5B). This result indicated a lower level of autophagic flux in the DBA/2 cells, agreeing with the observed slower rate of LD-stored CE hydrolysis (Figure 4B). In the absence of chloroquine we observed that p62 levels were 2.0-fold higher in DBA/2 vs. AKR macrophages (Figure 5B, p<0.01). This finding is also consistent with lower rates of degradation via autophagy in DBA/2 vs. AKR cells.
Figure 5. DBA/2 vs. AKR macrophages have impaired autophagic flux.
(A) LC3-II to LC3-I protein ratio in AKR (open bar) and DBA/2 (solid bar) cells loaded for 24h with 50 μg/mL AcLDL. Results represent the mean±SD of triplicates. (B) p62 protein expression in AcLDL loaded AKR (white bars) and DBA/2 (black bars) cells incubated for 4h with (solid bars) or without (crosshatched bars) chloroquine (Cq). Results represent the mean±SD of triplicate cell samples. 1 vs. 2, p<0.01 by ANNOVA posttest. (C) Bi-fluorescent LC3 assay for autophagic flux as describe in Methods. Fluorescent puncta in the red channel indicate LC3-II in autophagosomes and autolysosomes, while green puncta indicate autophagosome bound LC3-II only. (D) Mean puncta RFP/GFP fluorescence ratio per cell in AcLDL loaded AKR and DBA/2 macrophages in the absence or presence of a 1h amino-acid starvation. Higher ratios indicate autophagosome fusion with lysosomes. Different numbers above the data points represent p<0.05 by ANOVA posttest.
In order to independently assess autophagic flux, macrophages from both strains were transfected with the fluorescent reporter ptfLC3.14 This reporter encodes for a bi-fluorescent mRFP-EGFP-LC3 protein that is endogenously converted to LC3-II and associates with autophagosomes creating doubly green and red puncta. However, upon fusion with the lysosome, the pH-sensitive EGFP fluorescence diminishes, leaving puncta with red/green fluorescence ratio >1. Upon AcLDL loading of the transfected AKR macrophages, epifluorescent microscopy revealed that the median number of fluorescent puncta was similar in AKR (7 puncta) and DBA/2 (8 puncta) macrophages (p=0.61 by non-parametric t-test after counting puncta in 19 AKR and 21 DBA/2 transfected cells, Supplemental Figure III). Thus, autophagosome formation appeared similar in the two strains. Image analysis revealed a broad distribution of cells with mean fluorescent puncta red/green ratios ranging from ~1 to ~2.7. This is indicative of cells with non-lysosomal LC3 localization (red/green ratio ~1) and lysosomal LC3 localization (red/green ratio >1), the latter consistent with relatively rapid autophagic flux and EGFP emission decreased in the low pH lysosome environment. However, DBA/2 foam cells all had puncta with red/green ratios of ~1 (p<0.05 vs. AKR cells by ANOVA posttest), indicative of relatively slow autophagic flux (Figures 5 C, D). Thus, this assay suggests that the impaired step in autophagic clearance of LD-stored CE in DBA/2 cells is the fusion of autophagosomes with lysosomes. Our finding of higher basal levels of p62 in DBA/2 vs. AKR foam cells (Figure 5B) is consistent with the proposed impairment in the DBA/2 strain at the level of autolysosome formation. In order to validate this bi-fluorescent reporter assay, we induced autophagic flux in both strains by amino starvation. Image analysis revealed increased cellular red/green puncta ratios in both strains (p<0.05 vs. non-starved cells of the same strain by ANOVA posttest. Figure 5D). Thus, both p62 western blot and LC3 bi-fluorescent reporter revealed slower autophagic flux in DBA/2 vs. AKR macrophages.
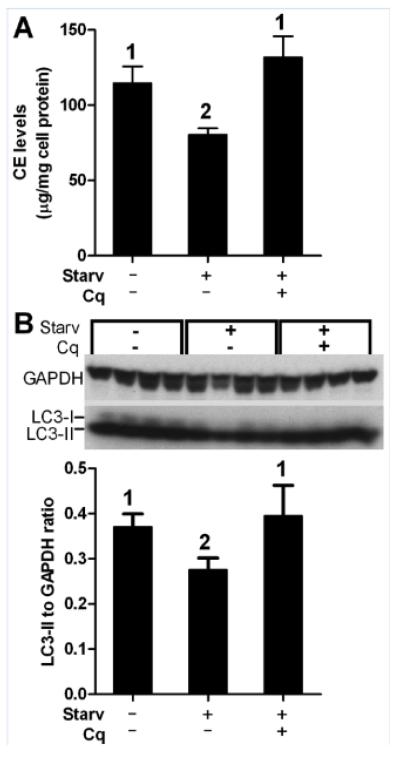
We then subjected DBA/2 pre-loaded macrophages to amino acid starvation, a commonly used activator of autophagy. A 2h chase in the presence of apoAI and ACATi was performed in complete DMEM or in amino acid-free EBSS; and, CE levels were reduced by 29.8% in the EBSS treated compared to the control cells (80.4±4.3 and 114.5±11.3 μg/mg cell protein, respectively, p<0.05 by ANOVA posttest, Figure 6A). When we added chloroquine during the EBSS chase in order to block autophagy at the level of autolysosome function, CE levels went back up to 131.6±14.1 μg/mg cell protein (not significant vs. non-starved controls), indicating that the EBSS induced LD-stored CE hydrolysis was dependent upon autolysosome function. LC3-II levels also decreased by 26% upon amino acid starvation (p<0.05 by ANOVA posttest), which was reversed to basal levels by the addition of chloroquine (Figure 6B). Thus it appears that DBA/2 cells have decreased basal autophagic flux leading to slower LD-stored CE turnover, but that upon induction of autophagy by amino acid starvation, DBA/2 macrophages are capable of clearing LD-stored CE more efficiently.
Figure 6. Starvation-induced autophagy decreased CE content in DBA/2 macrophages.
AcLDL loaded DBA/2 cells chased in DMEM (control), amino acid-free EBSS (starv) to induce autophagy, or EBSS plus chloroquine (Cq) were evaluated for (A) CE mass (mean±SD of triplicates) and (B) LC3-II protein expression by western blot followed by densitometry normalized to GAPDH (mean±SD of quadruplicates). Different numbers above the bars represent p<0.05 by ANOVA posttest.
Discussion
We characterized altered cholesterol metabolism in AKR vs. DBA/2 apoE-deficient bone marrow macrophages. Upon cholesterol loading with AcLDL, DBA/2 macrophages, compared with AKR macrophages, accumulated more CE due to decreased LD-stored CE hydrolysis leading to less cholesterol efflux to apoAI and HDL. There were no apparent strain differences in AcLDL uptake or ACAT activity, the later measured both in live cells and cell lysates. The AKR strain is known to harbor a mutation in the ACAT gene leading to exon skipping that results in a 33 amino-acids deletion at the N-terminus of the protein.7 However, Meiner et al. reported that this protein change is not associated with a change in adrenal ACAT activity7, and our results confirm equivalent activity in macrophages carrying this truncated ACAT isoform.
Recently, Ouimet et al. demonstrated that lysosomal acid lipase is the predominant activity responsible for the hydrolysis of CE in foam cell LD5; and we confirmed this finding by the use of lalistat 1, a specific lysosomal acid lipase inhibitor. We found that AKR and DBA/2 cells had equal amounts of lysosomal acid lipase activity; thus, the activity or expression of this enzyme does not appear to be responsible for our observed strain effect on LD-stored CE hydrolysis. However, alterations in human lysosomal acid lipase, encoded by the LIPA gene, may play a role in coronary artery disease (CAD) susceptibility. Three genome wide association studies have identified LIPA intronic SNPs rs141244 or rs2246949 (which are in complete linkage disequilibrium with each other and thus co-inherited on the same allele) as associated with CAD.15–17 Although the effect of the risk allele was not very strong, with CAD odds ratio of ~1.1, these findings were all highly significant, exceeding the conservative genome wide threshold of significance of p<10−8. LIPA gene expression was ascertained in whole blood or monocytes by microarrays, and the CAD risk allele of the LIPA gene was found to be associated with higher expression of LIPA mRNA.16, 17 The direction of this effect on LIPA gene expression does not match with our findings, in which increased lysosomal lipase activity would be expected to promote CE turnover, cholesterol efflux, and foam cell regression. However, LIPA mRNA levels in blood cells are far removed from lysosomal acid lipase activity in cholesterol engorged arterial foam cells; thus, it is unknown if these SNPs play a role in human atheroma CE metabolism, and the mechanism responsible for their CAD association has not been determined. Nevertheless, we ruled out lysosomal acid lipase activity differences as being responsible for our observed strain effects on LD-stored CE turnover.
The role of autophagy in lipid droplet metabolism was first demonstrated by Singh et al. in oleate treated hepatocytes and fibroblasts, in which autophagy was shown to decrease cellular triglyceride levels, lipid droplet size and number, and promote fatty acid oxidation.18 Ouimet et al. extended this observation to foam cells, in which mobilization of macrophage foam cell LD-stored CE was shown to be via autophagy.5 Three recent studies have highlighted the role of autophagy in atherosclerosis. Atg5 is one of the genes required for formation of the autophagic isolation membrane and engulfment. Macrophage specific Atg5-deficient mice were created via Cre-lox methodology resulting in mice with a proinflammatory macrophage phenotype.19, 20 Razani et al. compared apoE-deficient mice with or without macrophage Atg5-deficiency and found that that Atg5 knockout led to increased aortic p62 levels, indicative of decreased autophagic flux, increased aortic root and arch lesion area, and decreased cholesterol efflux to apoAI from peritoneal macrophages.13 Furthermore, they demonstrated increased inflammasome activation in Atg5-deficient macrophages, as well as increased cholesterol crystals in the lesions of the double knockout mice, potentially creating a positive feedback loop to increase inflammation in these autophagy-impaired mice.13 Liao et al. bred the macrophage Atg5-deficient mice onto the LDLr-deficient background, and also observed larger aortic root lesions with larger necrotic cores associated with increased lesional macrophage oxidative stress and apoptosis, and decreased clearance of apoptotic cells.20 Le Guezennec et al. identified the Wip1 phosphatase as a mTOR dependent inhibitor of autophagy, thus knocking out Wip1 led to decreased mTOR activity and increased autophagy.21 They found that Wip1-apoE double knockout mice were less susceptible to diet induced weight gain, and their macrophages accumulated less CE when loaded with modified LDL, and had increased cholesterol efflux to apoAI and HDL compared to the apoE single knockout mice. Furthermore, the double knockouts had significantly smaller aortic root and entire aorta lesion areas associated with increased macrophage autophagic flux.21 The above mouse studies are consistent with our results, namely that macrophage autophagy is impaired in the DBA/2 atherosclerosis sensitive strain that displayed greater accumulation of CE in LD and reduced efflux capacity compared to the atherosclerosis resistant AKR macrophages. We were able to show decreased CE levels in the DBA/2 macrophages in vitro upon inducing autophagy by amino acid starvation. Recently, p38 MAPK was found to promote macrophage CE accumulation by inhibition of autophagy22; thus, autophagy regulation in either direction appears competent to play a physiological role in macrophage CE metabolism.
The gold standard for the presence of autophagy is the identification of double membranes by electron microscopy. The lack of this evidence is a limitation of our study; however, this method yields a static measure of the number of autophagosomes while the methods we used have been validated for measuring autophagic flux. In addition the presence of double membrane bound LD have been previously shown in mouse macrophage foam cells, proving the presence of this pathway in macrophages.5
The immunosuppressant Rapamycin and its derivatives have been successfully employed in human stents given after angioplasty to prevent restenosis. Rapamycin inhibits mTOR kinase and thus blocks downstream effector activation leading to inhibition of cell proliferation. The antiproliferative activity of Rapamycin led to it being tested in mouse models of atherosclerosis, where it potently reduced lesion is apoE-deficient as well as LDLr-deficient mice, despite leading to worse hyperlipidemia.22–24 Rapamycin inhibition of mTOR kinase leads to Atg13 dephosphorylation allowing autophagosome formation and induction of autophagic flux.25 Thus, it is attractive to speculate that some of Rapamycin's antiatherogenic activity stems from induction of autophagy in arterial foam cells, promoting hydrolysis of LD-stored CE, cholesterol efflux, and foam cell regression.
In conclusion, we demonstrated that LD-stored CE hydrolysis is mediated by lysosomal acid lipase and our data support the notion that the delivery of LD-stored CE to the lysosome is mediated by autophagy. We found that the higher CE levels in DBA/2 macrophages were caused by a decrease in CE hydrolysis from LD due to slower basal autophagic flux in these cells leading to lower rates of cholesterol efflux. Thus, the physiological regulation of autophagy plays an important role in foam cell CE metabolism, which may play a role in the increased atherosclerosis susceptibility of the DBA/2 vs. the AKR mouse strain. The identification of the genes and mechanisms responsible for this strain effect on macrophage autophagy is of great interest and may aid in identifying new therapeutic targets.
Supplementary Material
Significance.
Atherosclerosis is a complex disease with genetic and environmental risk factors, and much of our knowledge about the pathogenesis of this disease has come from the study of animal models. Previously we observed 10-fold larger aortic root lesions in apoE-deficient mice on the DBA/2 vs. the AKR genetic background. Here, we identified an intermediate phenotype, the excess storage of cholesterol esters in cultured macrophages derived from DBA/2 mice. We determined that the mechanism for this phenotype is the slower hydrolysis of cholesterol esters stored in lipid droplets, which is in turn mediated by a lower rate of lipid droplet autophagy, specifically at the autophagosome to lysosome fusion step. This novel finding that physiological factors regulate autophagy to impact foam cell cholesterol ester metabolism demonstrates that this pathway may play an important role in atherosclerosis susceptibility. This knowledge may be leveraged to find novel diagnostics and therapeutics for atherosclerosis.
Acknowledgments
Sources of funding This work was supported by the NIH National Heart Lung & Blood Institute grants R01 HL098193 and P01 HL098055.
Abbreviations
- ACAT
Acyl-CoA cholesterol acyl transferase
- ACATi
ACAT inhibitor
- AcLDL
acetylated low density lipoprotein
- CE
cholesterol esters
- FBS
fetal bovine serum
- FC
free cholesterol
- HDL
high density lipoprotein
- LD
lipid droplets
- TC
total cholesterol
Footnotes
Disclosures None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.W.H.O. Global atlas on cardiovascular disease: prevention and control. WHO, World Heart Federation, World Stroke Organization; 2011. [Google Scholar]
- 2.Breslow JL. Mouse models of atherosclerosis. Science. 1996;272:685–688. doi: 10.1126/science.272.5262.685. [DOI] [PubMed] [Google Scholar]
- 3.Smith JD, James D, Dansky HM, Wittkowski KM, Moore KJ, Breslow JL. In silico quantitative trait locus map for atherosclerosis susceptibility in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2003;23:117–122. doi: 10.1161/01.atv.0000047461.18902.80. [DOI] [PubMed] [Google Scholar]
- 4.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 5.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu B, Potter CS, Silva KA, Liang Y, Reinholdt LG, Alley LM, Rowe LB, Roopenian DC, Awgulewitsch A, Sundberg JP. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice. J Invest Dermatol. 2010;130:2666–2668. doi: 10.1038/jid.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meiner VL, Welch CL, Cases S, Myers HM, Sande E, Lusis AJ, Farese RV., Jr Adrenocortical lipid depletion gene (ald) in AKR mice is associated with an acyl-CoA:cholesterol acyltransferase (ACAT) mutation. J Biol Chem. 1998;273:1064–1069. doi: 10.1074/jbc.273.2.1064. [DOI] [PubMed] [Google Scholar]
- 8.Rosenbaum AI, Rujoi M, Huang AY, Du H, Grabowski GA, Maxfield FR. Chemical screen to reduce sterol accumulation in Niemann-Pick C disease cells identifies novel lysosomal acid lipase inhibitors. Biochim Biophys Acta. 2009;1791:1155–1165. doi: 10.1016/j.bbalip.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbaum AI, Cosner CC, Mariani CJ, Maxfield FR, Wiest O, Helquist P. Thiadiazole carbamates: potent inhibitors of lysosomal acid lipase and potential Niemann-Pick type C disease therapeutics. J Med Chem. 2010;53:5281–5289. doi: 10.1021/jm100499s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 13.Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15:534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 15.The IBC 50K CAD Consortium Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Coronary Artery Disease (CAD) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 17.Wild PS, Zeller T, Schillert A, et al. A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ Cardiovasc Genet. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, Mizushima NN, Denison MR, Virgin HWt. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 20.Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, Robbins J, Martinez J, Tabas I. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15:545–553. doi: 10.1016/j.cmet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Guezennec X, Brichkina A, Huang YF, Kostromina E, Han W, Bulavin DV. Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab. 2012;16:68–80. doi: 10.1016/j.cmet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Mei S, Gu H, Ward A, Yang X, Guo H, He K, Liu Z, Cao W. p38 mitogen-activated protein kinase (MAPK) promotes cholesterol ester accumulation in macrophages through inhibition of macroautophagy. J Biol Chem. 2012;287:11761–11768. doi: 10.1074/jbc.M111.333575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elloso MM, Azrolan N, Sehgal SN, Hsu PL, Phiel KL, Kopec CA, Basso MD, Adelman SJ. Protective effect of the immunosuppressant sirolimus against aortic atherosclerosis in apo E-deficient mice. Am J Transplant. 2003;3:562–569. doi: 10.1034/j.1600-6143.2003.00094.x. [DOI] [PubMed] [Google Scholar]
- 24.Mueller MA, Beutner F, Teupser D, Ceglarek U, Thiery J. Prevention of atherosclerosis by the mTOR inhibitor everolimus in LDLR−/− mice despite severe hypercholesterolemia. Atherosclerosis. 2008;198:39–48. doi: 10.1016/j.atherosclerosis.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Weichhart T. Mammalian target of rapamycin: a signaling kinase for every aspect of cellular life. Methods Mol Biol. 2012;821:1–14. doi: 10.1007/978-1-61779-430-8_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.